Ribose_Nuclei_Acids7.
Localization of mRNAs in Cytoplasm:
Most of the RNAs synthesized in the nucleus undergo processing in one or the other form to their final products. Then they are identified and packaged and exported to their final destinations. The diagram below depicts the fate of pre mRNA, nc RNAs, tRNAs and rRNAs.

www.sequenceontology.org
Unlike bacterial systems, intracellular region of the eukaryotic cells/systems is highly compartmentalized and specialized in structure and function.
- Most of the mRNAs move out of the nucleus with cap bound CBP (cap binding protein) and often the mRNA bound to ribosomes. As soon as mRNAs emerge out the nucleus they associate with more ribosomes and translate.
As the 5’ end of the mRNA is translated the N-terminal end of the polypeptide chain emerges out of large ribosomal tunnel at exit point. The nature of first 20 to 30 amino acids in the protein chain determines the destination of the protein.
If the amino acids are hydrophobic in nature and if they are destined to ER or to extra cellular region, they are carried on to ER surface. If the protein in as secretary proteins or destined to plasma membrane, Golgi membranes, lysosome or any endocytotic vesicles such as transport vesicles, all of them are docked on to Endoplasmic reticulum through SRP complex and docking proteins.
- The other mRNAs, which generate proteins for mitochondria, peroxisome, plastids and nucleus, are synthesized outside or say free from endoplasmic membranes in the cytosol. However there are experimental proofs to show, that the said mRNAs are sequestered into a maze of Actin and Microtrabaculae (?), where the poly-A tails are held on to the Actin cytoskeletal complex.

Structural organization of eukaryotic mRNA and the different points of possible regulation of translation through various trans-acting factors; http://www.biolcell.org/
Possible sites of interaction of transacting factors in the coding sequence are known. Regions of mRNA involved in subcellular localization and stability are also indicated.
5′-m7G, cap structure, eIF, eukaryotic initiation factor; CPE- cytoplasm polyadenylation element; EDEN- embryonic deadenylation signal; DICE- differential control element; PABP- poly(A)-binding protein are some of the proteins associated are known
- Highly exemplified example for localization of mRNAs within the cell is young Myeloblast cells and developing oocyte cells in Drosophila. The young muscle cells, which are still unicellular, exhibit pseudopodia movements. Such movements require beta Actin derived microfilaments in the cortical regions where the filament bundles organize to push the membrane into a pseudopodia structure. By fluorescent-tagged antibodies it has been shown that mRNAs for beta-Actins are actually located in the pseudopodia region entangled in the meshwork of Actin filaments. As Myeloblast fuse to produce multinucleated muscle cells, beta Actin mRNAs disappear so also their location. Instead alpha Actin mRNAs produced are found to be located at the periphery of the nucleus, again discerned by fluorescent antibodies against Actins. In either case the said and required mRNAs are held in specific position by the network of Actins and its associated proteins.
- If the 3’ end of the Actin mRNA gene with its upstream UTR region is fused with beta Galactosidase at its 3’ end, and such recombinant genes are allowed to express in an eukaryotic cell, one finds the expression of beta Galactosidase in particular location which can be detected by staining with X-gal. Deep coloration in the region is due to cleaving of X-Gal by the localized beta Galactosidase enzyme. This localization is actin dependent.
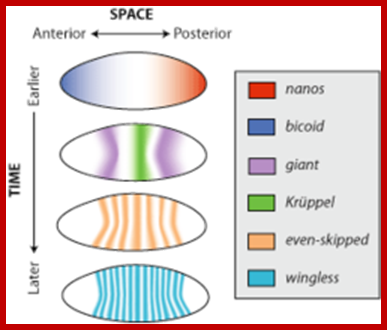
- In a developing Drosophila embryo, many transcripts especially mRNAs produced by nurse cells are transported across their cells into oocyte at very early stages of development. Some gene transcripts (mRNAs) are transported across the oocyte and localized; some at anterior end and some at posterior end.
- The Bicoid mRNAs are placed in the anterior position, Gurken in antero-dorsal region, Nanos and Oskar at posterior region. This specific directional transport of mRNA is aided by specific 3’UTR zip code sequences in mRNAs bound by specific proteins.
