Nuclear mRNA Transport:
In bacterial system, cytoplasm is not compartmentalized where transcription and translation processes are coupled. Eukaryote cells are highly compartmentalized and transcription and translation are uncoupled or separated in time and space.
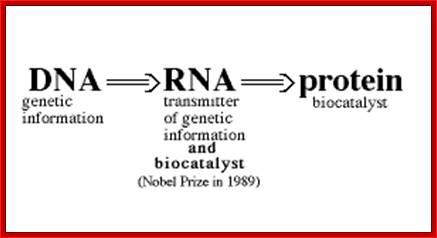
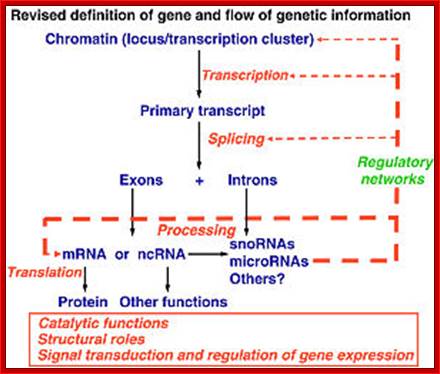
The silence of Lambs The power of Uncoded RNA; An Illustration of Central dogma of Molecular Biology annotated with the process ncRNA are involved in Credit WIKIPEDIA; http://pharmaceuticalintelligence.com/
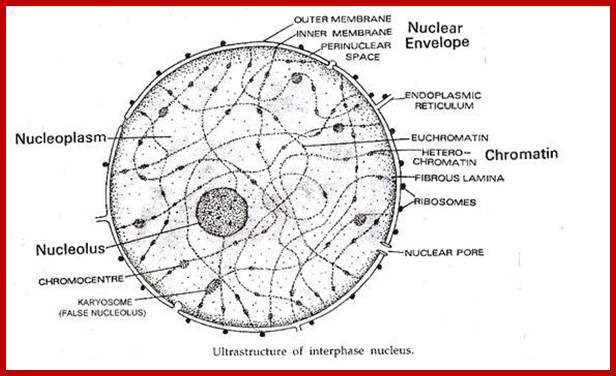
http://www.yourarticlelibrary.com/
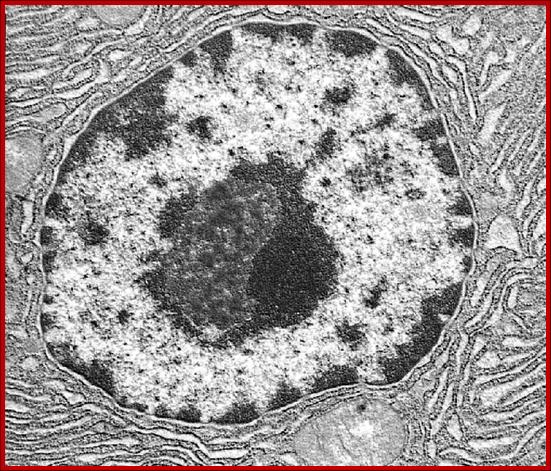
2D EM-image showing double-membrane Nuclear envelope, Nucleolus, condensed chromatin and diffused chromatin; http://os1.amc.nl/celbiologie/
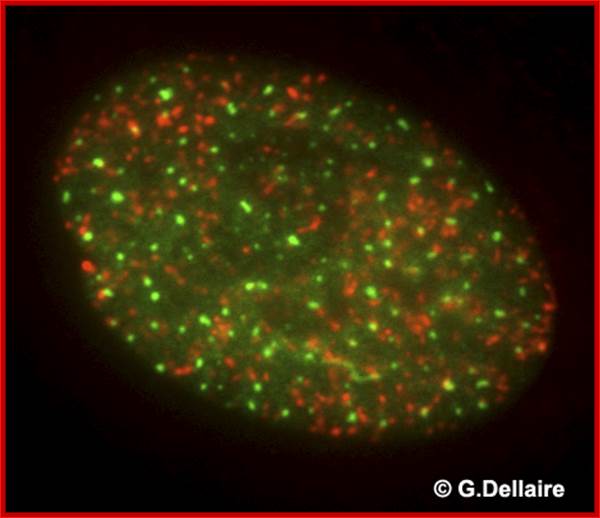
This is an image taken by wide field epifluorescence light microscopy of a human skin fibroblast 10 minutes after irradiation with 10 Gray of ionizing radiation. Promyelocytic leukemia nuclear bodies (PML NBs) are shown in green and repair foci of damaged chromatin containing phosphorylated histone H2AX are shown in red. Damaged chromatin does not co-localize with PML NBs immediately after DNA damage. http://dellairelab.medicine.dal.ca/; www.umassmed.edu/cellbio/
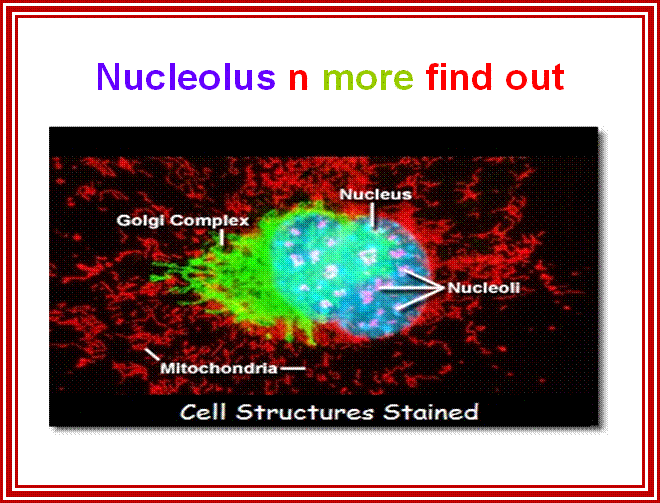
This EM is great for it shows all the structures with specific colors; http://labs.umassmed.edu/
http://www.webring.org/
Enriched in pre-mRNA splicing factors located in interchromatin region. Proteins and RNAQ recycle between speckles and other nuclear bodies, Mintz ,P, et al http://spectorlab.cshl.edu/
![]()
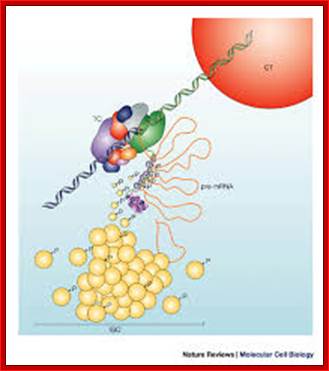
Nuclear speckles- A model for nuclear organelles; Nuclear speckles (interchromatin granule clusters; IGCs) form as the result of protein–protein interactions among pre-messenger RNA splicing factors and other constituents at the telophase/G1-phase transition. A basal level of factor exchange occurs between the speckles and the nucleoplasmic pool that is regulated by phosphorylation/dephosphorylation in a cell-type-specific manner. Modulation of the phosphorylation level of speckle proteins results in an increased release and recruitment to transcription sites. The model is not drawn to scale, and is modified with permission from Ref. 97 © Saunders (2002). CT, chromosome territory; IGC, interchromatin granule cluster; TC, transcription complex; pre-mRNA, pre-messenger RNA. Angus I. Lamond; http://www.nature.com/
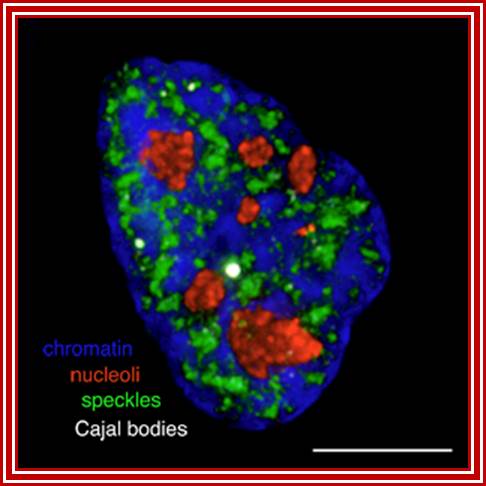
Chromatin-Blue, Nucleoli-Red, Speckles-green and cajal bodie-nuse colored; Group leader: Dr Judith Sleeman; https://www.st-andrews.ac.uk
|
Structure |
Dimensions |
|
Cajal Bodies |
0.2-2nm |
|
PIKA |
5um |
|
PML bodies |
0.2-1.0um |
|
Paraspeckles |
).202um |
|
Speckles |
20-25um |
http://micro.magnet.fsu.edu/
Most of the mRNAs in eukaryotes are synthesized as long precursor RNAs in the nucleus. Until these are processed to functional RNAs they are not transported out of the nucleus. Only processed ones are transported out.
First the pre-mRNAs are associated with certain proteins called hnRNPs which are involved in hnRNA processing; once processing is complete, mRNAs associate with specific proteins, such as Cap binding complex proteins, poly-A binding PAB-II, exon-joint complexes (EJC) and many others, which cover the whole length of mRNA.
- Transportation has to take place through nuclear pore complex (NPC) found in two-unit nuclear membrane. The pore complex Mol.wt is 120 x10^6 Daltons while the Mol.wt of ribosome is just 2million Daltons. The pore complex consists of a veritable structural proteins as well as transport proteins.
Transport is an active process and specific, not all RNAs are transported out, only those specified and marked are transported. None of the cytoplasmic RNAs that are transported into cytoplasm return to the nucleus with certain exceptions such as few snRNAs.
Similarly many inputs for DNA replication, RNA synthesis and many such components have to be imported via Nuclear pore complex (NPC).
Transportation of ribosomes, tRNA, snRNA, scRNAs and other NC RNAs and mRNA is highly regulated.
The presence of Sn RNPs at splice junction sites in unspliced mRNAs prevents them from transport. But mRNAs with specific EJC proteins found at exon-exon splice sites are transported. Mutation in one site does not allow transportation but mutations at both sites allow transportation. Only capped mRNAs are transported.
All mRNA coated with mRNPs show helically coiled forms. In Chironema titans, in a larvae at 11 th day, many genes are expressed in large scale, which appear as chromosomal puffs and Balbiani rings, but one of the transcripts, for a glue protein for the larva, is expressed in massive amount. The protein produced is responsible for the larva to glue to the surface of the substrate at the time of pupal transformation. This mRNA is transported in large amounts. The transcripts are long and covered with RNPs and coiled. As they transport across the pore complex, they unwind.
In every active cell huge precursor mRNAs are found. The mRNAs which contain polyadenylation site and poly9A) is added. They are then transported with their cap structure in fore-front and poly-A tail at 3’end. Transportation is selective and determined by certain nuclear proteins.
Adenoviral transcripts are preferably transported against host mRNAs, because the 5’ ends of these transcripts are associated with E1B and E4 proteins.
During transportation, mRNAs are bound to ribosomes at their 5’end. In the case of HIV, full length transcript is not transported until and unless it is associated with specific REV protein at its Rev Recognizing Elements (RRE) found at 3’ end. The primary transcript, if it is 9 KB, with first splicing; its size is reduced to 4kb. This is not transported. But the second splicing leads to 2kb mRNA; this 2kb mRNA is transported and translated. The product is REV protein, which moves back into the nucleus and binds to full length mRNA at (Rev Response Element) RRE element; then only the full length HIV mRNA is transported out.
U2 Sn RNA transcribed by RNAP-II is transported but U6Sn RNA transcribed by RNAP-III is not transported for it has no cap structure. Sn RNAs like U1, U2, U4 and U5 are capped. They move out of the nucleus, there they are further methylated at cap sites (Tri methylation), associate with certain protein such as SM (aptamers) and then they return to the nucleus.
Many of the mRNPs transported with mRNA into cytoplasm are displaced with cytoplasmic mRNPs and nuclear mRNPs return to nucleus, a good example is PAB-II.
Presence of poly-A or its absence does not matter for mRNA transport from nucleus to cytoplasm
Many different kinds of Thalassimia are mainly due to their failure to transport their mRNAs out of the nucleus, because of mutations at splicing positions or because of the presence of cryptic splicing sites elsewhere.
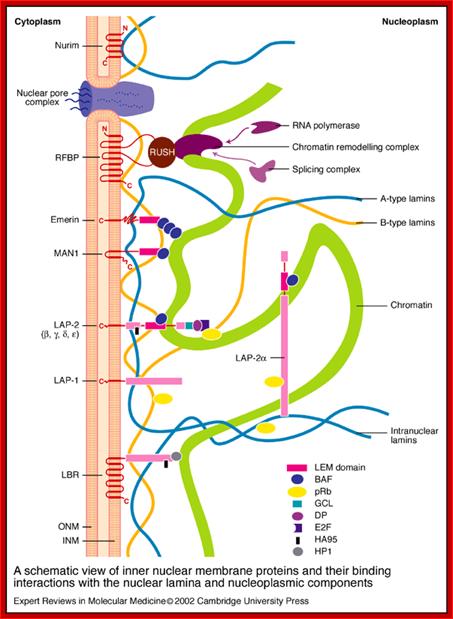
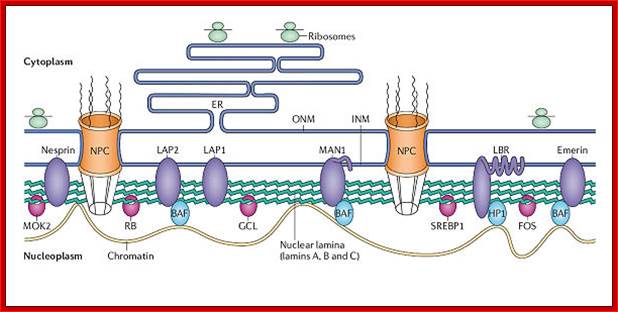
A schematic view of inner nuclear membrane proteins and their binding interactions with the nuclear lamina and nucleoplasmic components; Stephen L. Maidment and Juliet A. Ellis; http://journals.cambridge.org
A schematic view of inner nuclear membrane proteins and their binding interactions with the nuclear lamina and nucleoplasmic components. The outer and inner nuclear membranes (ONM and INM, respectively) are shown in cross-section, with a nuclear pore complex spanning the two membranes. The exact interactions and organization of the inner nuclear membrane, nuclear lamina and chromatin are unknown and are hypothetically depicted in this figure. Twelve inner nuclear membrane proteins have been characterized in the mammalian nuclear envelope. These include: the multi-spanning membrane proteins nurim, lamin B receptor (LBR), ring-finger-binding protein (RFBP); the double-spanning membrane protein MAN1; and the single-spanning membrane proteins emerin, lamina-associated protein 2 (LAP-2) isoforms (b, g, d, e) and LAP-1 isoforms (A, B, C). All the LAP-2 isoforms, emerin and MAN1 share a homologous N-terminal domain called the LEM domain, which binds to BAF (‘barrier to auto-integration factor’). Interactions occur between the inner nuclear membrane proteins and the A-type lamins (shown in blue) and B-type lamins (shown in orange), which are helical filamentous proteins of the nuclear lamina and nucleoplasm. Intranuclear lamins bind to the soluble LAP-2 isoform LAP-2a. Transcriptional regulators crosslink inner nuclear membrane proteins and chromatin. These include: the retinoblastoma protein pRB; the ‘germ-cell-less’ protein GCL; the transcription factor E2F; and RNA polymerase, RNA splicing complex and DP protein. Heterochromatin-binding proteins include HP1, BAF and HA95. Reviews in Molecular Medicine © Cambridge University Press

The vertebrate nuclear envelope. The two-membrane sheets of the nuclear envelope are separated by a luminal space and are continuous with the bulk endoplasmic reticulum (ER) network. The outer nuclear membrane (ONM) and the inner nuclear membrane (INM) are fused at nuclear pores, where nuclear pore complexes are integrated to regulate bidirectional transport between the cytoplasm and the nucleoplasm. The INM is distinctly characterized by a set of integral membrane proteins that connect the nuclear envelope to chromatin by interacting directly or indirectly via chromatin-associated proteins and the nuclear lamina. The nuclear lamina is additionally connected to the cytoplasmic cytoskeleton by the interaction of LINC complex proteins of the ONM and INM across the NE lumen; http://php.med.unsw.edu.au/
The NE consists of the inner and outer nuclear membrane (INM and ONM), nuclear pore complexes (NPCs) and the lamina. The ONM is continuous with the endoplasmic reticulum (ER). NPCs cross the INM, ONM, the lamina and are associated with chromatin. A-type lamins (LA and LC) and B-type lamins (LB1 and LB2) in the lamina bind to INM proteins such as emerin, lamina-associated polypeptide 2β (LAP2β), lamin B receptor (LBR), and SUN domain proteins (SUN1 and SUN2) in the INM. All of the lamins and some of the INM proteins interact with chromatin. SUN1 and SUN2 bind to the KASH domain of nesprins in the luminal region between the INM and ONM to form the LINC complex. Nesprins in the ONM bind to cytoskeletal filaments such as actin, microtubules, and intermediate filaments (IFs) directly or indirectly through plectin or kinesin. Actin and IFs are associated with the plasma membrane through integrin complexes.
