Messenger RNAs (mRNAs):
Jacques Monod and François Jacob, were the first to suggest mRNA acts between the Gene and the polypeptide. Then Sidney Brenner and Mathew Meselson discovered mRNAs (1961). Among the different species of RNAs, it is only mRNAs that are linearly related to DNA and to polypeptide chain as an intermediary. It is this species of RNA that carries encoded message from the master molecule in the form of codons. It is these molecules that translate the information by decoding it into polypeptide chains. It is ultimately the protein, having a unique structural and functional properties determine the structure and function of the cell that is why proteins are deemed to be molecular work-horses. Proteins are the quintessence of the gene function at molecular level. DNA is considered as the master molecule, but without protein it has no function and without DNA protein has no existence.
Here the triple or tripartite relationship, between linear DNA, linear mRNA and linear polypeptide chain (in the form of nucleotides sequence and amino acid sequence respectively), is referred to as co-linearity. Two important functions that occur between a gene and the polypeptide chain are transcription and translation, which are separated in space and time.
Prokaryotic mRNAs:
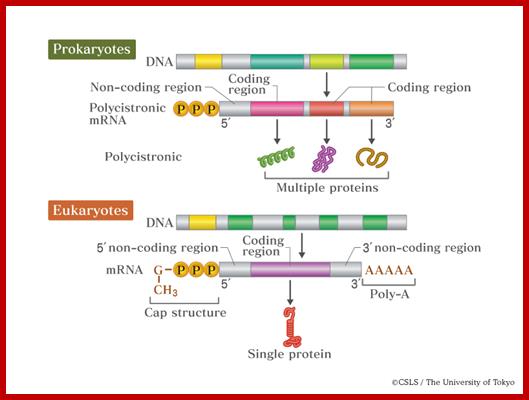
Translation is the process cells use to synthesize polypeptides using the codon sequence in the mRNA as a template. mRNA; In the bacterial cell, several related polypeptide sequences are carried on the same mRNA template. Each sequence in the mRNA strand has its own ribosome binding site, start and stop codons at the end. Since transcription takes place in the cytoplasm, translation can begin even before transcription is complete. A cistron starts with AUG and ends in UAA. http://www.quora.com/;http://academic.pgcc.edu/;http://csls-text.c.u-tokyo.ac.jp/
![[prokaryotic%2520mrna%255B5%255D.gif]](Ribose_Nucleic_Acid5-Messenger_RNAs-mRNAs_files/image002.jpg)
Eukaryote mRNA; this kind of mRNA one finds rarely; most of the time the coding region is split into Introns and Exons; http://lh5.ggpht.com/

Each of the cistrons has its own SD sequence and Ter sequence at the end of the cistron; Z cistron starts with AUG and ends in UAA; intercistronic spacer-AAAUCAA- The Y cistron starts with AUG and ends in UAA-intercistronic Z & Y spacer-UAACGGAGUGAUC- and A cistron starts with UUG ends in UAA. http://www.quora.com/; ;http://lh5.ggpht.com/-ijk9yxnlLuU
5’p-p-p---s/d--AUG-------UAA---s/d---AUG------UGA--s/d---AUG------UAG---3’
General Features of PK mRNAs:
- Prokaryotic mRNAs are made up of polynucleotide chains of a specific sequence endowed with 5’ to 3’ polarity.
- Though it is linear, it can exhibit several but short, secondary structures either at 5’end or 3’ end.
- Most of the mRNAs are polycistronic and monocistronic number is less.
- The number of cistrons per mRNA varies from one to fifteen.
- Each of the cistrons code for one polypeptide and contain all the features of a gene with is own ORF i.e. initiator and terminator codons. But these cistrons together have a common promoter operator region in their genes. They are transcribed as one transcript but translated individually as separate components.
- Size of the mRNAs is more or less proportional to the gene from which it generates.
- The 5’end contains three phosphate groups, characteristic of prokaryotic mRNAs. Even if one phosphate is missing, it is degraded.
- The 3’ end does not contain any poly-A tail. Poly-A tail is added during it degradation. But 3’ region may contain inverted repeats which produces stem loop structure which provides stability to mRNAs.
- Chloroplast and mitochondrial mRNAs are transcribed into polycistronic RNAs and they are then processed into monocistronic mRNAs.
- Chloroplast mRNAs from different plant species are edited by substituting C to U changes or addition or deletion of nucleotides. But mitochondrial mRNAs in some species do possess short 50 ntd long poly-A tail.