- The Bicoid transcripts are positioned in the anterior and enmeshed with cortical Actin filaments. As the development progresses into segmentation one can discern segmentation specific mRNAs found positioned in the cortical region in their respective locations. Again their tails in the network of cortical Actin filaments holds these transcripts.
- It is not only destinations but also enrichment of specific mRNA at a given locus.
- Transcript localization has been shown to control key biological processes, such as Endoplasmic reticulum, embryonic axis specification, cell fate determination, cell polarity, motility, and synaptic plasticity,(diversity of mRNA localization), Krause and Henry M.
- It has become clear that subcellular mRNA transport is highly complex and includes several components: (i) sequences within the RNA molecule, referred to as cis-acting elements, that may adopt secondary, tertiary, or even quaternary structures; (ii) a whole array of proteins, trans-acting factors, which mediate RNA sorting by binding either directly or indirectly to the mRNA to be transported; (iii) mechanisms of translational silencing and derepression; and (iv) components of the cytoskeleton as railway tracks and anchor sites of the ribonucleoproteins (reviewed in refs. Bashirullah etal 1998).
- The 3’UTR signals contain mRNA localization signals in the form of cis acting sequences and secondary structure bound by trans-acting specific proteins. Loss of 3’UTR disrupts mRNA localization.
- Localization produces a spatial organization of the synthesis of specific proteins within cells close to where they function.
- Transfer of free mRNA into ER is not SRP dependent, but uses separate translocon system for transportation free mRNA whose products are as good as SRP mediated products.
- Some of the proteins associated with specific mRNA for specific localization, Examples- Puf3p (P=Pumilo) mitochondria, Puf1p/2p membrane association, Puf4p ribosomal RNA processing and Puf5p has chromatin regulatory functions. RBP analysis using RIP techniques scientists have identified more than 12000 different RBP-mRNA interactions.
- RPB based RIP chip analysis shows specific proteins bind to specific mRNA and their destination is also determined.
- Microscopy based high throughput ISH screening is useful to find localization of specific mRNAs.
- During transport mRNAs are silenced and activated when they reach their specific destination.
- Destination of mRNA in the cell is specific for specific function; delivery of such specific mRNA to specific cellular sites is a par excellence process and pre-designed too.
- Localizing mRNAs are packaged into ribonucleoprotein complexes (RNP complexes) that engage with cytoskeletal motors for directed transport along cytoskeletal tracks (BOX 1) and ensure their translational silencing.
- There is an emerging consensus that in many cases the mRNA signals that determine intracellular destination are more complex and difficult to define than was first anticipated. Furthermore, the transacting factors that interpret the mRNA signals are numerous and their combinations change during the life of an mRNA, perhaps allowing the selection of many sub-destinations in the cell. Lastly, an emerging theme over the past few years is that many proteins that determine the destinations of mRNAs are recruited on nascent transcripts in the nucleus.
- Considerable intracellular sorting is achieved by molecular motors transporting cargo along the cytoskeleton, a system analogous to a trains transporting cargo on train tracks. This requires RNA ticket, which train and the ticket inspector check which passenger chose which train (ILLan Davis). Passengers’ i.e. mRNAs should be identified and loaded on to specific train on specific track with specific motor engine.
Schematic representation of early oogenesis (A) and stage 9 egg chamber (B); The germline and specified somatic cells are indicated individually. In both panels, anterior is to the left; http://www.nature.com/
Gene expression patterns are regulated both spatially and temporally in embryos of Drosophila melanogaster; http://en.wikipedia.org/
http://en.wikipedia.org/wiki/Positioning of the mRNAs is prevalent in eukaryotic systems like drosophila and other vertebrates.