Structure and function of the nuclear lamina. The nuclear lamina lies on the inner surface of the inner nuclear membrane (INM), where it serves to maintain nuclear stability, organize chromatin and bind nuclear pore complexes (NPCs) and a steadily growing list of nuclear envelope proteins (purple) and transcription factors (pink). Nuclear envelope proteins that are bound to the lamina include nesprin, emerin, lamina-associated proteins 1 and 2 (LAP1 and LAP2), the lamin B receptor (LBR) and MAN1. Transcription factors that bind to the lamina include the retinoblastoma transcriptional regulator (RB), germ cell-less (GCL), sterol response element binding protein (SREBP1), FOS and MOK2. Barrier to autointegration factor (BAF) is a chromatin-associated protein that also binds to the nuclear lamina and several of the aforementioned nuclear envelope proteins. Heterochromatin protein 1 (HP1) binds both chromatin and the LBR. ONM, outer nuclear membrane. Coutinho et al. Immunity & Ageing 2009; http://webcache.googleusercontent.com/
![]()
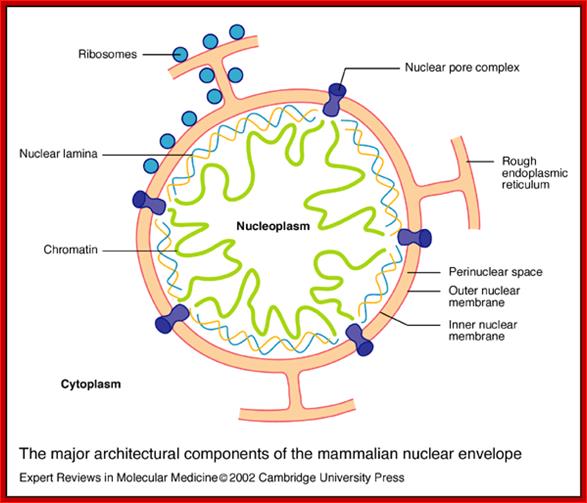
The major architectural components of the mammalian nuclear envelope; Stephen L. Maidment and Juliet A. Ellishttp://journals.cambridge.org/
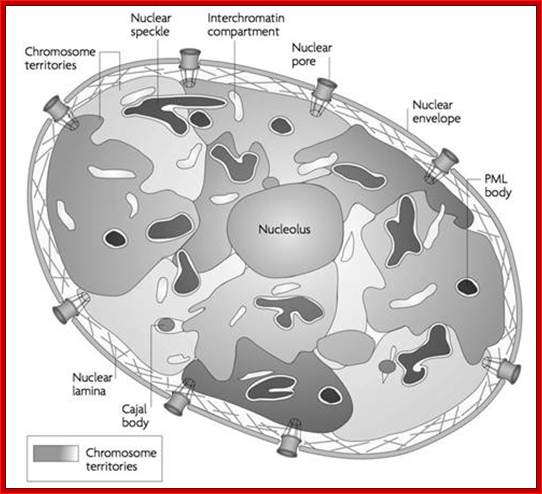
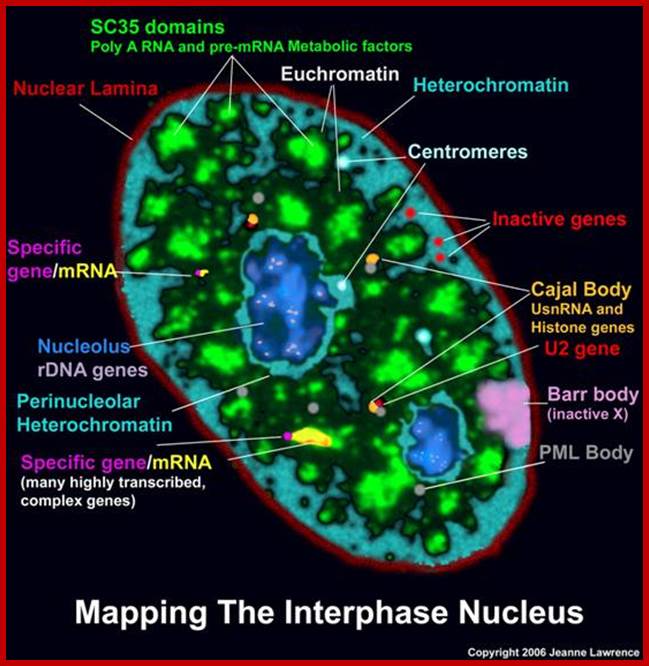
Nucleus is composed of Nuclear membrane (Karyotheca), Nucleoplasm (Karyolymph), Nucleolus and chromatin network; Nuclear speckles, PML bodies, Cajal bodies, and can see chromosomal territories and nuclear lamins http://www.sivabio.50webs.com/
The major architectural components of the mammalian nuclear envelope:
The nuclear envelope consists of two membranes: the outer nuclear membrane is contiguous with the rough endoplasmic reticulum and is coated with ribosomes; the inner nuclear membrane contains unique integral membrane proteins. The space between the two membranes is referred to as the perinuclear space. The two membranes (together making up the nuclear envelope) are spanned by nuclear pore complexes, which are involved in regulating the transport of materials between the nucleus and the cytoplasm. The two membranes join at the nuclear pore membrane, which surrounds the nuclear pore complexes. Underlying the inner nuclear membrane is the nuclear lamina, which is a dense filamentous network. Major components of the nuclear lamina are the nuclear lamins A and B, which are a group of filamentous proteins that interact with both inner nuclear membrane proteins and chromatin. The nuclear interior (nucleoplasm) contains soluble proteins; Stephan L.
While transportation, Nuclear pore complexes show sphincter like motions-contraction and expansion; and they almost behave and functions like a cargo carrier motor protein complex.
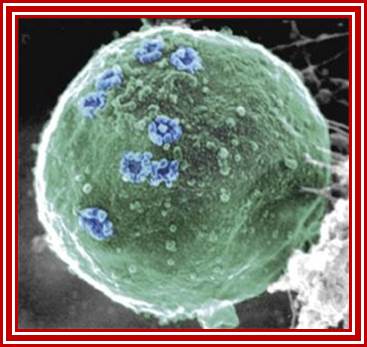
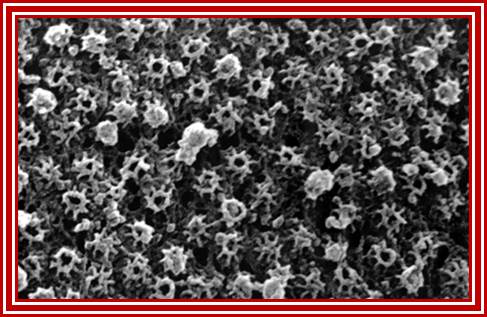
Nuclear Pore Complexes (blue) on nuclei of baker’s yeast visualized by scanning electron microscopy. The NPC has a diameter of 100nm.; Elena Kiseleva); http://www.nature.com/

Gate Keeper for the Nucleus; Nature Magazine front cover Photo.
The nuclear pore complex plays a crucial role in the cell, as gatekeeper for traffic between the cytoplasm and the interior of the nucleus. It is a large supramolecular complex made up of multiple copies of about 30 different proteins - 456 protein molecules in all. Cell biologists would love to know how each of the pore molecules are placed, but so far this has eluded conventional structural studies. Now, a new proteomics-based technique has provided a detailed view of the architecture of the yeast nuclear pore complex. Half of the complex is made of a core scaffold forming a network coating the surface of the nuclear envelope membrane within which the complex is embedded. The selective transport barrier is formed by the many proteins lining the inner face of the scaffold. Despite its size, there are only a few structural modules in the complex; this underlying simplicity provides possible pointers to an evolutionary origin from a 'primordial' nuclear pore complex. [Articles pp.683, 695; News & Views p. 621; www.nature.com/podcast; www.nature.com/news] In the cover graphic, the 100-nm diameter pores are shown in the silver-grey nuclear envelope;Diagrammatic sketch view of the NPC; http://www.nature.com/
Acknowledgement: Electron micrograph kindly provided by Elena Kiseleva, Institute of Cytology and Genetics, Novosibirsk. Reference: "RNA Export is Mediated by Structural Reorganization of the Nuclear Pore Basket", Elena Kiseleva, Martin W. Goldberg, Bertil Daneholt and Terence D. Allen, Journal of Molecular Biology, 260:289-477;http://www.nobelprize.org/
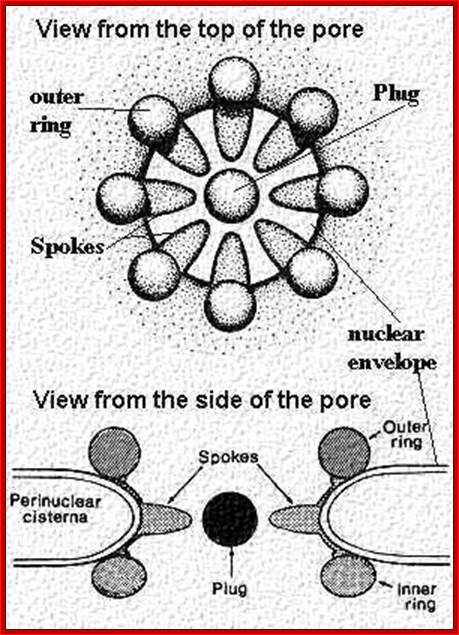
Surface view of the NPC cytoplasmic side-octamer structure visible under TEM; http://www.visualphotos.com/ Nuclear envelope; http://www.cytochemistry.net/
Nuclear transport is facilitated:
There are specific protein receptors which bind to specific proteins and specific RNAs; then they are locked on to transporters; these are then transported across the NPC in energy dependent manner; the energy provider is GTP and it is associated with GTP/GDP binding protein called Ran.
Nuclear pore complex is one of the most complex structures, details of which is yet to be established; similarly the transport proteins involved are yet to be understood, for each cargo there is a specific protein.
Nuclear pore complex:
Nuclear pore complex can be dissociated. At the time of cell division, when nuclear membrane dismembers, nuclear pore complexes perse are released. When nuclear membrane reassociate at the end of telophase they reassemble and probably with more or less the same number of NPCs. The number of NPC differs depending upon the status of the cell. Active cell contain more NPCs than resting cells.
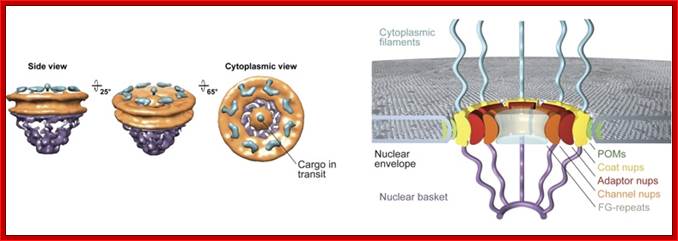
Sole gateway for transport across the nucleus is NPC; http://ahweb.caltech.edu
The NPC is massive structure with many proteins; the total mass is about ~125 mega Daltons (MDa) in vertebrates and 66 MDa in yeast. It has an octagonally organized symmetric cylindrical structure around the axis of transport. NPCs are embedded in the two nuclear membranes. The yeast NPC has been well characterized and it is thought to contain an upper limit of 30 distinct types of proteins (termed nucleoporins, or nups). The number is lower than the number of proteins ~75 in ribosomes whose mol.wt is~ 4MDa. The NPC looks like an open ended cylinder with broader perimeter at its cytoplasmic and nucleoplasmic sides. NPC is approximately 145 nm in diameter and 80 nm length across the nuclear envelope. The yeast NPC is smaller, approximately 96 nm in diameter and 35 nm in length. The central channel is about 69nm; which can expand and contract when required. This in comparison to another transporter aquaporin 6.5nm size shown below, the NPC is truly large.
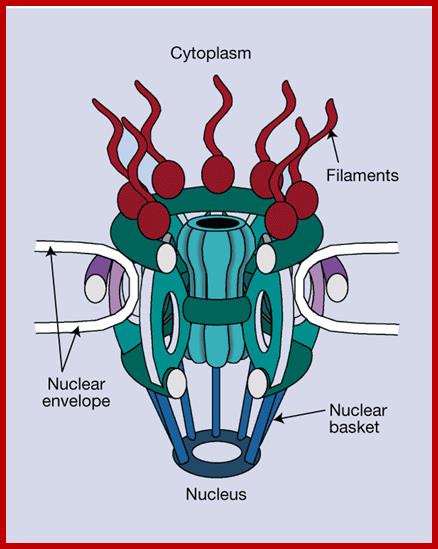
Rout et al. show that it consists of only about 30 different proteins, forming a basic subunit that is repeated 16 times. There are additional, filamentous structures on both sides of the pore. On the nuclear side, these link together to form the nuclear basket.;
Günter Blobel and Richard W. Wozniak; http://www.nature.com/
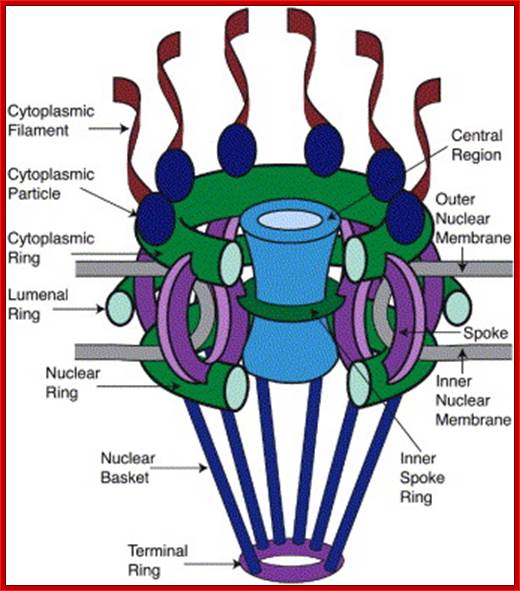
Schematic Diagram of NPC SubstructuresThe structure has an apparent 8-fold rotational symmetry perpendicular to the plane of the nuclear membrane; however, certain facing portions are removed in this diagram to reveal the architecture of the central region. Labels for all the structures found in vertebrate NPCs are included, with the cytoplasmic face on top. Adapted from the model in Rout and Wente, 1994.