- Recently scholars have discovered that chloroplast mRNAs are added with a poly-A tail, which provides signal for degradation.
- The polypeptide chain it produces is directly proportional to the coding size of ORF of the mRNA. Longer the protein longer is the mRNA’s ORF and vice versa.
- Average size of mRNA in bacterial systems is 1000 ntds to 1500.
- Longest mRNA is 110,418 ntds codes for 36805 a.as.? The shortest is 54 ntds codes for 17a.as.
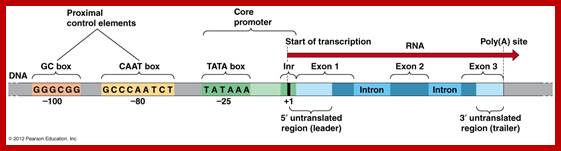
- All mRNAs consists of a 5’ non-coding, called 5’UTR (~150ntds), called leader sequence, whose length and sequence varies from one species of mRNA to the other.
- All mRNAs contain another noncoding region at 3’ end called 3’UTR ~150-200 ntds; in some they have regulatory role.
- Both 5’UTR and 3’UTR are required for efficient translation (!).
http://www.csirnetlifesciences.com/

https://www.researchgate.net


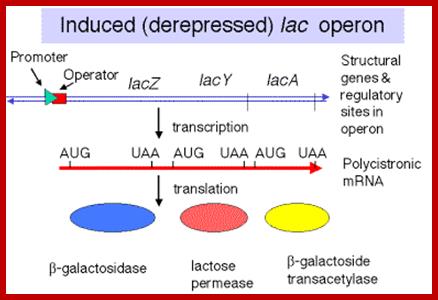
http://www.bx.psu.edu/
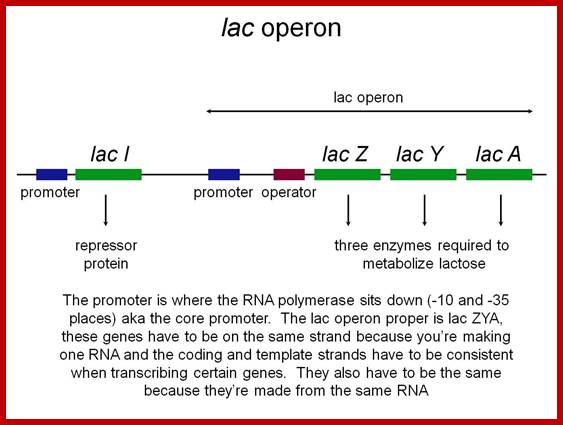
Polycistronic mRNA with spacer between cistrons: The mRNA transcript is of Lac Z operon; between Z cistron and Y cistron there is spacer consisting S/D sequence. Each of the cistrons end in a terminator codon and the next cistron has initiator codon AUG or GUG. At the end of the cistron the ribosomes dissociates and reassociate at the next cistron using S/D sequence and starts translation of the next cistron.mol-biol4masters-copy from web
- The leader sequence in every bacterial mRNAs contain an all-important structured sequence called Shine-Delgarno sequence-AGGAGGU, seven to nine nucleotides upstream of the start codon AUG. This 5’UTR sequence base pairs with a sequence at 3’ end of 30s ribosomal RNA, thus ribosomes bind to 5’region of mRNAs during initiation of protein synthesis..
- Some viral mRNAs (e.g Picorna viral RNA) contain Internal Ribosomal Entry sequences (IREs) ex. Rhinovirus, foot and mouth disease virus, Hepatitis A virus, Swine flu virus, Rous sarcoma virus, HIV, FGF, NF-kB,4G TF and Apaf-1.
- Internal ribosomal entry sites can be found at 5’ UTR or found at intercistronic spacer.
- The 5’UTR at first coding cistron contains S/D sequence essential for the binding of small ribosomal subunit for the initiation of translation. AU rich sequence upstream of S/D is synergistic in translation.
- Intercistronic spacers contain S/D sequences, if Au (UAUUUUAAUAAUUAA) rich sequence is introduced upstream of the intercistronic spacer translation efficiency increases.
- Each of the intercistronic spacer size varies from ~20 ntds to 40-50 ntds. These spacers contain S/D sequences for the 30s ribosomal subunit to bind and reinitiate translation of the succeeding cistron.
- The coding region of each cistron (most of the bacterial mRNAs are polycistronic) starts at the initiator codon AUG (mostly) or GUG (rarely) of each cistron and ends in a terminator codon like UAG (Opel), UGA (Amber) or UAA (Ochre) of that cistron.