mRNA Export - An Integrative Component of Gene Expression; ttp://www.cipsm.de

Mechanisms of translational regulation in Drosophila; A speculative model for translational regulation of OSK mRNA: James E. Wilhelm*1 and Craig A. Smibert; http://submit.biolcell.org/
Microtubule filaments are mostly responsible for the transport of mRNAs to specific location; in few cases it has been clearly established.
There are maternal mRNA in Drosophila, as they are transported into the egg cell, specific mRNAs are transported to specific destination; for Ex. Bicoid mRNA are positioned in the anterior half, Oskar and Nanos are localized in the posterior region of the egg; similarly Gurken mRNA in the antero-dorsal region. The transportation to specific sites depends on the kind TCS sequence in 3’UTR region of mRNAs and they require the binding of specific proteins, for example steufen protein in the case of Oskar mRNA, shy protein in the case of Veg1 mRNA of Xenopus. The diagram shows the localization elements belonging to different mRNA species such as Veg1 of Xenopus, Bicoid of drosophila and Ash mRNAs.

a | The Vg1 3' UTR contains a 340-nucleotide zip code that is sufficient for mRNA localization, termed the Vg1LE, or Vg1 localization element. The functional units in this type of element are short repetitive sequences (either UAUUUCUAC or UUCAC)59, 104. b | The bicoid localization element (BLE) in the bicoid 3' UTR is an example of a zip code with a modular architecture. bicoid mRNA undergoes several sequential transport steps, each involving different, partially overlapping, regions in the highly structured 3' UTR. (Light blue, early localization; dark blue, early and late localization; purple, mRNA anchoring.) c | ASH1 mRNA is an example of a zip code element that lies in the coding region (E1, E2a, E2b) and in the 3' UTR (E3). The E3 element also represents an example of a structure-based zip code, because the displayed stem loop structure but not the primary sequence is important for the function of the zip code; mRNA localization; message on the move;Types of mRNA zip codes.; http://www.nature.com/
The transport proteins act like tracts which in turn bind to motor proteins that are anchored to MTs or Actin filaments. The motor proteins can be Dyneins and Myosin II. They are directed by bound motor proteins, which use microtubules and actins for the transport in specific direction and specific location. Actin filaments alone cannot localize mRNAs to positions; there must be some other factors involved in positioning mRNAs to their respective sites, they are called positional factors or localization factors.
Differential routes for mRNA localization in yeast and transport of ER components to daughter cells:
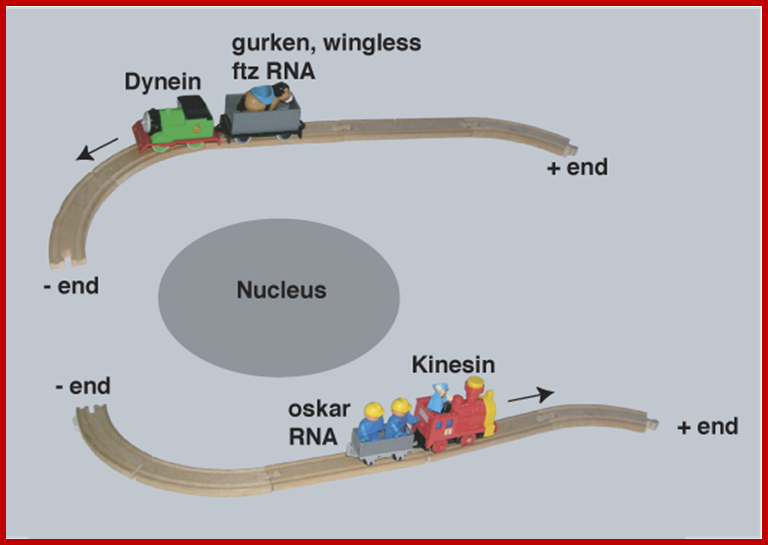
Intracellular mRNA transport illustrated with a train analogy. The train tracks represent the polarized microtubule cytoskeleton, engines represent molecular motors (dynein and Kinesin) and the passengers represent different RNA cargo (Gurken,wingless Ftz and Oskar mRNA); Ilan Davis:http://www.bioch.ox.ac.uk
Once transcribed, in yeast bud-tip one finds localized mRNAs, typified by Ash1 mRNA, may be identified by She2p in the nucleus; thereby marking them as targets for mRNA localization. In the cytoplasm, the She2p/mRNA complex associates with She3p and Myo4p forming a heterotrimeric transport complex. Subsequently, the transport complex delivers the mRNA cargoes to daughter cells on polarized actin cables. The mRNA cargoes are retained at the bud tip by an anchoring complex (AC) that may contain components of the actin cap structure; translational machinery as well as Khd1p. Myo4p and She3p deliver a second cargo, ER, to daughter cells. mRNAs such as OXA1 that are found associated with mitochondria are likely to be targeted by a diffusion/entrapment mechanism.

The intracellular localisation of mRNAs is a general mechanism to target proteins to the regions of a cell where they are required, and plays an important role in the polarisation of many cell types. A striking example of this phenomenon is provided by the Drosophila oocyte, where the localisation of bicoid, oskar, and gurken mRNAs to three distinct positions within the cell determines the polarity of the anterior-posterior and dorsal-ventral axes of the embryo. Using the powerful genetics of Drosophila , we are using a combination of molecular, cell-biological and genetic techniques to investigate the mechanism of mRNA localisation. In addition, we are studying how the two axes of the oocyte become polarised to define the destination of these transcripts, in order to understand the origin of polarity in Drosophila development; St Johnston Lab Home Page; http://www2.gurdon.cam.ac.uk/