NPC and transport:
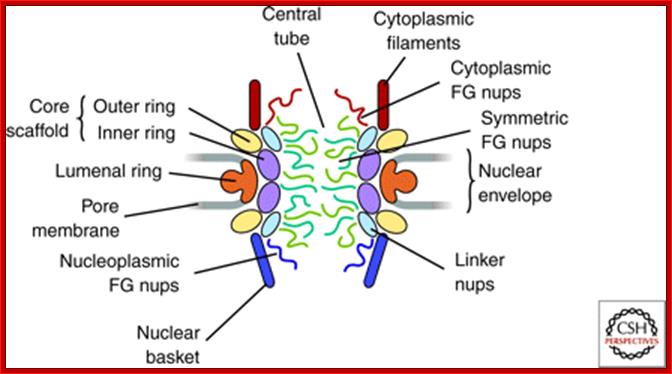
NPC is the most complex structure; .The NPC is Structural Organized into three major domains - the nuclear basket, central core and cytoplasmic filaments. B. Nups form distinct structural and functional domains in the NPC. The lumenal, inner and outer rings anchor and stabilize the structure. Linker Nups form a bridge between this core and the FG Nups making up the transport barrier in the central tube; http://www.mc.vanderbilt.edu/
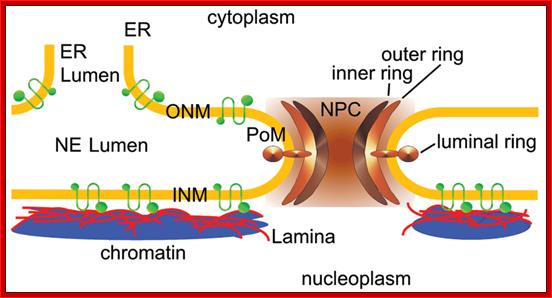
Nuclear pore complex bind to ONM and INM with luminal ring; and observe ER membrane continuous with outer ONM; the INM bound by Lamina cytoskeleton which is also associated with chromatin. Poonam Malik, et al;http://www.landesbioscience.com/
The layered organization of NPC complexes in the form of cylinder is described below:
The NPC associated proteins are called NUPs. There are 8 composite ring of proteins at the cytoplasmic surface and similar ring of eight at the inner surface of the nuclear membrane. From the outer ring each protein emanate filaments into cytoplasm; similarly eight filament produce from the inner ring of proteins, but they are connected to each other produce basket like structure. The membrane embedded part of NPC is associated with nuclear lamins (network of intermediate filaments). A central channel is positioned in the center of the NPC that extends from inner surface of the NPC to the outer surface act as the contractile structure for the movement of proteins, RNAs and metabolites of 40Kd passively. Other larger ones are transported actively, where the channel acts like a sphincter- expands and contracts. Spokes connect the outer ring with central channel proteins. NPC is a dynamic protein complex. The central channel is loaded with importins at the outer surface and exportins at the inner surface of the NPC; they are called karyophorins (importins and exportins).

Rings of NPC with their constituents has been displayed; there are overlaid one on the other; Albert Laboratory; R. Kalhor, H. Tjong, N. Jayathilaka, F. Alber L. Chen http://www.cmb.usc.edu/
Nuclear pore complexes (NPCs) are proteinaceous assemblies of approximately 50MDa that selectively transport cargoes across the nuclear envelope. To determine the molecular architecture of the yeast NPC (Fig. 2), we collected a diverse set of biophysical and proteomic data, and developed a method for using these data to localize the NPC’s 456 constituent proteins. Our structure reveals that half of the NPC is made up of a core scaffold, which is structurally analogous to vesicle-coating complexes. This scaffold forms an interlaced network that coats the entire curved surface of the nuclear envelope membrane within which the NPC is embedded. The selective barrier for transport is formed by large numbers of proteins with disordered regions that line the inner face of the scaffold. The NPC consists of only a few structural modules that resemble each other in terms of the configuration of their homologous constituents, the most striking of these being a 16-fold repetition of ‘columns’. These findings provide clues to the evolutionary origins of the NPC. R. Kalhor, H. Tjong, N. Jayathilaka, F. Alber L. Chen
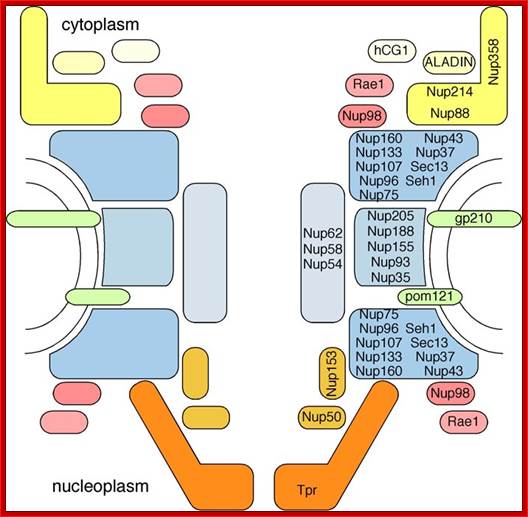
Schematic diagram of the NPC architecture and its subcomplexes modules; Thomas U. Schwartz; ttps://biology.mit.edu
Chromosomal domains associated with inner Nuclear membrane: Although, often depicted as a static structure upon which proteinaceous factors bind to control gene expression, the genome is actually highly mobile and capable of exploring the complex domain architecture of the nucleus, which in turn controls genome maintenance and gene expression. Numerous genes relocate from the nuclear periphery to the nuclear interior upon activation and are hypothesized to interact with pre‐assembled sites of transcription. In contrast to the nuclear interior, the nuclear periphery is widely regarded as transcriptionally silent. This is reflected by the preferential association of heterochromatin with the inner nuclear envelope (NE). However, some activated genes are recruited to the nuclear periphery through interactions with nuclear pore complexes (NPCs), and NPC components are capable of preventing the spread of silent chromatin into adjacent regions of active chromatin, leading to the speculation that NPCs may facilitate the transition of chromatin between transcriptional states. Thus, the NE might better be considered as a discontinuous platform that promotes both gene activation and repression. As such, it is perhaps not surprising that many disease states are frequently associated with alterations in the NE. Here, we review the effects of the NE and its constituents on chromatin organization and gene expression. Copyright © 2010 John Wiley & Sons, Inc.
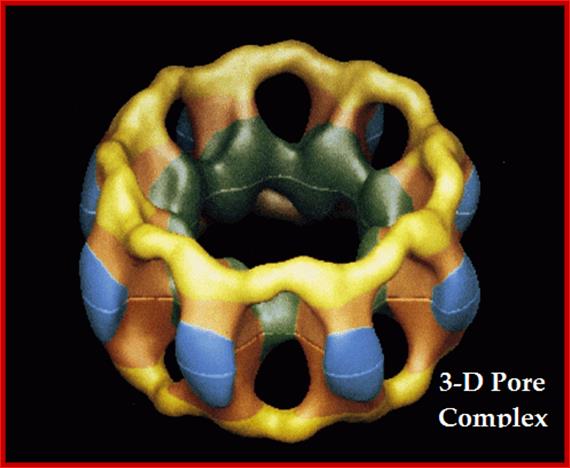
NPCs are the most distinctive structural components of the NE as resolved by EM. NPC structure was initially dissected using transmission EM (TEM), but later expanded to scanning transmission as well as to scanning EM and, most recently, to cryo-electron tomography (CET). The first EM study on the NE in 1950 revealed that it is perforated by pores (Callan and Tomlin1950). Gall ( 1967) showed for the first time that NPCs exhibit an octagonal structure; Lim, Roderick Y. Hhttp://scolaris.beta.semantico.com/
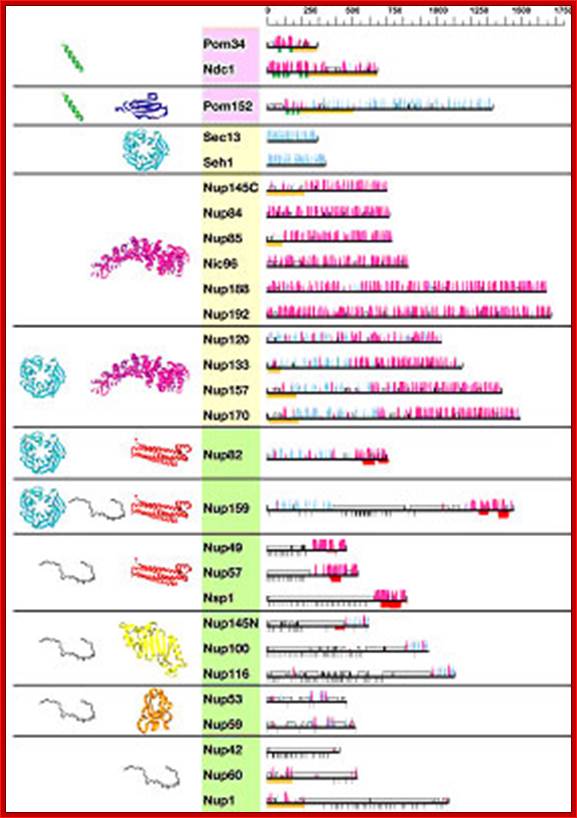
Nucleoporin homologs of yeast and vertebrates (not complete): http://www.pnas.org/
Predicted secondary structure and fold types of the nups; The names of the nups are boxed according to the group they define: transmembrane (pink), scaffold (orange), and FG repeat (green). The black horizontal lines on the right represent the sequence of each yeast nup. The predicted α-helices (magenta) and β-strands (cyan) are indicated by bars above each line. The height of the bars is proportional to the confidence of the prediction (39). Predicted transmembrane helices are shown in green, coiled coils are shown in red, FG repeats are shown in black, and unstructured regions are shown by an empty box. An orange block underlines regions of >50 residues to which a fold type could not be assigned. Representative models of the nup domains are colored according to the fold type and are shown on the left. Models are not to scale for visualization reasons. There are eight fold types. First, a TMH segment (green) is a hydrophobic 15- to 30-residue helical segment that spans the membrane. Second, cadherin domains (dark blue) have ≈110 residues that fold into a seven-stranded β-sandwich structure. Third, β-propellers (cyan) contain several blades arranged radially around a central axis, each blade consisting of a four-stranded antiparallel β-sheet. Fourth, α-solenoid domains are composed of numerous pairs of antiparallel α-helices stacked to form a solenoid. Fifth, coiled coils (red) generally display seven-residue repeats where the first and fourth residues of an α-helix are often hydrophobic. The coiled-coil structure is formed by helices (generally two) twisting together to bury their hydrophobic seams. Sixth, disordered FG-repeat segments are indicated schematically by a black curve. Seventh, the autoproteolytic domain of Nup98 (yellow) adopts a half-open, β-sandwich-like fold dominated by a large β-sheet with helices capping two of its ends. Finally, the RRM (orange) is a two-layer α/β sandwich typically found in proteins involved in RNA binding; http://www.pnas.org/
Predicted secondary structural maps of NUPs: Thin horizontal lines represent the primary sequence of each protein; secondary structure predictions are shown as columns above each line for -strands (cyan) and -helices (magenta), with arrows showing proteolytically sensitive sites. Ribbon representation of nup models: -sheets (propellers), -helices ( -solenoids) are colored magenta, cadherin domains are dark blue, the autoproteolytic domain of Nup98 are yellow, the RRM is orange, predicted transmembrane helices are shown in green, coiled-coils in red, FG repeats in black, and unstructured regions are represented by an empty box. Further complex organization is shown in another figure below.
|
NPC substructure |
Yeast components |
Vertebrate components |
|
Outer Ring |
Nup84 subcomplex (Nup84, Nup85, Nup120, Nup133, Nup145C, Sec13, Seh1) |
Nup107-160 complex (Nup160, Nup133, Nup107, Nup96, Nup75, Seh1, Sec13, Aladin, Nup43, Nup37) |
|
Inner Ring |
Nup170 subcomplex (Nup170, Nup157, Nup188, Nup192, Nup59, Nup53) |
Nup155 subcomplex (Nup155, Nup205, Nup188, Nup35) |
|
Cytoplasmic FG Nups and Filaments |
Nup159, Nup42 |
Nup358, Nup214, Nlp1 |
|
Lumenal Ring |
Ndc1, Pom152, Pom34 |
Gp210, Ndc1, Pom121 |
|
Symmetric FG Nups |
Nsp1, Nup57, Nup49, Nup145N, Nup116, Nup100 |
Nup62, Nup58/45, Nup54, Nup98 |
|
Linker Nups |
Nup82, Nic96 |
Nup88, Nup93 |
|
Nucleoplasmic FG Nups and Filaments |
Nup1, Nup60, Mlp1, Mlp2 |
Nup153, Tpr |
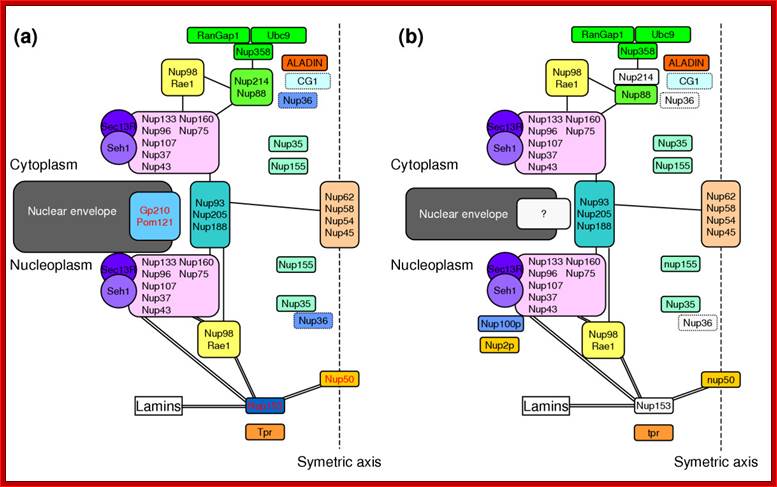
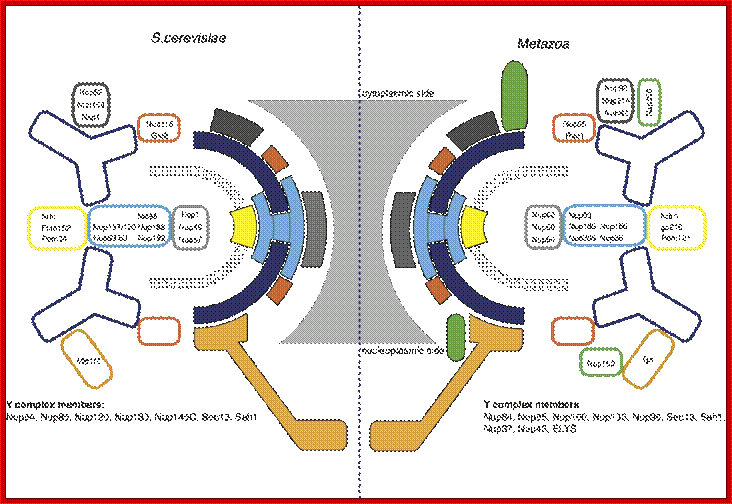
The structure of the nuclear pore complex: Schematic representation of the position of the major nucleoporins sub complexes in (a) unikonts and (b) bikonts. The schematic organization of the NPC in unikonts is based on the schematic organizations of NPC in vertebrates published by Powers and Dasso, completed accordingly with recent works. Boxes delimited by dashed lines indicate proteins having unknown or no precise localization within or around the NPC. Light gray boxes represent nucleoporins present in unikonts but having no homologs in bikonts. Protein names in black in (a) indicate proteins having homologs in fungi, whereas those in red indicate proteins having no homologs but structural analogues in fungi. Lines between sub complexes indicate putative interactions whereas double lines indicate undisputable interactions. Bapteste et al. Genome Biology 2005 6:R85 doi:10.1186/gb-2005-6-10-r85; Inner Nuclear Membrane Proteins are Actively Imported: Category: Pure Biology;by Alex Palazzo
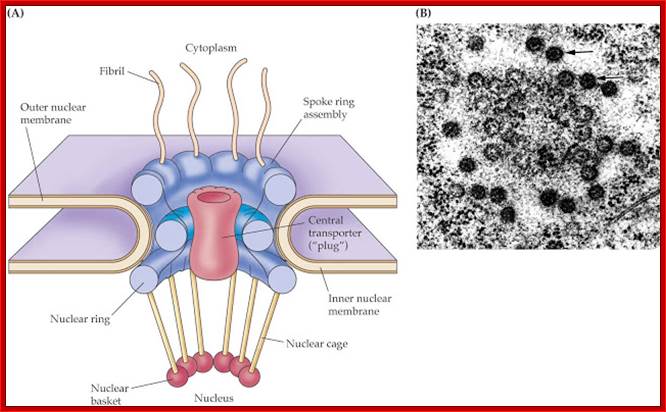
Model of the nuclear pore complex: The complex consists of an assembly of eight spokes attached to rings on the cytoplasmic and nuclear sides of the nuclear envelope. The spoke-ring assembly surrounds a central channel containing the central transporter. Cytoplasmic filaments extend from the cytoplasmic ring, and filaments forming the nuclear basket extend from the nuclear ring. http://www.biologyexams4u.com/
Visualization of nuclear pore complexes by electron microscopy reveals a structure with eight fold symmetry organized around a large central channel, which is the route through which proteins and RNAs cross the nuclear envelope. Detailed structural studies, including computer-based nuclear analysis, have led to the development of three-dimensional models of the nuclear pore complex. These studies indicate that the nuclear pore complex consists of an assembly of eight spokes arranged around a central channel. The spokes are connected to rings at the nuclear and cytoplasmic surfaces, and the spoke-ring assembly is anchored within the nuclear envelope at sites of fusion between the inner and outer nuclear membranes. Protein filaments extend from both the cytoplasmic and nuclear rings, forming a distinct basketlike structure on the nuclear side. The central channel is approximately 40 nm in diameter, which is wide enough to accommodate the largest particles able to cross the nuclear envelope. It contains a structure called the central transporter, through which the active transport of macromolecules is thought to occur. http://www.biologyexams4u.com
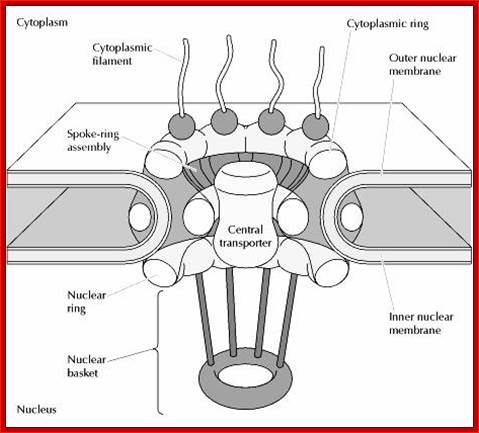
http://www.sivabio.50webs.com/
NPC is like a most sophisticated basket of motor-protein complex; its structural organization befuddles even the sophisticated bioengineers assisted by computer software. There are many layers of rings of proteins which fit into each other, which has been described elsewhere. The motor proteins are Ran-GTP and Ran-GDP assisted by GAPs and GEFs. Karyophorins at the outer surface of the basket and inner surface of the basket play a role in transport. The FG domain repeats play an elusive role in transport inwards and also outwards.