- Sometimes the terminator codon over laps with the next initiator codon UGAUG (UGAUG). How the ribosome dissociates and reinitiates is very interesting aspect of molecular biology.
- The region between initiator codon and terminator codon is often called open reading frame (ORF). Within each cistron (ORF) there is no scope for any codon to be non-coding, but there can be missense codon or there can be premature termination codon due to mutation. If the reading frame changes by one or more nucleotides by insertion or deletion the overall meaning of the message changes.
- Often one finds overlapping reading frames in mRNAs, UAA-
AUG , as translation ends the ribosomes next initiates the next cistron
- The ORF refers to the region between initiator codon and terminator codon, but the initiator codon should be in proper sequence context for the initiating translation.
- In the 5’ region there may be several initiator codons (i.e. AUGs, very rarely GUG or UUGs), but only one of the AUGs will be in right context, in the sense the sequences around the Initiator codon is important for correct and proper initiation of translation. Instead of AUG, if others are used for initiation the efficiency goes down. If it initiates at wrong AUG translation will be terminated for it certainly encounters a terminator codon.
- In the case of prokaryotes, the coding region is split into cistrons with initiating and terminator sequences. In between two such coding regions one finds a short spacer region with S/D sequences, this increases the efficiency of translation.
- At the 5’ end, about 10 nucleotides up stream of AUG codon it consists of a sequence called Shine Delgarno (AGGAGGU), which facilitates the binding of mRNA to 30s ribosomal 3’ end of 16s rRNA-(3′-AUUCCUCCAC…5′).
- Ribosomes bind to S/D sequence and the Ribosome binding site (RBS) is also found on either side of the initiator codon AUG.
- In some 5’UTR also contains stem loops which act as Riboswitches.
- In E.coli leaderless mRNAs recognize and bind to 5’ terminal AUG. Cross-linking assays indicate that a subset of 30S subunit r-proteins, located at either end of the mRNA tunnel, contribute to tRNA-independent contacts and/or interactions with a leaderless mRNA's start codon.
- Presence of 5’triphosphates in mRNA is very important for the binding of ribosomes to leaderless mRNA directly to AUG of mRNA to form ternary complex.
- 30S subunits containing IF2 stimulate ribosome binding and translation of leaderless mRNAs. Based on data, that 70S ribosomes bind leaderless mRNAs in vitro, suggest the possibility of a novel pathway for translation of leaderless mRNAs. This does not require S/D sequences?
- It is interesting to note that 70S ribosomes bind with higher affinity, or stability, than did 30S subunits to leaderless mRNAs containing AUG or GUG start codons. This is also true to archaea and eukaryotic mRNAs.
- The 5’leader sequences assume stem loop structures for attenuation mechanism of translation ex. Tryptophan mRNA. Attenuation is a regulatory feature found throughout Archaea and Bacteria causing premature termination of transcription.
- This spacer region is used for the ribosome dissociation at the end of coding sequence and reassociation with the next cistron and again to initiates translation. The size of the inter-cistronic space varies, but mostly very short. Often the terminator codon sequence overlaps with next cistron which contains AUG. This facilitates the reinitiation smoothly (e.g. UGAUG).
- There are few bacterial sRNAs bind to ribosome binding sites either to activate translation or to repress translation, often they bind to Hfq proteins for their activity.
5’---UAGGAGG—-AUG-----II-AUG---II-AUG----UGA--ter
In prokaryotes transcription and translation events are coupled. Half-life of mRNAs is very short ranging between 2-5 minutes. Most of prokaryotic mRNAs don’t have poly-A tail, even if they have the tail will be 15-20ntds long. This poly-A adenylation induces degradation of mRNAs, in fact it is added during degradation.
Only some mRNA from Yersinia contain bicistronic and monocistronic mRNAs for Major Cold Shock protein (MCSP) induced by cold shock. Bacterial and yeast mRNA lack introns.