http://www.mun.ca/biology/
The localization of Bicoid mRNA and bicoid proteins- Drosophila oocyte;. Bicoid mRNA has been labeled with MS2-GFP and Oskar mRNA with RFP- http://www.mun.ca/
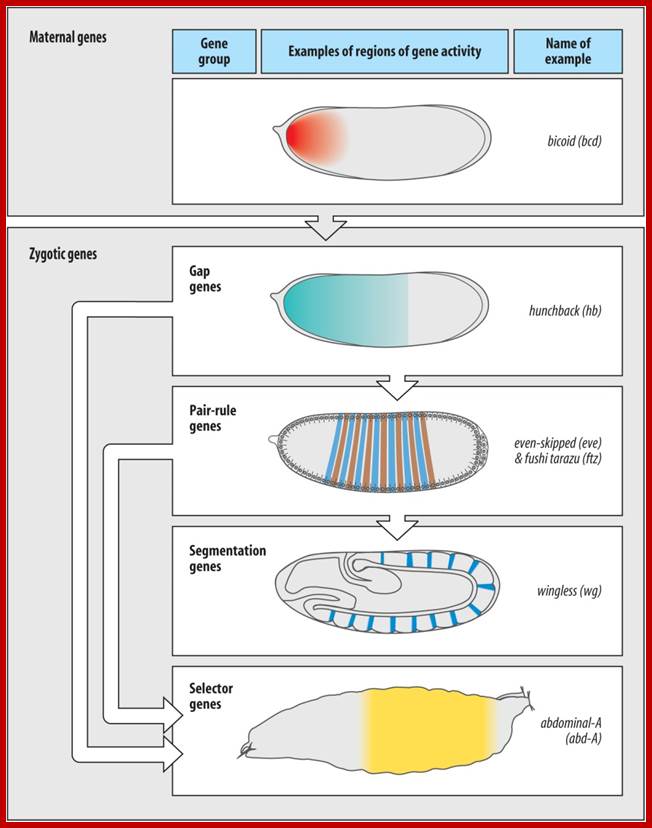 Three Three classes of maternal genes set up the A/P axis
Three Three classes of maternal genes set up the A/P axis
Maternally expressed genes distinguish the
anterior from the posterior. ;http://www.mun.ca/biology

http://www.mun.ca/biology

Bicoid and Bcd-target gene expression along the anteroposterior axis of the Drosophila embryo.: Localization of the bcd mRNA is shown in blue. The Bcd protein gradient is represented in red and the expression of otd, hb and kni represented in green. Embryos are shown anterior to the left and dorsal; http://scienceblogs.com/
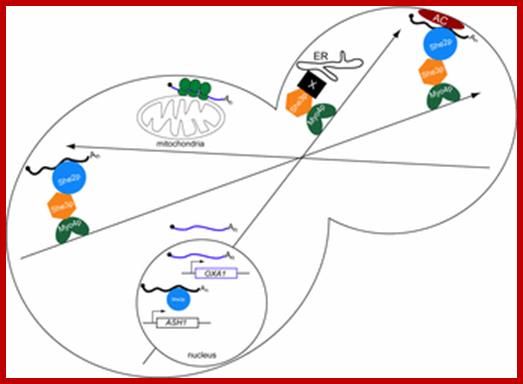
RNA localization in yeast: moving towards a mechanism; Differential routes for mRNA localization in yeast and transport of ER to daughter cells; Higher eukaryotes utilize both microtubules and microfilaments for polarized growth. Long distance transport is often accomplished using microtubules, whereas short-range transport and anchoring often utilize microfilaments. Actin cables serve as the highway for delivering molecular components (e.g. secretory vesicles, vacuole membrane, peroxisomes and Golgi cisternae) to buds (Pruyne and Bretscher, 2000a; Pruyne and Bretscher, 2000b),Graydon B. Gonsalvez*, Carl R. Urbinati† and Roy M. Long†;http://www.biolcell.org/

Central lab-Mobtreal University. Model for the transport and localization of bud-localized mRNAs in yeast. Image modified from Chartrand et al., Annu. Rev. Cell. Dev. Biol., vol. 17. (2001).
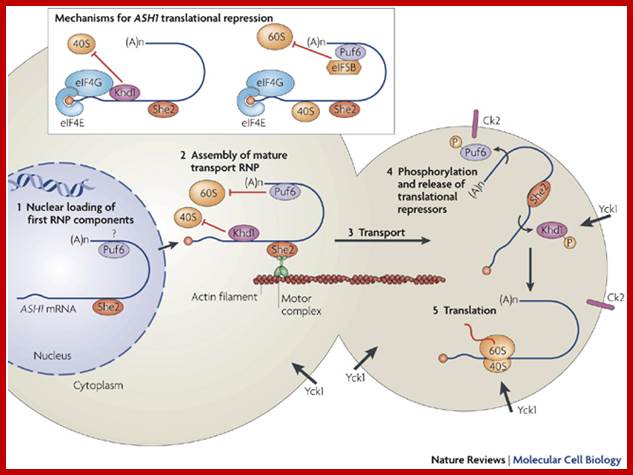
Several trans-acting factors involved in the localization of the ASH1 mRNA in yeast have been identified, allowing researchers to develop a complete working model of this pathway (Figure ). In this model, the ASH1 transcript is first recognized in the nucleus by RNA-binding proteins, especially She2p which recognizes the localization elements, or “zip codes”, in the localized transcripts. Once in the cytoplasm, the localization machinery assembles on the tagged mRNA as a complex called the “locasome". This complex includes the molecular motor Myo4p and the bridging protein She3p. The locasome is transported along the actin cytoskeleton to the bud tip where it becomes anchored. Once localized, the mRNA is translated and the Ash1 protein appears at the bud tip and diffuses back to the daughter cell nucleus.
Model for the regulation of Khd1p-mediated translational repression of the ASH1 mRNA: The ASH1 mRNA is recruited by the Khd1p-eIF4G1 complex, which represses its translation (1), and associates with the localization machinery (2). The resulting RNP complex moves towards the bud on the actin cytoskeleton (3). Upon reaching the bud, Khd1p is phosphorylated by the membrane-anchored Yck1p (4) which lead to the local activation of ASH1 mRNA translation (5). Image taken from Paquin et al.(2007).