Simple fold composition and modular architecture of the nuclear pore complex:
Simple fold composition and modular architecture of the nuclear pore complex; http://www.pnas.org/
“Simplicity of the fold composition and modular architecture of the NPC- (a) The schematic structure and hierarchy of the NPC. Most of the nups consist of sequence repeats (step I). The nups assemble into multiple copies to form each half-spoke (step II) that dimerize to form the spokes (step III), which are themselves repeated eight times to form the complete NPC (step IV). (b) The architecture of the NPC ring, viewed in the plane of the NE, is segregated into the membrane (pink), scaffold (orange), and FG (green) groups. The domain fold types assigned to each group is indicated on the left side of the schema. The schema illustrates the coarse organization of the NPC and is not a precise map of the three dimension nup locations”. Simple fold composition and modular architecture of the nuclear pore complex; http://www.pnas.org/
Protein Transport: A protein destined for the nucleus and/or cytoplasm contains a specific sequence which can be recognized directly by importin/exportin or through an adaptor protein. For example, importin, among others, is a well characterized importin protein. It cannot recognize the specific sequence but can be assisted by importin which is an adaptor. By contrast, the exportin for the heterogeneous nuclear ribonucleoprotein (hnRNP) can recognize directly the specific sequence in hnRNP. These are named "transportins".
The function of exportins and importins is regulated by a G protein called "Ran". There are two types of G proteins: heterotrimeric G proteins and monomeric G proteins (or small G proteins). The latter is similar to Ras, Rho, Rab or Arf. Like other G proteins, Ran can switch between GTP-bound and GDP-bound states. Transition from the GTP-bound to the GDP-bound state is catalyzed by a GTPase-activating protein (GAP) which induces hydrolysis of the bound GTP. The reverse transition is catalyzed by guanine nucleotide exchange factor (GEF) which induces exchange between the bound GDP and the cellular GTP.
The GEF of Ran (denoted by RanGEF) is located predominantly in the nucleus while RanGAP is located almost exclusively in the cytoplasm. Therefore, in the nucleus Ran will be mainly in the GTP-bound state due to the action of RanGEF while cytoplasmic Ran will be mainly loaded with GDP. This asymmetric distribution has led to the following model for the function of exportins and importins.
It is thought that binding between an exportin or importin and its cargo depends on their interaction with Ran: RanGTP enhances binding between an exportin and its cargo but stimulates release of importer’s cargo; RanGDP has the opposite effect, namely, it stimulates the release of exportins cargo, but enhances the binding between an importin and its cargo. Therefore, the exportin and its cargo may move together with RanGTP inside the nucleus, but the cargo will be released as soon as the complex moves into the cytoplasm (through nuclear pores), since RanGTP will be converted to RanGDP in the cytoplasm. By contrast, the importin and its cargo may move together with RanGDP in the cytoplasm, but the cargo will be released in the nucleus since RanGDP will be converted to RanGTP in the nucleus.
Localization Signals with respect to the Nucleus:
The number of proteins and other raw materials, for the metabolism of nucleic acids, imported is countless (?). They are of various sizes and dimensions. Nucleus does not possess the function of translation machinery (protein synthesis).
Nuclear Import Signals: Two major types of signals have been identified for the nuclear import of proteins: SV40 type and bipartite type. The former was first found in the large T antigen of the SV40 virus. It has the following sequence. PKKKRKV- This type of signal is characterized by a few consecutive basic residues and in many cases also contains a proline residue.
The bipartite type was first identified in Xenopus nucleoplasmin with the following NLS: KRPAATKKAGQAKKKK; Its characteristic pattern is: two basic residues, 10 spacer residues, and another basic region consisting of at least 3 basic residues out of 5 residues.
Nuclear Export Signal (NES): The signal for nuclear export is a leucine-rich domain which can be recognized by a class of exportins called exportin 1 or Crm1. An example is given below:
The NES ‘LQLPPLERLTL’ sequence is found in the rev protein of HIV-1. Some proteins which need to shuttle between the nucleus and cytoplasm contain both NLS and NES. Examples include the rev protein of HIV-1 and the nuclear factor of activated T cells (NFAT). The rev protein plays a key role in the regulation of viral expression. NFAT is the target of immunosuppressive drugs widely used in organ transplantation.
Karyophorins nuclear transport receptors:
A large number of soluble transport receptors mediating either nuclear import or nuclear export have been identified. Most of these receptors belong to one large family of proteins, all of which share homology with the protein import receptor importin β (also named karyopherin β). Members of this family have been classified as importins or exportins on the basis of the direction they carry their cargo. To date, the family includes 14 members in the yeast Saccharomyces cerevisiae and at least 22 members in humans. Importins and exportins are regulated by the small GTPase Ran, which is thought to be highly enriched in the nucleus in its GTP-bound form. Importins recognize their substrates in the cytoplasm and transport them through nuclear pores into the nucleus. In the nucleoplasm, RanGTP binds to importins, inducing the release of import cargoes. In contrast, exportins interact with their substrates only in the nucleus in the presence of RanGTP and release them after GTP hydrolysis in the cytoplasm, causing disassembly of the export complex. Thus, common features of all importin-β-like transport factors are their ability to shuttle between the nucleus and the cytoplasm, their interaction with RanGTP as well as their ability to recognize specific transport substrates.(Anne-Christine Ström1 and Karsten Weis).
Karyopherins are a group of proteins involved in transporting molecules through the pores of the nuclear envelope (the membrane around a cell's nucleus). Karyopherins, which may act as importins or exportins, are part of the Importin-β super-family which all share a similar three-dimensional structure. They belong to the Nuclear Pore Complex Family in TCDB.
For example:
Importin beta + adapter + cargo protein (cytoplasm)-->(karyoplasm), importin beta + adapter + RanGTP (karyoplasm)-->(cytoplasm),Ran-GTP --> RanGDP + P----
Genes: KPNA1, KPNA2, KPNA3, KPNA4, KPNA5, KPNA6 andKPNB1

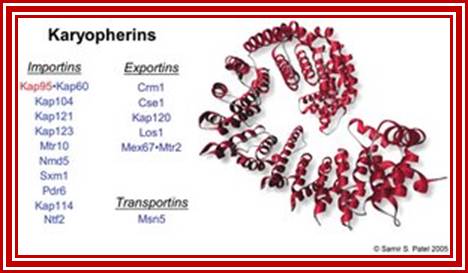
Yeast contains 14 karyophorins, most of them involved in import functions. Karyophorins are divided in to importins and exportins and transportins (bidirectional); Samir Patel; http://www.rcsb.org/Nature v399 pp, 229, 1999; David Goodsell;http://www.rcsb.org/
Structure of importin β. (a) Importin β is composed of 19 helical-repeat motifs (HEAT repeats). Each consists of an A and a B helix connected by a short turn, which in HEAT-8 is replaced by an acidic loop critical for the regulation of substrate binding and release. The HEAT repeats 1-8 are required for high-affinity binding to RanGTP. The importin-β-binding (IBB) domain of importin α interacts mainly with residues located in repeats 7-19 of importin β. The binding site for nucleoporins of the NPC is located between residues 152 and 352, corresponding to repeats 4-8. On the basis of the crystal structure, the A helices of HEAT repeats 5 and 6 and a region between HEAT repeats 6 and 7 are thought to be critical for recognition of the FxFG motif. N, amino terminus; C, carboxyl terminus. (b) Structure of importin β bound to the IBB domain of importin α. Importin β (yellow) forms a super helical structure that wraps like a snail around the IBB domain (blue). The 19 HEAT repeats share a common core of 21 residues, comprising the A helix with about three turns and the B helix with about four turns. The helices (green), critical for the interaction with FxFG repeat nucleoporins. Important residues for interaction with RanGTP are in red. Note the acidic loop, which contacts both RanGTP and the IBB domain (white arrow).
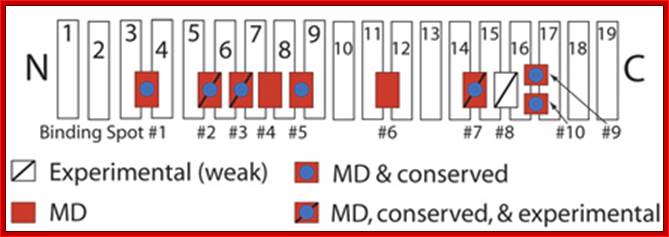
Schematic picture of importin-β showing binding spots for
FG-Nups on its surface:
Those spots discovered experimentally are labeled with a slash, while those uncovered in our simulations are red.
http://www.ks.uiuc.edu/
Importin-β: Authors have simulated a crystal structure of the transport receptor importin-β with cargo, fully solvated and ionized, in the presence of FG-Nups, and performed a sequence alignment using eight importin-β species. Using this combination of molecular dynamics simulations and sequence alignment, we were able to confirm the existence of 3 out of 4 binding spots which were known previously from experiments. Furthermore, they were able to identify six novel binding spots for FG-Nups which were previously unknown. (One of these spots was identified independently by experimentalists). The simulations provide the first atomic-scale look at these novel binding spots for FG-Nups, and suggest that the extent of binding on the surface of importin-β is much larger than previously realized. The additional binding spots on the surface of importin-β suggest that the molecule has a more extensive ability to interact with the NPC than previously expected and also implies that other transport receptors similar in size harbor an equally large number of binding spots
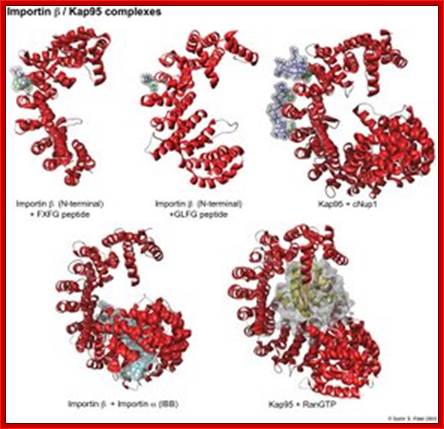
Karyopherins: They are defined by 3 types of binding partners: nucleoporins, cargo, and RanGTP. In red is the structure of importin or Kap95. As shown in green, phenylalanines are central for the binding of nups to the outer grooves of the karyopherin. RanGTP (yellow/orange) and cargo (in this case the IBB domain of importin , shown in light blue) form more extensive interactions with the interior of the kap. RanGTP causes large conformational changes in the tertiary structure of Kap95 (bottom right). Structure References: (1) Bayliss, R., Littlewood, T., Stewart, M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell v102 pp. 99-108, 2000. (2) Bayliss, R., Littlewood, T., Strawn, L.A., Wente, S.R., Stewart, M. GLFG and FxFG nucleoporins bind to overlapping sites of importin-beta. J Biol Chem v277 pp. 50597-50606, 2002. (3) Liu, S.M., Stewart, M. Structural basis for the high-affinity binding of nucleoporin Nup1 to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J Mol Biol v349 pp. 515-525. (4) Cingolani, G., Petosa, C., Weis, K., Muller, C.W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature v399 pp. 221-229, 1999. (5) Lee, S. J., Matsuura, Y., Liu, S.M., Stewart, M. Structural basis for nuclear import complex dissociation by RanGTP (naturev435)
Operation of the NPC: The “FG’ translocation machinery: (reviewed in Rout and Wente 1994; Allen et al. 2001; Tran and Wente 2006). (Bayliss et al. 2002; Grant et al. 2003; Isgro and Schulten 2005; Liu and Stewart 2005), (Yang et al. 2004; Kubitscheck et al. 2005; Yang and Musser 2006)
Although the cargo binding mechanisms of the transport factors discussed earlier vary, almost all binding sites with the NPC are found in the same class of Nups. These Nups are collectively termed FG Nups because they contain “FG repeat regions.” The NPC core framework provides the correct positioning of these FG Nups so that they flank and fill the central tube. Approximately a third of all Nups contain FG repeat regions, which consist of multiple small hydrophobic clusters containing an FG (Phe-Gly) dipeptide (usually FG, FXFG, or GLFG) separated by ∼20–70 residue hydrophilic linkers Binding of these repeats is cooperative, with two to four repeats associating with each transport factor molecule. With 5–50 repeats per FG Nup and ∼200 FG Nups per NPC, there is a potential for >1000 transport factor binding sites per NPC. Imaging of the trajectories of a single translocating molecule through the NPC is consistent with movement between multiple binding sites (presumably the many FG repeats) within the NPC. FG repeat regions are natively disordered and having no secondary structure they instead form filaments that can diffusively writhe around their attachment sites at the NPC. Although some will be diffuse as random coils, others may have enough internal cohesion to form more compact “molten globules”
Involvement of Nucleoporins: (D'Angelo and Hetzer, 2008)
Nucleoporins, as constituents of the nuclear pore complex (NPC), are mediating transport of macromolecules between the cyto- and the nucleoplasm. Moreover, anchor functions for microtubules, chromatin, protein modifying enzymes and transcription factors have been ascribed to the NPC. The NPC is a macromolecular assembly of approx. 30 different proteins in multiple copies, which are organized in different sub complexes with transport regulating or pore scaffolding functions.