Kelly A. Doroshenk, Andrew J. Crofts, Haruhiko Washida, Mio Satoh-Cruz, Naoko Crofts, Yongil Yang, Robert T. Morris, Thomas W. Okita, Masako Fukuda, Toshihiro Kumamaru and Hikaru Satoh; http://www.landesbioscience.com
In plants storage –protein mRNAs transport; Although evidence supports the existence of two regulated RNA localization pathways to the ER that are dependent upon RNA cis‑localization sequences and cytoskeleton, little is known about the trans‑acting factors required for transport. Much remains to be discovered about plant RBPs in general and the identification and subsequent characterization of cytoskeleton‑associated RBPs and accessory proteins involved in seed storage protein RNA localization will further our knowledge of cytoplasmic gene expression events.

mRNA localization is a widely used mechanism for the spatial and temporal control of gene expression. In higher eukaryotes, the mRNA‑transport machinery has a complexity that renders a mechanistic understanding at the molecular level difficult. During the last 15 years, studies in fungi have proven to be attractive alternatives. They are experimentally more accessible and the organization of their mRNA‑transport particles is less complex. Here, our current understanding of mRNA transport in fungi will be summarized. The highly specific mRNA localization observed in the budding yeast Saccharomyces cerevisiae will be compared with mRNA localization in the human pathogen Candida albicans and the less specific transport of transcripts in the plant pathogen Ustilago maydis. Dierk Niessing;
![]()
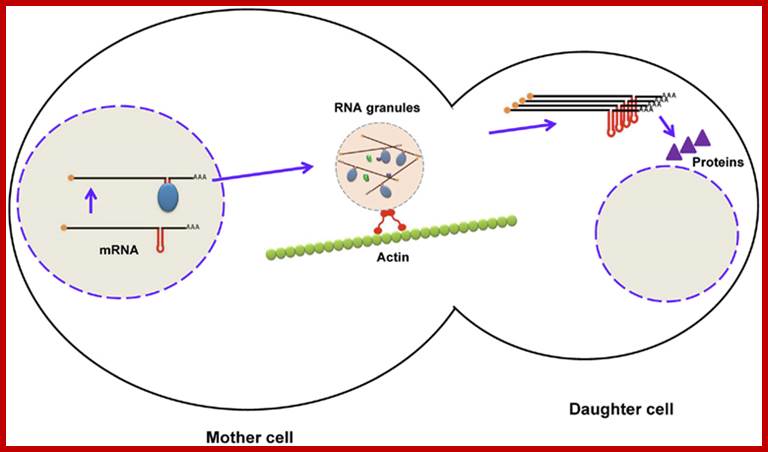
Localization of mRNA in budding yeast using zipcodes. During maturation of mRNA (ASH1), RNA-binding proteins bind to zipcodes and form ribonucleoprotein complexes. After export to the cytoplasm, some proteins are added to form RNA granules, which are transported to the daughter cell along the actin cytoskeleton. After mRNA reaches the distal pole, it is translated into Ash1p (a transcription factor), which enters the nucleus of the daughter cell. Firoz Ahmedhttp://journal.frontiersin.org;

In budding yeast, localization of ASH1 mRNA to the bud tip by myosin-mediated transport on actin cables (see A in figure) targets Ash1p to the daughter cell, where it is required to repress mating type switching (reviewed by Gonsalvez et al., 2005). Thus, mating type switching occurs only in the mother cell, thereby ensuring that both yeast mating types are present in the population. Germ layer specification in Xenopus embryos relies on localized mRNAs, including Vg1 and VegT (reviewed by King et al., 2005) (see B in figure), which are transported to the vegetal pole of the oocyte by kinesin motors and anchored to the cortex by an actin-dependent mechanism. After fertilization, Vg1 and VegT RNAs are inherited by the embryonic vegetal cells, where Vg1 protein, a TGFβ homolog, participates in mesoderm specification, while VegT, a transcription factor, regulates endoderm specification and mesoderm induction. mRNA localization plays an important role in the polarization of somatic cells, such as fibroblasts and neurons (see C,D in figure). Localization of β-actin mRNA to the leading edge of migrating fibroblasts provides a high local concentration of actin monomers that drives assembly of the actin filaments needed for forward movement. Similarly, β-actin mRNA localization to growth cones in developing axons promotes the motility required for axon guidance (reviewed by Condeelis and Singer, 2005). Dendritic localization of RNAs like calcium/calmodulin-dependent protein kinase IIα (CaMKIIα) mRNA in hippocampal neurons facilitates a rapid response to synaptic activity in the form of local protein translation and contributes to learning and memory-related synaptic plasticity (reviewed by Martin et al., 2000).. Agata N. Becalska and Elizabeth R. Gavis*:

Ira Clark1 [Author Vitae], Edward Giniger2,Hannele Ruohola-Baker3,Lily Yeh Jan1, Yuh Nung Jan1,
Figure: Speculative model for polar plasma assembly, derived in part from arguments presented in text. The genetic pathway shown in (a) has been established by a wide body of work summarized in the introduction and in recent reviews. We suggest that the upstream genes cappuccino, spire, Notch and Delta, affect the organization of, or transport along, the microtubule cytoskeleton in oocytes at stages 8 to 9. (b) Schematic diagram of a stage 9 egg chamber, with most of the nurse cells removed for purposes of illustration. We speculate that association with plus-end-directed microtubule motors could promote transport of Staufen (Stau) protein (blue squares) and osk RNA (yellow triangles) along oocyte microtubules to the oocyte posterior, as also suggested in; we emphasize, however, that an association of osk and Stau with microtubule motors has not been demonstrated. We hypothesize that kin:βgal may mimic such a process by transporting itself along microtubules to the posterior.
Assembly of the polar plasm involves the sequential localization of several messenger RNAs and proteins to the posterior of the oocyte, beginning with the localization of oskar mRNA and Staufen protein during stages 8 and 9 of oogenesis. The mechanism by which these two early components accumulate at the posterior is not known. We have investigated whether directed transport along microtubules could be used to accomplish this localization.
Conclusion: In Drosophila, association with a plus-end-directed microtubule motor can promote posterior localization of a reporter protein during oogenesis. The genetic requirements for this localization and its sensitivity to colchicine, both of which are shared with the posterior transport of oskar mRNA and Staufen protein, suggest that similar mechanisms may function in both processes.

Graydon B. Gonsalvez*, Carl R. Urbinati† and Roy M. Long †RNA localization in the above figure, is a widely utilized strategy employed by cells to spatially restrict protein function. In Saccharomyces cerevisiae asymmetric sorting of mRNA to the bud has been reported for at least 24 mRNAs. The mechanism by which the mRNAs are trafficked to the bud, illustrated by ASH1 mRNA, involves recognition of cis-acting localization elements present in the mRNA by the RNA-binding protein, She2p. The She2p/mRNA complex subsequently associates with the myosin motor protein, Myo4p, through an adapter, She3p. This ribonucleoprotein complex is transported to the distal tip of the bud along polarized actin cables. While the mechanism by which ASH1 mRNA is anchored at the bud tip is unknown, current data point to a role for translation in this process, and the rate of translation of Ash1p during the transport phase is regulated by the cis-acting localization elements. Subcellular sorting of mRNA in yeast is not limited to the bud; certain mRNAs corresponding to nuclear-encoded mitochondrial proteins are specifically sorted to the proximity of mitochondria. Analogous to ASH1 mRNA localization, mitochondrial sorting requires cis-acting elements present in the mRNA, though trans-acting factors involved with this process remain to be identified. This review aims to discuss mechanistic details of mRNA localization in S. cerevisiae.

mRNA transport in Drosophila neurons: Patricia S. Estesa, Michele O’Sheaa, Sara Clasena, Daniela C. Zarnescu. Motor proteins Dynein and Kinesins transport RNAs on cable microtubules in opposite direction, Dynein from + end to _ end and Kinesins from (-) end to ( + ) end.
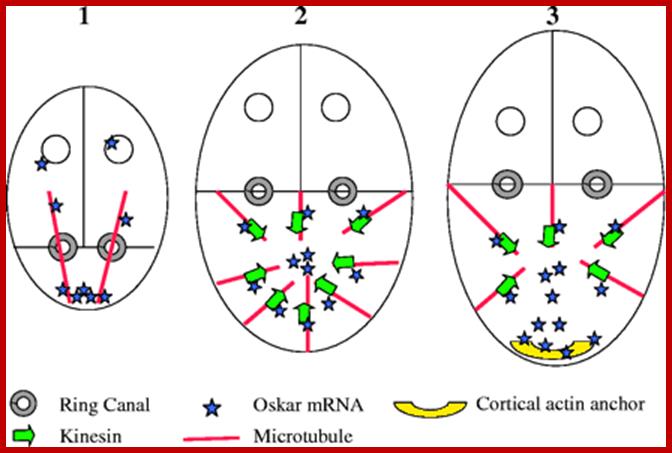
Cell science; Transport and anchoring of oskar mRNA in Drosophila oogenesis. (1) In early oogenesis (stage 2-6), the MTs extend from the oocyte posterior to the nurse cells. Then, oskar mRNA is transported from the nurse cells, on the MTs, via ring canals, to the posterior cortex of the oocyte. (2) In mid oogenesis (stage 8), there is a re-polarization of the oocyte MTs, the MTOC at the oocyte posterior disassembles, and new MTs are nucleated from most of the oocyte cortex. The plus-end-directed motor kinesin transports oskar mRNA away from the cortex and towards the oocyte interior. (3) Subsequent destabilization of MTs at the oocyte posterior (late stage 8 and early stage 9) uncovers the posterior actin anchor, leading to the entrapment and concentration of oskar mRNA at the posterior pole [adapted from Cha et al cha et al 2004].