The stable architectural core is shown. Schematic representation of the organization of nucleoporins into in blue;Brohawn et al., Structure, 2009;http://schwartzlab.mit.edu/
Combining structural data with computational methods authors established the evolutionary relationship between the NPC and vesicle coats, notably COP II, Several architectural nucleoporins share a common helical 65 kDa domain, the ancestral coatomer element ACE1, with Sec31, an essential component of the COP II coat. (Faculty of Biology-Genetics)
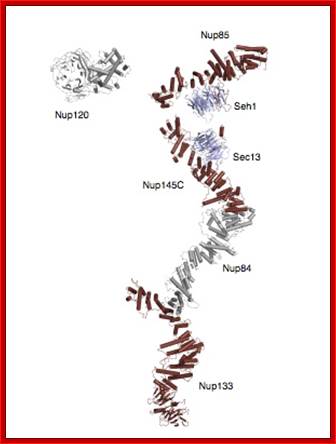
Composite crystal structure of the heptameric Y-complex (~ 0.6 MDa), one of the major components of the NPC. Its branched topology, together with other evidence, suggests a lattice-like NPC scaffold. Based on several recent papers published by the lab. http://schwartzlab.mit.edu/
Researchers construct a device that mimics one of nature's key transport machines
January 6, 2009
Artificial transportation: A schematic representation of the genuine (top) and artificial (bottom) nuclear pore complexes. By experimenting with a nuclear pore complex “mimic,” researchers have shown how transport factors (red), which help proteins move through the complex, are assisted by proteins called FG-nucleoporins (twisting lines).
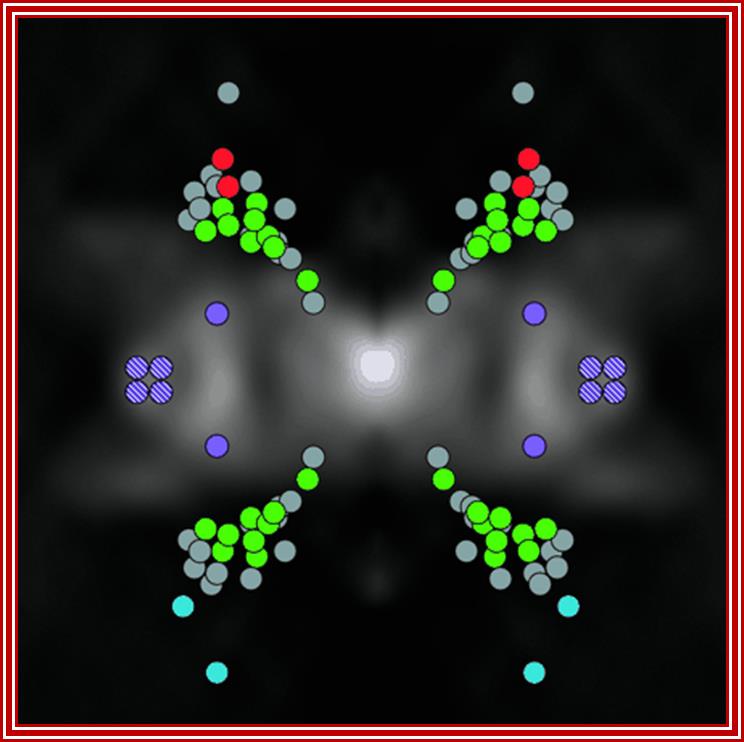
The position of nucleoporins (nups) in the yeast nuclear pore complex (NPC). A proteomic and biochemical analysis of the NPC revealed 29 different components exhibiting simple stoichiometries. Each of the components was tagged and its position determined by statistical analysis of the distribution of anti-tag, gold-labeled antibodies. A cross-section of the structure of the yeast NPC is shown in the background Shown (in green) are FG nups found on both sides of the NPC; FG nups found towards the periphery and exclusively on the nuclear side (blue) or the cytoplasmic side of the NPC (red); non-FG nups (gray); integral membrane proteins Pom34p (purple) and Pom152p and Ndc1p (purple stripes). See related article in this issue by RouterRout et al., 635–651(kindly provided by Christopher Akey; Yang et al., 1998. Mol. Cell.
To help protect its genes, a cell is highly selective about what it allows to move in and out of its nucleus. Yet that choosiness is regulated by just a thin barrier, perforated with tiny transport machines called nuclear pore complexes: protein-coated holes surrounded by flimsy, unfolded protein strands. Now, by building an artificial mimic of this membrane barrier and its pores, scientists have discovered a key to its selectivity and, in the process, have found a practical tool for drug development.(PhysOrg.com).
A general view of Nuclear Export and Import :
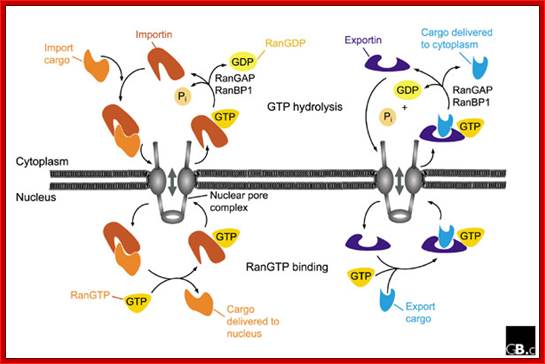
Fig: A schematic representation of nuclear import and export cycles through the NPC. Typically, an import cargo is first recognized by its importin in the cytoplasm. The cargo-loaded importin translocates through the NPC into the nucleus, where the cargo is dissociated from the importin by binding of importin to RanGTP. The importin-RanGTP complex recycles back to the cytoplasm, where RanGTP hydrolysis is stimulated by RanGAP and RanBP1; this frees the importin for the next round of import. Binding of cargoes to exportins is regulated in a converse manner. Exportins bind their export substrates in the nucleus, forming a trimeric cargo-exportin-RanGTP complex. This complex is exported from the nucleus and dissociated in the cytoplasm by hydrolysis of RanGTP to RanGDP and inorganic phosphate (Pi). This releases the export substrate, and the exportin is recycled back into the nucleus. https://openi.nlm.nih.gov
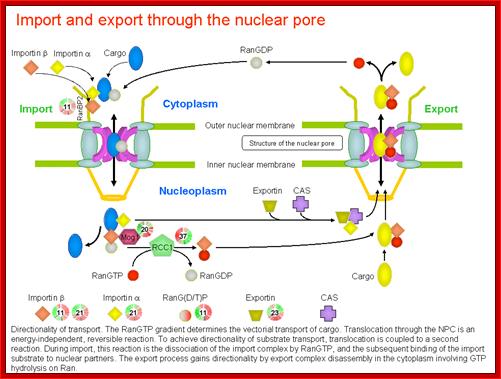
Directionality of transport. The RanGTP deterine the vectoria transport of cargo. Translocation is energy dependent and reversible reaction.
http://priweb.cc.huji.ac.il/malaria/maps/import_export.html.
The Ran cycle. Ran cycles between its GTP- and GDP-bound form dependent on its subcellular localization. The different forms of Ran confer directionality to transport by dictating where karyopherins bind and release their cargoes. See “The Energetics of Transport” for details. D, Ran-GDP;T, Ran-GTP; The Nuclear Pore Complex as a Transport Machine; http://www.jbc.org/
The Ran cycle. Ran cycles between its GTP- and GDP-bound forms dependent on its subcellular localization; the different forms of Ran confer directionality to transport by dictating where karyopherins bind and release their cargoes. See “The Energetics of Transport” for details. D Ran-GDP,T-Ran_GTP
Import of proteins:
Any cargo with a nuclear localization signal (NLS) exposed will be destined for quick and efficient transport through the pore. Several NLS sequences are known, generally containing a conserved polypeptide sequence with basic residues such as PKKKRKV. Any material with an NLS will be taken up by importins to the nucleus.
The classical scheme of NLS-protein importation begins with Importin-α first binding to the NLS sequence, and acts as a bridge for Importin-β to attach. The importinβ—importinα—cargo complex is then directed towards the nuclear pore and diffuses through it. Once the complex is in the nucleus, RanGTP binds to Importin-β and displaces it from the complex. Then the cellular apoptosis susceptibility protein (CAS), an exportin which in the nucleus is bound to RanGTP, displaces Importin-α from the cargo. The NLS-protein is thus free in the nucleoplasm. The Importinβ-RanGTP and Importinα-CAS-RanGTP complex diffuses back to the cytoplasm where GTPs are hydrolyzed to GDP leading to the release of Importinβ and Importinα which become available for a new NLS-protein import round.
Although cargo passes through the pore with the assistance of chaperone proteins, the translocation through the pore itself is not energy dependent. However, the whole import cycle needs the hydrolysis of 2 GTPs and is thus energy dependent and has to be considered as active transport. The import cycle is powered by the nucleo-cytoplasmic RanGTP gradient. This gradient arises from the exclusive nuclear localization of RanGEFs, proteins that exchange GDP to GTP on Ran molecules. Thus there is an elevated RanGTP concentration in the nucleus compared to the cytoplasm.
Simple view of Importing and exporting- the nucleus:
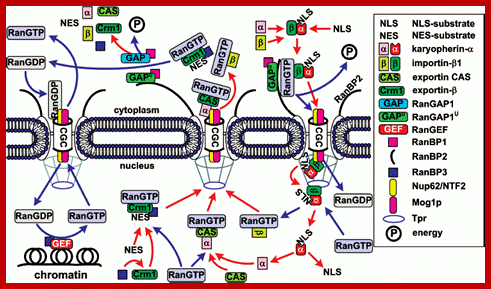
General scheme of nuclear import of ñNLS/NES-containing
proteins. The conventional model of transition of cNLS/NES-bearing proteins
through NPC includes the following stages. 1) Formation of the import
karyopherin-alpha/importin-beta1/cNLS-protein complex in the
presence of RanGAP1U and
low concentration of RanGTP in the cytoplasm; docking of the import transport
complex to cytoplasmic filaments (the top right corner of the figure). 2)
Transition across the central channel (CGC). 3) RanGTP-stimulated release of
karyopherin-alpha/cNLS-protein to the nucleoplasm. 4) Retention of
importin-beta1 in NPC and transport of importin-beta1 into
cytoplasm in complex with RanGTP (bottom right corner of the figure). 5) After
the dissociation of the karyopherin-alpha/cNLS-protein complex,
karyopherin-alpha is
exported from the nucleus in complex with CAS/RanGTP. NES-containing proteins
are exported in complex with Crm1/RanGTP (bottom central part of the figure).
6) On the way out of the NPC, all the exported transport complexes dissociate
due to GTP hydrolysis in Ran stimulated by RanGAP1 or RanGAP1U (top central part of the figure). 7)
Nuclear import of RanGDP is mediated by the NTF2 factor (top left corner of the
figure). 8) When in the nucleus, RanGEF (Ran guanine exchange factor)
transforms Ran from its GDP-bound form into the GTP-bound one. The minimal
cytoplasmic pool of RanGTP is maintained due to the transport factor Mog1
(bottom left corner of the figure). The details of the scheme are given in the
text. http://protein.bio.msu.ru
Figure: Highly schematic view of the mechanism of active transport through nuclear pores;
The proteins and structures involved in the active transport process are not known. A diverse set of related cytosolic proteins, however, is required for the initial binding of nuclear proteins to the complex. These proteins, called nucleoporins, contain a simple sugar ( N-acetylglucosamine) that aided their identification through the use of lectins and specific antibodies. The fibrils that project from the pore complex and are thought to help guide nuclear proteins to the center of the pore are not shown.
Nuclear import of glucocorticoid receptor protein:

Fig: The nuclear import of the glucocorticoid receptor
The glucocorticoids receptor is a gene regulatory protein that, in the non-hormone-treated cell, is bound in the cytosol to the chaperone protein hsp90. When activated by the binding of the appropriate steroid hormone, it is released from hsp90 and is directed into the nucleus by a nuclear localization signal; once in the nucleus, it binds to specific DNA sequences and regulates the transcription of a discrete set of genes (discussed in Chapters 9 and 15).
Every known protein (s) to be imported into the nucleus is marked by signal sequence called NLS-nuclear localizing sequence. The T-antigen first protein identified was of SV40 with its signature PKKKRKV. This protein is called cargo; the receptor importin binds to the cargo using the signature sequence; then it couples with motor protein called Ran-GDP. This trimeric complex diffuses through (in concentration gradient manner) the proteins found in the channel outer surface called FG motif containing proteins. Once the complex enters nucleoplasm the Ran-GDP interacts with GTP exchange actor called GEF. This releases the cargo into the nuclear sap and the receptor-Ran-GTP moves out of the NPC, where it it is displaced with GDP by GTP activating factor called GAP. Thus the receptor and Ran-GDP is ready for another round of import.
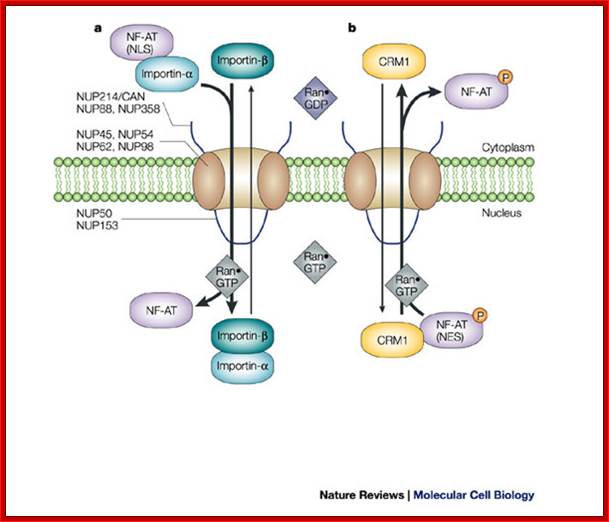
Classic NLS- and NES-mediated nuclear transport across the NPC.; Lan Xu & Joan Massagué; http://www.nature.com/
a | Import.