Abstract. We (above authors) developed and analyzed a stochastic model of directed intermittent search for a hidden target on a one-dimensional track. A particle injected at one end of the track randomly switches between a stationary search phase and a mobile, non-search phase that is biased in the anterograde direction. There is a finite possibility that the particle fails to find the target due to an absorbing boundary at the other end of the track or due to competition with other targets. Such a scenario is exemplified by the motor-driven transport of mRNA granules to synaptic targets along a dendrite. We first calculate the hitting probability and conditional mean first passage time (MFPT) for finding a single target. We show that an optimal search strategy does not exist, although for a fixed hitting probability, a unidirectional rather than a partially biased search strategy generates a smaller MFPT. We then extend our analysis to the case of multiple targets, and determine how the hitting probability and MFPT depend on the number of targets; Paul Bressloff1 and Jay Newby;
|
Inside living cells organelles and other cell constituents are constantly moving. While for many small molecules and particles diffusion is sufficient to transport them to their destination, larger organelles require active transport. This is provided by the system of molecular motor proteins that transport cargos along the cytoskeletal tracks (see schematic in figure 1). The importance of active transport is most evident in the case of neurons, where the axon extremity of the cell can be tens of centimeters long and of the order of a micrometer thin. For a large cargo to diffuse from one end to the other could take more than a lifetime! Examples of such cargos include vesicles, mRNA particles, mitochondria, lipid droplets, golgi bodies, melanosomes and pathogens like virus particles. Motor function is manifest in a wide range of cellular activities such as chromosome movement during cell division, ribonucleoprotein particle delivery in the developing Drosophila oocyte, and the long-distance axonal transport of vesicles between nerve cell bodies and the synapse, to name a few examples. Failure of this transport machinery leads to defects in many cases. Impaired axonal transport in motor neurons, for example, is associated motor neuron degeneration observed in many diseases. |

Molecular Motors; University of Texas at Austin; http://chaos.utexas.edu/
Translational control of localized mRNAs: restricting protein synthesis in space and time; Florence Besse & Anne Ephrussi.
Trans-acting factors, such as She2, first associate with ASH1 mRNA in the nucleus (step 1), and are subsequently exported together with the mRNA to the cytoplasm. A mature transport ribonucleoprotein particle (RNP) is then assembled (step 2) by further recruitment of motor proteins and translational repressors (Khd1 (also known as Hek2) and Pumilo-homology domain family-6 (Puf6)). Note that Puf6 strongly accumulates in the nucleus but has not been shown to associate with the mRNA in this compartment. During transport along actin filaments (step 3), ASH1 mRNA translation initiation is blocked by two complementary mechanisms (inset) that prevent assembly of the eukaryotic translation initiation factor-4F (eIF4F) complex and recruitment of the 40S ribosomal subunit (Khd1-mediated mechanism; left), and prevent recruitment of the 60S ribosomal subunit (Puf6-mediated mechanism; right). After reaching the bud tip, ASH1 RNP contacts the membrane-associated kinases Yck1 (type I casein kinase) and casein kinase-II (Ck2). Phosphorylation of Khd1 and Puf6 by Yck1 and Ck2, respectively, (step 4) induces their release from the complex, and leads to translational activation of ASH1 mRNA (step 5). (A)n, polyadenine.

Techniques for following the movement of single RNAs in living cells
Advanced Review
The ability to investigate gene expression has evolved from static approaches that analyze a population of cells to dynamic approaches that analyze individual living cells. During the last decade, a number of different fluorescent methods have been developed for monitoring the dynamics of single RNAs in living cells. Spatial–temporal analyses of single RNAs in living cells have provided novel insight into nuclear transport, RNA localization, and decay. Technical advances with these approaches allow for single molecule detection, providing an unprecedented view of RNA movement. In this article, we discuss the methods for observing single RNAs in living cells, highlighting the advantages and limitations of each method. WIREs RNA 2011 2 601–609 DOI: 10.1002/wrna.83
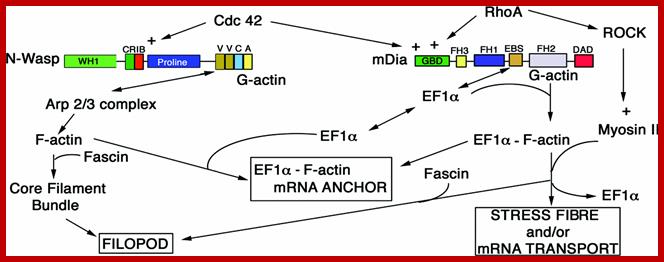
A schematic model for the relationship of the actin remodelling complex to the localization of b-actin mRNA;John Condeelis and Robert H. Singer; http://submit.biolcell.org/
b-Actin mRNA is localized near the leading edge in several cell types where actin polymerization is actively promoting forward protrusion. The localization of the b-actin mRNA near the leading edge is facilitated by a short sequence in the 3´UTR (untranslated region), the ‘zipcode’. Localization of the mRNA at this region is important physiologically. Treatment of chicken embryo fibroblasts with antisense oligonucleotides complementary to the localization sequence (zipcode) in the 3´UTR leads to delocalization of b-actin mRNA, alteration of cell phenotype and a decrease in cell motility. The dynamic image analysis system (DIAS) used to quantify movement of cells in the presence of sense and antisense oligonucleotides to the zipcode showed that net path length and average speed of antisense-treated cells were significantly lower than in sense-treated cells. This suggests that a decrease in persistence of direction of movement and not in velocity results from treatment of cells with zipcode-directed antisense oligonucleotides. We postulate that delocalization of b-actin mRNA results in delocalization of nucleation sites and b-actin protein from the leading edge followed by loss of cell polarity and directional movement. Hence the physiological consequences of b-actin mRNA delocalization affect the stability of the cell phenotype.
Jaffrey lab; at Weill Medical College of Cornell University;
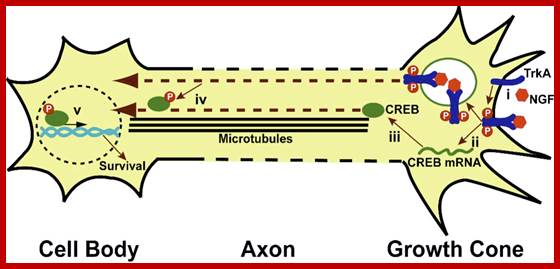
Local translation and retrograde transport of CREB mediates neuronal survival. (i) NGF binds and activates TrkA receptors. (ii) TrkA activation leads to translation of axonal CREB mRNA and (iii) the production of CREB protein. NGF-bound, activated TrkA receptors are internalized into endosomes and axonally translated CREB protein associates with this NGF-pTrkA signaling endosome, which is required for downstream activation of CREB signaling in the cell body. (iv) CREB is retrogradely transported to the nucleus. (v) Axonally-derived pCREB initiates the transcription leading to neuronal cell survival. Jaffrey Lab; at the Weill Medical College of Cornell University
|
|

Micro RNA in axon guidance; Archana N. Iyer et al.;http://journal.frontiersin.org/
Model of miRNA-mediated regulation of axon guidance; During pathfinding, tight regulation of mRNAs occurs to ensure protein expression of guidance molecules at the right time and place, and enable accurate growth cone steering. Within projection neurons, transcripts are translated into the cell body and are subsequently transported within the axon to the growth cone to mediate guidance cue-induced signaling. Alternatively, mRNAs associate into messenger ribonucleoprotein particles (mRNPs) to be transported to the growth cone, where they can be locally translated. Retrograde transport of transcripts from growth cones to cell soma also exists (not represented here). miRNAs are speculated to act at multiple level. They may regulate transcripts translation and stability (1) within the cell body as suggested for miR-124 and lin-4 (Baudet et al., 2012; Zhang et al., 2013) or (2) directly within growth cones as suggested for miR-134 (Han et al., 2011) and by the presence of RISC within this compartment (Table 3). (3) As speculated (Kosik, 2006), miRNAs may translocate along the axons alone or within mRNPs (shown here) and/or be transported as pre-miRNAs and locally produced within growth cones. Guidepost cells are important partners for projection neurons, as they provide them with positional information through the expression of guidance cues. The regulation of guidepost cell transcriptome is thus of crucial importance to ensure the correct patterning of these cells and also the delivery of the right guidance cue at the right place. miRNAs could act by directly regulating the expression of guidance cues within guidepost cells (4) or by indirectly regulating molecules involved in the patterning of these cells (5), as suggested for miR-9 (Shibata et al., 2011).