Importin-![]() associates with importin-
associates with importin-![]() . Importin-
. Importin-![]() binds to the exposed classic nuclear
localization signal (NLS) motif of cargo proteins, such as the active,
dephosphorylated form of nuclear factor of activated T cells (NF-AT). The
sequence of binding events has not been established for most cargoes. Through
the importin-
binds to the exposed classic nuclear
localization signal (NLS) motif of cargo proteins, such as the active,
dephosphorylated form of nuclear factor of activated T cells (NF-AT). The
sequence of binding events has not been established for most cargoes. Through
the importin-![]() -mediated interaction with
nucleoporins (NUPs), this complex transverses across the nuclear pore complex
(NPC) and is disassembled on binding Ran
-mediated interaction with
nucleoporins (NUPs), this complex transverses across the nuclear pore complex
(NPC) and is disassembled on binding Ran![]() GTP in the nucleus. NF-AT is retained
in the nucleus, whereas importin-
GTP in the nucleus. NF-AT is retained
in the nucleus, whereas importin-![]() and importin-
and importin-![]() shuttle back to the cytoplasm. b | Export. The phosphorylated form of
NF-AT has a nuclear export signal (NES) that is recognized by the nuclear
export receptor CRM1 (chromosome region maintenance 1), which mediates export
across the NPC by directly interacting with nucleoporins. Ran
shuttle back to the cytoplasm. b | Export. The phosphorylated form of
NF-AT has a nuclear export signal (NES) that is recognized by the nuclear
export receptor CRM1 (chromosome region maintenance 1), which mediates export
across the NPC by directly interacting with nucleoporins. Ran![]() GTP enhances the interaction of CRM1
with NES-containing proteins and facilitates nuclear export. Several
phenylalanine-glycine (FG)-repeat-containing nucleoporins are shown, some of
which are located only on the cytoplasmic or nucleoplasmic structures of the
NPC, whereas others are localized symmetrically. NUP214/CAN and NUP153 are
discussed in the main text; Lan Xu &
Joan Massagué;
GTP enhances the interaction of CRM1
with NES-containing proteins and facilitates nuclear export. Several
phenylalanine-glycine (FG)-repeat-containing nucleoporins are shown, some of
which are located only on the cytoplasmic or nucleoplasmic structures of the
NPC, whereas others are localized symmetrically. NUP214/CAN and NUP153 are
discussed in the main text; Lan Xu &
Joan Massagué;
Export of proteins:
Some nuclear proteins need to be exported from the nucleus to the cytoplasm, as do ribosome subunits and messenger RNA bound Rnps. Thus there is an export mechanism similar to the import mechanism.
In the classical export scheme, proteins with an nuclear export sequence (NES) can bind in the nucleus to form a heterotrimeric complex with an exportin and RanGTP (for example the exportin CRM1). The complex can then diffuse to the cytoplasm where GTP is hydrolysed and the NES-protein is released. CRM1-RanGDP diffuses back to the nucleus where GDP is exchanged to GTP by RanGEFs. This process is also energy dependent as it consumes one GTP. Export with the exportin CRM1 can be inhibited by Leptomycin B.
Export of RNA:
There are different export pathways through the NPC for each RNA class that exists. RNA export is also signal mediated (NES); the NES is in RNA-binding proteins (except for tRNA which has no adapter). It is notable that all viral RNAs and cellular RNAs (tRNA, rRNA, U snRNA, microRNA) except mRNA are dependent on RanGTP. Conseved mRNA export factors are necessary for mRNA nuclear export. Export factors are Mex67/Tap (large subunit) and Mtr2/p15 (small subunit).
Chaperoning ribonucleoprotein-sn RNA-snRNP-Export and import:
The
small nuclear ribonucleoprotein biogenesis pathway: RNA polymerase II
transcribes snRNAs as precursors containing an m7G cap and short 3'-end
extensions in the nucleus. The cap-binding complex (CBC) binds nascent snRNAs
and mediates the recruitment of the phosphorylated adaptor for RNA export
(PHAX), exportin 1 (Xpo1) and Ras-related nuclear protein GTP (RanGTP) for
nuclear export. In the cytoplasm, the seven Sm proteins bind first to the
chloride conductance regulatory protein (pICln) and the protein arginine
methyltransferase 5 (PRMT5) complex—which symmetrically dimethylates SmB, SmD1
and SmD3—and then to the survival motor neuron (SMN) complex. The SMN complex
bound to the Sm proteins interacts with snRNAs and mediates Sm core assembly.
Hypermethylation of the 5' cap of snRNAs by trimethylguanosine synthase 1
(TGS1) occurs after Sm core formation; both the SMN complex and snurportin,
which associates with the 5' cap of snRNAs, then bind to importin-![]() and
mediate snRNP nuclear import. The SMN complex and snRNPs transiently localize
to Cajal bodies, in which snRNPs undergo further maturation steps including
methylation and pseudo-uridylation by small Cajal body-specific
ribonucleoproteins (scaRNPs), before functioning in pre-messenger RNA splicing.
Depending on the cell type and developmental stage, the SMN complex also
localizes to Gems. snRNP, small nuclear ribonucleoprotein (Livio Pellizzoni).
Transport of snRNA-snRNPs is again explained below.
and
mediate snRNP nuclear import. The SMN complex and snRNPs transiently localize
to Cajal bodies, in which snRNPs undergo further maturation steps including
methylation and pseudo-uridylation by small Cajal body-specific
ribonucleoproteins (scaRNPs), before functioning in pre-messenger RNA splicing.
Depending on the cell type and developmental stage, the SMN complex also
localizes to Gems. snRNP, small nuclear ribonucleoprotein (Livio Pellizzoni).
Transport of snRNA-snRNPs is again explained below.
Small nuclear ribonucleoproteins (snRNPs) are active in recognizing and removing introns from pre-mRNA in the nucleus. Each snRNP particle is composed of small nuclear RNA (snRNA) of approximately 150 nucleotides, several Sm proteins and a number of specific proteins that are unique for each snRNP. Survival motor neuron (SMN) functions in the cytoplasm to assemble Sm proteins onto the snRNAs to produce an active snRNP particle. In the cytoplasm, the seven Sm proteins bind to the chloride conductance regulatory protein (pICln). In vitro studies show that pICIn first binds the Sm proteins as two separate complexes, with one complex including SmB and SmD3, and the other including SmD1 and SmD2 (Ref. 44). The latter subsequently binds SmE, SmF and SmG44 The protein arginine methyltransferase 5 (PRMT5) complex and PRMT7 methylate the Sm proteins SmB, SmD1 and SmD3, Sm proteins are released from the pICln–PRMT5 complex, and they bind the SMN complex. b | The SMN complex is composed of SMN, GEMIN2–8 and UNR-interacting protein (UNRIP). SMN is shown as an oligomer as it has been found to self-associate, and it has been suggested that oligomerization is crucial for SMN function (see main text). The exact numbers of SMN monomers in an SMN complex is unknown (it has been suggested to be an octomer). The gemins are shown as single units for simplicity, as the exact stoichiometry of the SMN complex has not been determined. c snRNA is transcribed in the nucleus and then binds the export proteins phosphorylated adaptor for RNA export (PHAX), cap-binding complex (CBC), exportin (XPO1) and Ras-related nuclear protein GTP (RAN), which transport it to the cytoplasm25. In vertebrates, the snRNA is brought into the Sm protein-bound SMN complex by binding to GEMIN5. d | The SMN complex places the Sm proteins onto the snRNA. The 7-methylguanosine (m7G) cap of the snRNA is hypermethylated by trimethylguanosine synthetase 1 (TGS1)25, allowing the SMN complex with the snRNA to bind snurportin and importin, which mediates transport of the SMN complex with an assembled snRNP into the nucleus. e | In the nucleus, the SMN complex and snRNPs localize to the Cajal body and the snRNPs undergo further maturation. Depending on the cell type and developmental stage, SMN can localize as a separate body adjacent to the Cajal body. scaRNP, Cajal body-specific RNP. Figure is modified, with permission, from Ref. 25 © (2007) Macmillan Publishers Ltd. All rights reserved; these are used for teaching in the class room.
Post capping and splicing of pre-mRNA-mRNPs export:
Processed mRNAs are bound by specific factors such as mRNPs, cap binding protein (CBP) and Exon-exon splice joint proteins called EJC and some SR proteins. This complex mRNA-and proteins are transported. A good example of mRNA transport is HIV processed mRNA. This mRNA has specific signature sequences Rev-elements, to which Rev Proteins bind. These sequences are called NES. Most of the mRNA-mRNP export is independent of Ran-GTP-GDP process. mRNA-mRNP exporters bind to heterodimeric exportin factors called NXF1 or TAP, they join with a small protein called Next1.
Multiple NXF1/Next1 proteins bind all along the length of mRNA. In this process NFX1/Next1 act like karyophorins. They interact with FG-nucleophorins and diffuse out. Nucleophorins –cargo requires RNA helicase at cytoplasmic side for the dissociation of TAP1 complex from the mRNA-mRNPs.. The helicase involved is Dbp5 which is ATP dependent motor protein (contains DEAD box). After the release of mRNA-mRNPs the NXF1/Next1 diffuse back into nucleoplasm.
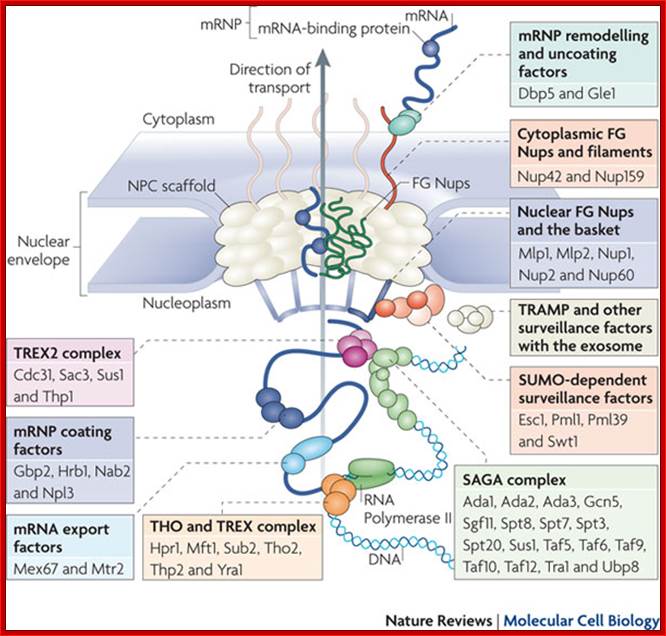
The movement of mRNAs from the transcription site inside the nucleus towards the cytoplasmic protein synthesis machinery involves several extensively coupled steps, in which overlapping factors coordinate a closely orchestrated 'dance' by accompanying the transcripts173. The precise order of events is poorly understood, but a consensus model for how this process might take place is depicted174. The SAGA chromatin remodelling complex is recruited to the promoter of a subset of inducible genes and promotes their transcription. Crosstalk between SAGA and the nuclear pore complex (NPC)-associated TREX2 complex might aid relocation of transcriptionally active genes to the nuclear periphery, resulting in the phenomenon of gene gating49 (whereby transcription and export machineries cooperate to 'gate' genes to the nuclear pore). Active transcription of gated genes produces nascent transcripts that recruit shuttling mRNA-coating factors175, THO, TREX and, subsequently, the mRNA export factors Mex67 and mRNA transport protein 2 (Mtr2), resulting in the formation of an export-competent ribonucleoprotein (mRNP)176. Thus, the association of the maturing mRNPs with components of the basket is strengthened in preparation for nuclear translocation, favouring mRNP surveillance mechanisms carried out by a basket-associated machinery104. After translocation, the release of mRNA export factors from mRNPs is induced by the combined action of DEAD box protein 5 (Dbp5p) and Gle1, which are docked to NPC cytoplasmic filaments through interactions with nucleoporin of 42 kDa (Nup42) and Nup159, respectively, and are thought to act as mRNP remodelling factors53. It is presumed that this process drives the directionality of mRNP export56 while priming mRNAs for translation initiation. Ada, transcriptional adaptor; Cdc31, cell division cycle protein 31; Esc1, establishes silent chromatin protein 1; GBP2, GTP-binding protein 2; Hpr1, hyper-recombination protein 1; Mlp, myosin-like protein; Mft1, mitochondrial fusion targeting protein 1; Nab2, nuclear polyadenylated RNA-binding protein 2; Npl3, nucleolar protein 3; Pml, pre-mRNA leakage; Sgf11, SAGA-associated factor 11; Spt, Suppressor of Ty; Swt1, synthetically lethal with TREX protein 1; Taf, TBP-associated factor; Tra1, transcription-associated protein 1; Ubp8, ubiquitin-specific processing protease 8.; http://www.nature.com
Majournal.cshlp.org

Yeast Nuclear Pore Comples and transport through it. Diagrammatic representation of mRNA export, adapted from Strambio-de-Castillia et al. (2010). The SAGA complex is recruited to the promoter of a subset of inducible genes and promotes their transcription. SAGA and the NPC-associated TREX-2 complex may help the genes move to the vicinity of the NPC. The nascent transcripts recruit shuttling mRNA-coating factors, THO, TREX, and, subsequently, the mRNA export factors Mex67p and Mtr2p, resulting in the formation of an export-competent mRNP (Rodriguez-Navarro and Hurt 2011); the association of the maturing mRNPs with components of the nuclear basket is strengthened in preparation for nuclear translocation, while nuclear basket-associated TRAMP and exosome complex-associated mRNP surveillance mechanisms ensure that the mRNP is correctly assembled for export (Fasken and Corbett 2009). After translocation through the NPC, the release of mRNA export factors from mRNPs is induced by the combined action of Dbp5p and Gle1p, which are docked to NPC cytoplasmic filaments via interaction with Nup42p and Nup159p, respectively, and are thought to act as mRNP-remodelling factors (Carmody and Wente 2009). It is presumed that this process drives the directionality of mRNP export while at the same time priming mRNAs for translation initiation. www.genetics.org
Nuclear Oskar RNP assembly is required for proper cytoplasmic localization and translational control:
Proper posterior localization of Oskar mRNA in Drosophila requires splicing of the first intron and the functions of the core EJC factors eIF4A3 and the Mago–Y14 heterodimer. It is proposed that the EJC helps recruits other factors, such as hrp48 and Barentsz, to form an RNP competent for localization. Additional factors involved in either localization or translational repression of Oskar mRNA include Bruno, which binds to the Bruno response elements (BRE) in the Oskar 3´ UTR, Staufen, Orb and p68. Once in the cytoplasm Oskar mRNP localization complexes assemble into large granules in which the mRNA is translationally repressed. These particles are then transported to the posterior pole where the mRNA is subsequently translated. assemble into large granules in which the mRNA is translationally repressed. These particles are then transported to the posterior pole where the mRNA is subsequently translated.
Nuclear events may affect localization of other mRNAs:
The cytoplasmic localization of other mRNAs may also depend upon the binding of specific factors in the nucleus (Farina and Singer, 2002). Drosophila hrp48 not only controls Oskar but also gurken mRNA localization and translation. In addition, the hnRNPA/B-like factor, Squid, governs the localization and translation of gurken mRNA (Goodrich et al., 2004). In Xenopus oocytes, the homologue of hnRNP I, VgRBP60, may influence the localization of the Vg1 mRNA at the vegetal pole (Cote et al., 1999). In rat oligodendrocytes, hnRNPA2 regulates localization and translation of the myelin basic proteins (MBP) mRNAs (Hoek et al., 1998). Export and localization of MBP mRNAs also depends upon the activity of the Quaking proteins (Larocque et al., 2002). In addition, other shuttling proteins such as the zipcode-binding protein-1 (ZBP1), required for localization of -actin mRNA in chick fibroblasts, may bind their mRNAs in the nucleus (Farina and Singer, 2002).
Transport of tRNA by Exportin-t and export of micro RNAs
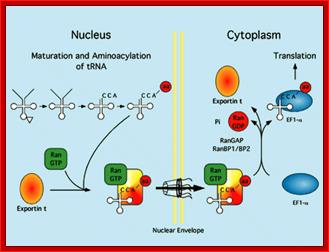
Export mechanism;Harvey Black; http://www.the-scientist.com/
Export mechanism: The tRNA must mature inside the nucleus before export. tRNAs are aminoacylated, and only tRNAs charged with an amino acid are exported efficiently. Export occurs when the tRNA is carried through the nuclear pore by a complex of exportin-t and Ran-GTP. Exportin –t binds to tRNA which in turn complexes with Ran-GTP. This complex diffuse through the channel filled with FG proteins, which actually generate hydrophobic environment. Harvey Black
Nuclear‑Cytoplasmic tRNA Trafficking:
tRNA trafficking between the nucleus and cytoplasm is a complex process that plays an essential role in protein synthesis, regulation of cell division, nutrient‑related regulation of gene expression, replication of the HIV genome and neuronal development. In eukaryotes tRNAs are made as precursors and undergo a number of processing steps in the nucleus. A small group of tRNAs require the removal of an intron, which in Saccharomyces cerevisiae, requires export of the immature tRNA to the cytoplasmic splicing machinery, followed by nuclear import and further processing. The functionality of the processed tRNAs is then tested by aminoacylation in the nucleolus. tRNAs deemed functional are translocated out of the nucleolus and delivered to the nuclear tRNA export receptors for transport across the nuclear pore complex (NPC). tRNAs exiting the cytoplasmic face of the NPC are collected and delivered to the protein translation apparatus for use in protein synthesis. Studies in S. cerevisiae and Xenopus laevis have established that nuclear‑cytoplasmic trafficking of tRNA involves multiple pathways. While the details of these tRNA export pathways are poorly understood, some broad concepts have emerged. These suggest that proteins acting as tRNA chaperones guide the transfer of tRNA molecules through the various stages of processing, translocation to the nuclear tRNA export machinery, transport through the NPC and delivery to the cytoplasmic protein synthesis machinery. However, to date only few of the proteins involved in the cellular tRNA transport process have been identified in both S. cerevisiae and mammalian cells. This chapter will review the current understanding of nuclear‑cytoplasmic tRNA dynamics and the role of the proteins that facilitate this process (Dev Mangroo, Shawn C. Chafe, Manoja B.K. Eswara, Aaron D. Johnstone, Andrew T. McGuire and Jacqueline B. Pierce)
Nuclear RNA export:
The Ran-GTP gradient governs the directionality of nucleocytoplasmic transport mediated by members of the karyopherin family of nuclear transport factors. The key role played by the GTP-bound form of Ran in mediating cargo binding and release by karyopherins functioning in nuclear import (importins) or nuclear export (exportins) is illustrated. See text for more detailed discussion. Imp, importin; Exp, exportin.
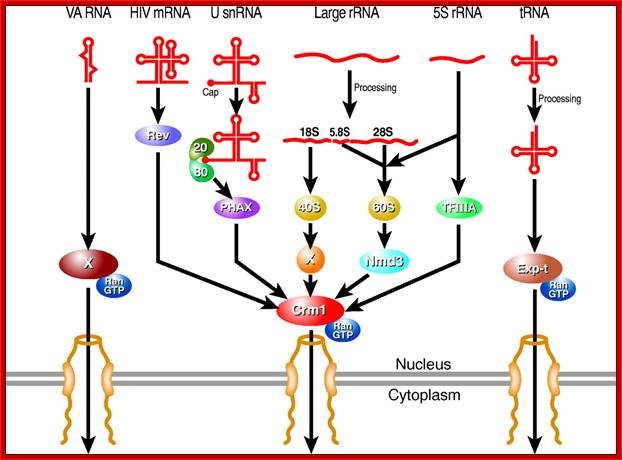
Karyopherin-mediated nuclear RNA export pathways. This schematic provides an overview of the key factors involved in mediating the nuclear export of different classes of non-coding RNA, as well as the minor class of mRNAs that use the Crm1 nuclear export factor. Hypothetical factors that remain to be identified are indicated by `X'. In addition to late HIV mRNAs, a small number of cellular mRNAs may also be exported from the nucleus in a Crm1-dependent manner. Candidate cellular Rev-like adapter proteins include APRIL, pp32 and NXF3. The nuclear export pathway used by VA RNAs is also used by other small, non-coding RNAs including Y RNAs and, possibly pre-miRNAs; Bryan R. Cullen; http://jcs.biologists.org/
Export of mRNA-mRNPs:
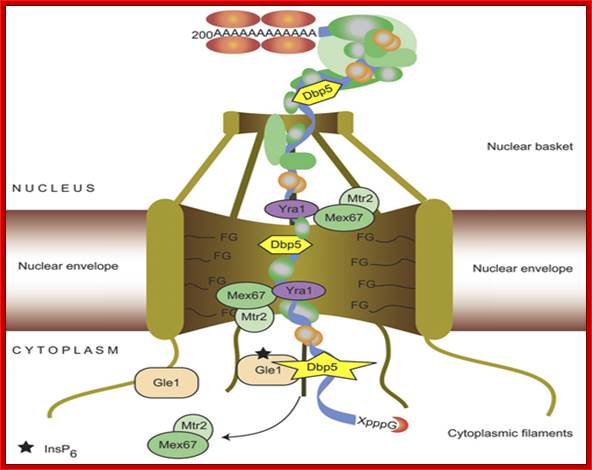
Nuclear Transport;Katja Sträßer;http://www.cipsm.de/
mRNA export is an
indispensable step during the complex process of eukaryotic gene expression:
the mRNA, created by RNA polymerase II in the nucleus, has to be transported to
the cytoplasm, where the ribosomes are responsible for the synthesis of the
encoded protein. This transport process is not restricted to the simple passage
of the mRNA through the nuclear pore complex, which spans the nuclear envelope,
but is embedded into the gene expression pathway. During transcription, the
message is capped, spliced and polyadenylated and mRNA export factors are
loaded onto the nascent transcript. This maturation and assembly of the mRNA
into a mature messenger ribonucleoprotein particle is controlled by nuclear
surveillance systems: the nuclear exosome and the Mlp1‑2 system prevent
the escape of aberrant transcripts to the cytoplasm. Only correctly assembled
mRNPs are transported through the nuclear pore to the cytoplasm by the mRNA
export receptor Mex67‑Mtr2/Tap‑p15, which is recruited to the mRNA
by interaction with the mRNP‑bound TREX complex and SR proteins. This
tight coupling between the single steps of the nuclear gene expression
guarantees an efficient and accurate transfer of the genetic information to the
cytoplasm

A model for the evolution of the EJC upon mRNA export:
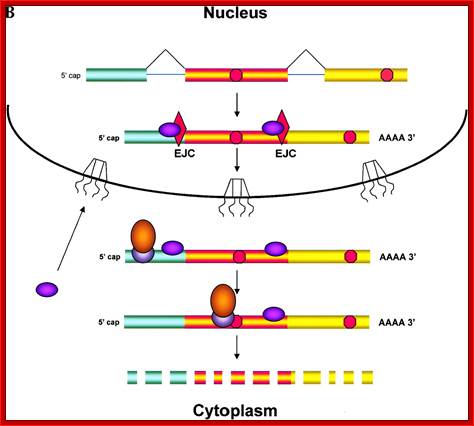
Pre-mRNA splicing deposits the EJC upstream of the splice junction. At this stage, the complex contains at least five proteins, SRm160, DEK, RNPS1, REF and Y14. After splicing, the TAP/p15 heterodimer and Upf3 join the complex in the nucleus. DEK probably dissociates (dashed arrow) before mRNA export to the cytoplasm, whereas SRm160 and the nucleocytoplasmic shuttling proteins RNPS1, REF, TAP/p15 and Upf3 are all likely to leave after mRNA export. Y14 remains stably associated with spliced mRNA in both compartments, while Upf2 only joins the complex in the cytoplasm. Question marks symbolize potential unidentified components
Fig: Nonsense mediated decay of mRNAs. The monitoring of mRNAs for premature termination codons (PTCs) is mediated by ribosomes during the “pioneering round of translation.” Several proteins associate with sequences near exon/intron borders of the spliced mRNA, forming the exon junction complex (EJC). Some EJC proteins are removed during export whereas others are removed during translation and thus serve as indicators of whether a particular region of the mRNA has been translated. (A) If no EJC proteins are on the mRNA when the pioneering ribosome encounters a chain termination codon, this codon is read as a normal termination signal, at the 3′ end of the coding region. (B) If the ribosome has not traversed all exon/intron borders, EJC protein(s) remain on the mRNA and serve as an indication that any termination codon encountered is a PTC. As a consequence, nonsense-mediated decay (NMD) is initiated, resulting in destruction of the mRNA. The pioneering round of translation is shown here as happening in the cytoplasm (or the perinuclear cytoplasm), and with the released proteins being imported into the nucleus.
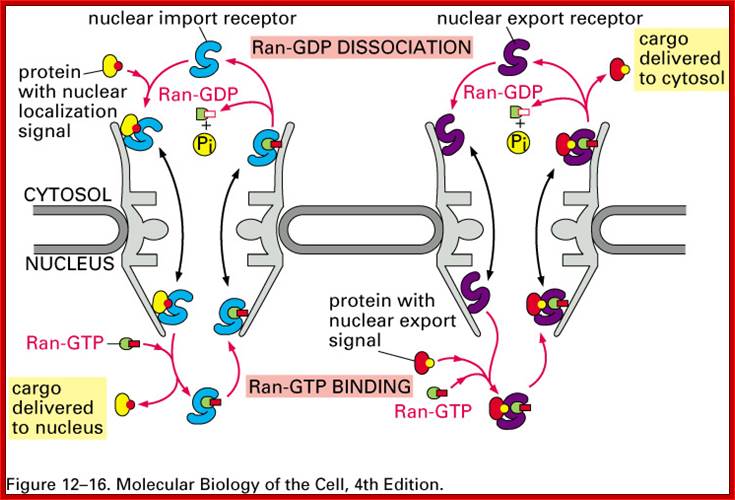
Import and export occur via similar, but non-identical, mechanisms. Molecular cargoes interact with carrier proteins known as importins or exportins. These are members of family of evolutionarily related proteins (the karyopherins). After binding cargo, the importin or exportin has an increased affinity for nuclear pore components, so the karyopherin/cargo complex becomes bound and is translocated across the pore. Molecular Biology of the cell :http://www.pha.jhu.edu/
Transcription, splicing polyadenylation and mRNA export are all coupled:
Gene Expression is a fundamental process of any living cell. In eukaryotic cells, the mRNA needs to be processed, i.e. capped, spliced, and polyadenylated, processes that occur already cotranscriptionally. In addition, the mRNA is packaged into a mature mRNP. Since nucleoplasm and cytoplasm are separated by the nuclear envelope, the mRNP needs to be exported through the nuclear pore complexes (NPCs) into the cytoplasm. Here, the information encoded in the mRNA is translated into a polypeptide chain by the ribosomes.
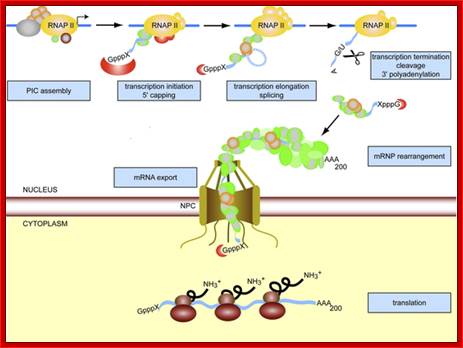
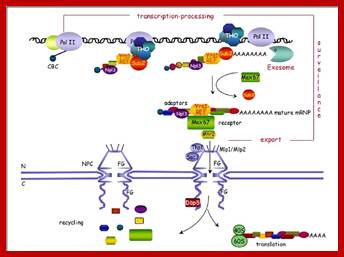
Transcription, Processing are linked to Export of the same.
A
defining feature of eukaryotic cells is the presence of a nuclear envelope
separating transcription and DNA replication in the nucleus from the site of
protein synthesis in the cytoplasm. The regulation of gene expression relies in
part on the controlled exchange of molecules between these two compartments.
Factors implicated in transcription regulation and DNA replication have to be
imported into the nucleus, whereas RNAs produced in the nucleus have to be
exported, either to fulfill their function in protein synthesis or to mature
into functional particles. This review summarizes studies performed over the
last 15 years that led to the identification of cellular factors mediating
nuclear export of the different classes of RNAs, including tRNAs, UsnRNAs,
micro-RNAs, ribosomal RNAs and mRNAs. We also discuss recent evidence
indicating that the nuclear transport step is intimately linked to RNA
synthesis, processing and mRNP assembly, thus ensuring that only properly matured
ribonucleoprotein (RNP) complexes reach the cytoplasm.
Nuclear export of RNA (PDF Download Available). Available from: https://www.researchgate.net/publication/8200196_Nuclear_export_of_RNA
[accessed Jun 10, 2017].
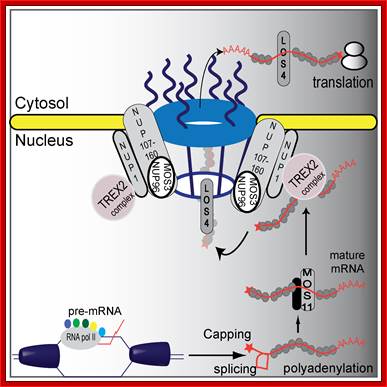
A proposed plant mRNA export pathway (in Plants). A new component in the mRNA export pathway; DNA helicase unwinds DNA and RNA polymerase II and co-activators proceed with transcription and mRNA maturation. Pre-mRNA is capped, polyadenylated, and spliced. Mature mRNA is then carried by MOS11 and its partners to TREX2, which associates with NUP1. LOS4 then binds the mRNA and passes the mature mRNA through the nuclear pores with the help of the NUP107–160 complex to the cytosol where it can be translated.
Hugo Germain equal contributor, Na Qu equal contributor, Yu Ti Cheng et al.a; http://www.plosgenetics.org/
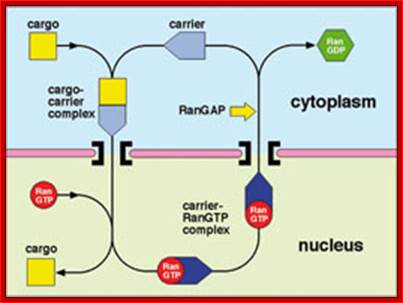
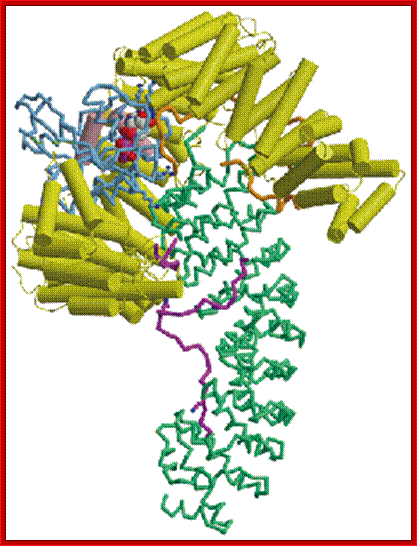
Molecular machinery of nuclear trafficking;;Cargo macromolecules are transported through the pores by specific carrier molecules and this process is often orchestrated by the Ran GTPase. the role of nuclear trafficking components, the TREX-2 complex, and the polyA-binding protein, Nab2p, in orchestrating the gene expression machinery, especially in the context of their acting as scaffolds for the formation of the large macromolecular complexes involved in transcription, splicing, mRNA processing and nuclear export. Murray sStewart;http://www2.mrc-lmb.cam.ac.uk/
In recent years, multiple links between the intranuclear steps of gene expression have been shown. One of the key players in this coupling is the so called TREX complex, which couples transcription and mRNA export. It consists of the heterotetrameric THO complex (consisting of the components Tho2, Hpr1, Mft1, and Thp2), the nuclear mRNA export factors Sub2 and Yra1, the SR-proteins Gbp2 and Hrb1, and Tex1. TREX travels along the transcribed gene with RNA polymerase II, binds the nascent mRNA, and “feeds” it into the mRNA export pathway via its subunits and mRNA export factors Sub2 and Yra1 (Fig. 2). Yra1 then directly binds the mRNA exporter Mex67-Mtr2. Mex67-Mtr2 in turn binds directly to the mRNA as well as to nuclear pore proteins and transports the mRNP through the nuclear pore complex to the cytoplasm.
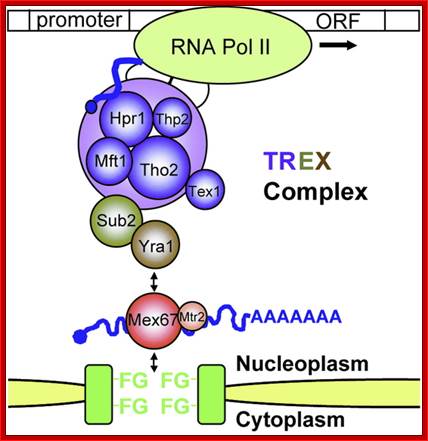
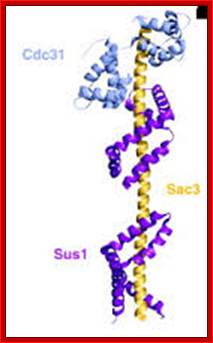
TREX Transcriptional-Export complex; http://trex-2.blogspot.com/
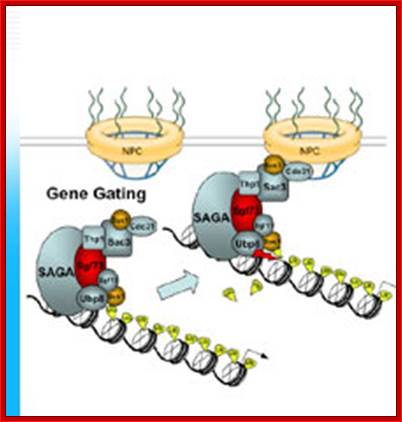
Most of the export pathways for RNA involve export receptors of the RanGTP-dependent importin β superfamily. It also requires receptors called Mex64-Mtr2; mRNA export is extensively coupled to transcription and to TREX (TRanscription-EXport) complexes, which play crucial roles in coupling co-transcriptional mRNP assembly with recruitment of mRNA export adaptors. Another exciting research achievement was the finding that transcription initiation at promoters is coupled to mRNA export and can involve ‘gene gating’ during which an activated gene becomes tethered at the inner side of the NPC.Nuclear export of mRNA and its coupling to transcription; Prof. Dr. Ed C. Hurt http://www.uni-heidelberg.de
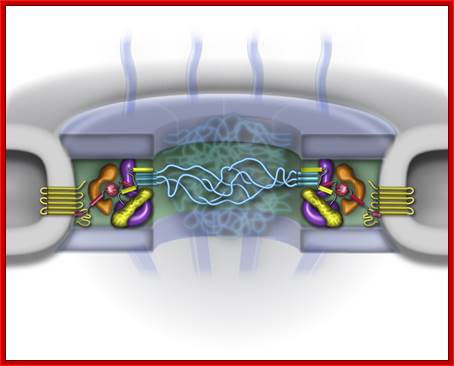
Nuclear pore complexes.The nuclear pore complex (NPC) is huge macromolecular assembly (~30 different nucleoporins, ~50 MDa) in the nuclear envelope that transports cargo between the nucleus and cytoplasm. We study the NPC in yeast and apply genetic, biochemical ,cell biological and structural approaches to unravel the structure and function of this gigantic transport machine. Milestones in this research were the cloning of the first nucleoporin (termed Nsp1), the development of genetic screens on the basis of synthetic lethality to genetically dissect the NPC, the biochemical analysis of the NPC by using powerful affinity-purification methods for nucleoporins and NPC modules, the reconstitution of a seven-subunit containing NPC module and the discovery of a novel structural principle to generate higher order structures within the NPC scaffold with the identification of the dynein light chain as a molecular glue. Future studies involve the structural and functional analysis of the nuclear pore complex including reconstitution of NPC modules into the higher order NPC structure. Prof. Dr. Ed C. Hurt;http://www.uni-heidelberg.de/
Export of RNA from the nucleus to
the cytoplasm is essential for gene expression. Most of the export pathways for
RNA involve export receptors of the RanGTP-dependent importin β
superfamily. We have found that export of mRNA requires another type of
transport receptor called Mex67-Mtr2, which opened in-depth research on the mechanism
of mRNA export and allowed the dissection of the conserved mRNA export
machinery. Moreover, the mRNA export receptor functions via coupling to mRNA
binding proteins, which serve as adapters.
Another conceptual advance was the discovery that mRNA export is extensively
coupled to transcription and to TREX (TRanscription-EXport) complexes, which
play crucial roles in coupling co-transcriptional mRNP assembly with
recruitment of mRNA export adaptors. Another exciting research achievement was
the finding that transcription initiation at promoters is coupled to mRNA
export and can involve ‘gene gating’ during which an activated gene becomes
tethered at the inner side of the NPC. Future studies involve the mechanistic
understanding of how mRNP biogenesis and surveillance is coupled to mRNA
export.
Recently, authors have identified a novel function for Ctk1, a known transcription factor. The kinase Ctk1 phosphorylates the C-terminal domain (CTD) of Rpb1, the largest subunit of RNA polymerase II, during transcription elongation. We showed that Ctk1 has a second function in translation, a cytoplasmic step of gene expression. Here, Ctk1 phosphorylates Rps2, a protein of the small ribosomal subunit. This phosphorylation event increases translational fidelity.
mRNA/mRNP Transport Rate:
The in vivo dynamics of bulk mRNP transport and export, from transcription to the nuclear pore complex (NPC), occurred within a 5–40 minute time frame, with no NPC pile-up. mRNP export was rapid (about 0.5 s) and kinetically faster than nucleoplasmic diffusion. The DEAD box RNA helicase Dbp5 is essential for nucleocytoplasmic transport of mRNA–protein (mRNP) complexes. Dbp5 is present mainly in the cytoplasm and is enriched at the cytoplasmic side of nuclear pore complexes (NPCs), suggesting that it acts in the late part of mRNP export. Here, we visualize the assembly and transport of a specific mRNP particle, the Balbiani ring mRNP in the dipterans’ Chironomus tentans, and show that a Dbp5 homologue in C.tentans, Ct-Dbp5, binds to pre-mRNP co-transcriptionally and accompanies the mRNP to and through the nuclear pores and into the cytoplasm. We also demonstrate that Ct-Dbp5 accumulates in the nucleus and partly disappears from the NPC when nuclear export of mRNA is inhibited. The fact that Ct-Dbp5 is present along the exiting mRNP fibril extending from the nuclear pore into the cytoplasm supports the view that Ct-Dbp5 is involved in restructuring the mRNP prior to translation. Finally, the addition of the export factor Dbp5 to the growing transcript highlights the importance of the co-transcriptional loading process in determining the fate of mRNA.
In yeast, the nascent transcript generated by RNA polymerase II (Pol II) is co-transcriptionally folded and assembled into a pre-messenger ribonucleoprotein (pre-mRNP) by the THO/TREX complex (Yra1 and Sub2 are subunits of TREX, and THO is a sub complex) that accompanies the elongating Pol II. After co-transcriptional association with additional pre-mRNA factors (for example, the cap-binding complex (CBC) and RNA-binding proteins, shown in orange), the Mex67–Mtr2 mRNA export receptor is recruited to the mRNP via adaptor proteins such as Yra1. When emerging at the cytoplasmic side of the nuclear pore complex (NPC), the mRNP encounters remodeling machinery on the NPC fibrils that consists of the ATP-dependent RNA helicase Dbp5 and its activators Gle1 and the signalling molecule inositol hexakisphosphate (InsP6). After dissociation of Mex67–Mtr2 and other export factors from the RNA cargo, the mRNA may still contain a few shuttling RNA-binding proteins, such as Npl3, that influence translation. mRNA is recruited to the translation-initiation machinery via its 5' cap that is recognized by the initiation factor eIF4A and by additional initiation factors. Transport factors are re-imported, which in some cases requires phosphorylation. b | For metazoa, several mRNA export models have been described. Depicted are the splicing- and cap-dependent modes of human TREX recruitment to the mRNP. The downstream events in mRNA export, including the recruitment of the TAP–p15 mRNA export receptor, are similar for metazoa and yeast. Export receptors are indicated in yellow and adaptors in blue. For simplicity, the exon-junction complex (EJC) on the translocating and cytoplasmic mRNP is not shown. E, exon; I, Intron, TREX transcription coupled export complex.
Model for the functional coupling of pre-mRNA splicing and nonsense-mediated decay:
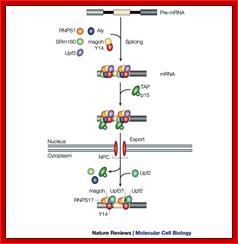

Model for coupling splicing to mRNA export and nonsense-mediated decay. In this model the splicing factor UAP56 (UAP, in yellow) associates with the pre-mRNA during spliceosome assembly along with other splicing factors including RNPS1 (R) and other SR proteins (a). UAP56 then recruits the mRNA export factor ALY along with NMD components including Y14 and UPF3 (b). All of these proteins form a complex that associates with the RNA near the exon–exon junction. UAP56 may then dissociate from ALY and be replaced by the export factor TAP, which mediates export of the mRNP through the nuclear pore complex (c). In the cytoplasm the export machinery is released and the NMD machinery remains associated with the exon–exon junctions. When the mRNA is wild type, the NMD machinery is displaced by the translating ribosome. In contrast, in the presence of a premature stop codon (represented by the red dot) (d) upstream from an exon–exon junction translation is arrested and the NMD machinery is not released but instead recruits additional components that trigger rapid degradation of the mRNA. nt, nucleotides. Maniatis and Robin Reed; http://www.nature.com
The exon–exon junction complex (EJC) assembles on messenger RNA as a result of splicing. TAP/p15 binds to components of the EJC and mediates the interaction of the mRNP with the nuclear-pore complex (NPC). During export, and immediately afterwards, the composition of the complex is changed as several of its components dissociate, leaving behind the remnants — the cytoplasmic EJC (cEJC), which includes Y14 and magoh, and possibly Upf3 and RNPS1 — at the same position as the complex was assembled in the nucleus. Upf2 probably binds to Upf3 after export (Gideon Dreyfuss, V. Narry Kim and Naoyuki Kataoka)
Shuttling of mRNPs-to and fro:
Yeast shuttling mRNA binding proteins Pab1p, the cap binding complex (CBC), Dbp5p, Hrp1p, Nab2p, Npl3p, Gbp2p and Hrb1p.bind to the mRNA during export from the nucleus to the cytoplasm. The export receptor heterodimer Mex67p-Mtr2p is recruited to the RNA by the export adapter protein Npl3p and docks the ribonucleoparticle to the nuclear pore complex (NPC).
From sites of transcription in the nucleus to the outreaches of the cytoplasm, mRNAs are associated with RNA-binding proteins. These proteins influence pre-mRNA processing as well as transport, localization, translation and stability of mRNAs. We want to learn which proteins are associated with the transported RNP, how transport is regulated and how the cell distinguishes between export incompetent and export competent mRNPs. Saccharomyces cerevisiae has been proven to be a useful model organism for eukaryotic cells and we use a combination of genetics, biochemistry and cell biology to gain insight into mRNA export out of the nucleus. Our studies led to the identification of two novel mRNA binding proteins, termed Gbp2p and Hrp1p, that accompany the mRNA to the cytoplasm. Both proteins are recruited early to the pre-mRNA by the THO complex, involved in transcription. They remain bound to the mRNA during export and leave the RNA in the cytoplasm. Their re-import into the nucleus is facilitated by the import receptor Mtr10p, which is a member of the transportin family. Both proteins contain serine/arginine (SR) rich domains, similar to an already known mRNA-export factor Npl3p. Npl3p recruits the export receptor Mex67p to the mature mRNP prior to its export. Binding of Npl3p to mRNA seems to be crucial for the formation of an export competent mRNP, as its dissociation results in an export incompetent mRNP. Interestingly, the RNA-binding affinity and the cellular localization of all three shuttling SR-proteins is influenced by the SR-type specific kinase Sky1p. In ongoing studies we analyze the function and regulation of these proteins further and investigate the following questions in detail:
- How can a cell distinguish between a pre-mRNA and an export competent mRNA molecule?
- Which RNA binding proteins accompany the mRNAs to the cytoplasm?
- What are the functions of each individual mRNA binding protein in this process?
- How is the mRNA export from the nucleus regulated?
The different RNA’s export pathways:

The major RNA export routes are shown (tRNA, microRNA (miRNA), small nuclear (sn)RNA, mRNA, ribosomal (r)RNA). In each case, the primary RNA transcript is shown, as well as the transport-competent RNA after it has undergone processing, maturation and assembly with export factors (export adaptors are shown in blue, export receptors are shown in yellow). Prominent structural motifs in pre-RNAs are indicated (single/double-stranded RNA, loops, exons and introns, 5' cap and 3' poly(A) tail). For the mRNA export route, the names of both metazoan and yeast proteins are indicated, and mRNAs are shown with additional adaptor proteins and RNA-binding factors (orange ovals). In the case of rRNA, the general exporter in eukaryotes, CRM1, and two auxiliary exporters, Mex67–Mtr2 and Arx1, that have only been studied in yeast are depicted. CBC, cap-binding complex; EXP-exporin (Alwin Köhler & Ed Hurt). http://www.nature.com
MYC controls multiple components of ribosome biogenesis; Jan van Riggelen, Alper Yetil & Dean W. Felsher; http://www.nature.com/
MYC directly regulates the expression of several ribosomal protein and RNA components, and auxiliary factors that are required for ribosomal RNA (rRNA) processing, ribosome assembly, the export of mature ribosomal subunits from the nucleus into the cytoplasm, as well as factors that control the initiation of mRNA translation. The transcriptional upregulation of these factors by MYC requires the recruitment of cofactors, remodelling of chromatin structure and involves all three types of RNA polymerase complexes (RNA pol I, II and III). MYC facilitates RNA pol I transcription from ribosomal DNA (rDNA) clusters that encode the 5.8S, 18S and 28S rRNAs, which requires upstream binding transcription factor (UBF) and selectivity factor (SL1). MYC activates transcription of 5S rRNA and transfer RNA (tRNA) through RNA pol III. In addition, MYC–MAX stimulates the transcription of protein components in an RNA pol II-dependent manner. This includes proteins of the small ribosomal subunit (RPS) and large ribosomal subunit (RPL), nucleolin (NCL) and nucleophosmin (NPM1), which are involved in rRNA processing and export, UBF that serves as a cofactor for RNA pol I-dependent transcription as well as eukaryotic initiation factors that control the initiation of translation. TFIIIB, transcription factor IIIB. Jan van Riggelen