In plastids (chloroplasts), primary transcripts undergo a complex series of mRNA maturation steps. These include processing of the 5′ and 3′ ends (RNA trimming), intron splicing, RNA editing, and cleavage of polycistronic precursor transcripts into monocistronic or oligo cistronic mRNAs (RNA cutting). On the other hand, chloroplast mRNA generated, most of them are polycistronic, but for translation they have to be cut to generate monocistronic mRNAs,( FiZhou, Daniel Karcher, Ralph Bock;2007). They have retained their ancestral characters. Plastids’ 5′ and 3′ end processing is catalyzed by nucleus-encoded prokaryotic-type ribonucleases. Whereas 5′ end maturation is catalyzed primarily by endoribonucleases, 3′ end formation is mediated by the concerted action of endoribonucleases and 3′→5′ exoribonucleases. Stem–loop-type RNA secondary structures within the 5′ and 3′ untranslated regions (UTRs) of plastid messenger RNAs provide important recognition elements for RNA processing enzymes, and, in addition, can serve as protective elements preventing rapid RNA degradation (Barkan and Goldschmidt-Clermont, 2000; Mayfield et al., 1995; Monde et al., 2000; Stern and Gruissem, 1987). One of the exceptions is the psbE operon, which comprises four small genes for polypeptides of photosystem II (psbE, psbF, psbL and psbJ;

(A) Transcription initiation of a gene cluster occurs from multiple promoters (bent arrow) upstream of open reading frames (ORFs) or within ORFs. Together with inefficient transcription termination, this setup generates numerous precursor transcripts that can include complete or incomplete ORFs. Introns and RNA stem–loop structures are depicted as light black rectangles and hairpins, respectively. (B) Precursor transcripts are processed by a combination of exo- and endo-ribonucleases. The precursor transcripts also can be polyadenylated by the addition of a Poly(A)-tail at the 3′-end of the transcripts. The sequence-specific RNA-binding proteins define functional RNAs followed by ribonuclease digestion. Introns and incomplete ORFs without sequence-specific RNA-binding proteins protection were digested by exo- or endo-ribonucleases. (C) RNA processing produces a pool of functional RNAs. http://www.nature.com/
Mitochondrial mRNAs are polycistronic, one H strand transcribes and the other is L strand transcribes. These long transcripts are cleaved to liberate tRNAs found in between and monocistronic mRNAs are released, sometimes the terminator codon of one cistron overlaps with the initiator codon of the next cistron. The released monocistronic mRNA are immediately polyadenylated up to ~50 (A) s.
Eukaryotic mRNAs:
- All mRNAs are single stranded polynucleotide chains with 5à3’ polarity and mostly monocistronic. Most of the eukaryotic mRNA get circularized for they are bound to eF4E and 3’ poly-A binding proteins; both bind to eIFG. Most EUK transcript are monocistronic and produce only one protein.
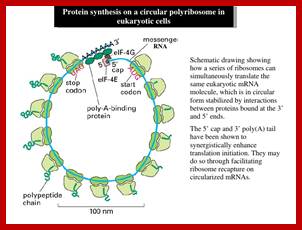
http://image.slidesharecdn.com/
Circularization provides easy way for ribosomes as they finish, they start another round of translation- this is a time programmed and efficient process. Some viral mRNA and some viral RNA genomes also show circularization.
http://biotech.christopher-vidal.com/
5’- 7’CH3 Gmp (Cap)-http://www.studyblue.com/
topsy.fr/hashtag.php;mol-biol4 masters (copy)
- Average size of eukaryotic mRNAs is 1500 to 2000 nucleotides. Smaller mRNAs are found in mammalian systems such as mRNAs for polypeptide hormones like Oxytocin, Glucagons and few such proteins. The longest mRNAs found are of proteins such as Dystrophin, Apolipoprotein, Fibronectin and Titin (till now Titin is considered as the largest protein and Crystallin (three types-alpha, beta and gamma) as long living protein in the lens of eyes.
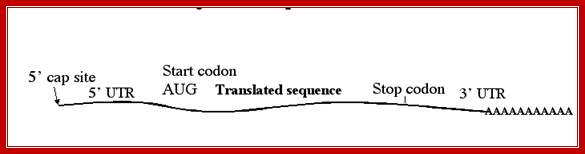
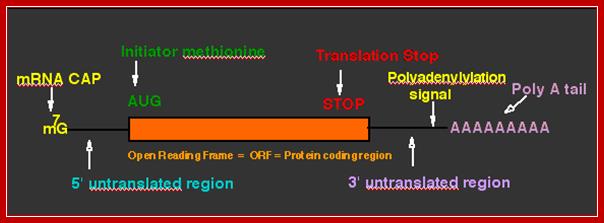
- The open reading frame (ORF) starts with an initiator codon and ends in a terminator codon.
topsy.fr/hashtag.php
- Upstream of the initiator codon of pre-mRNAs contain untranslatable sequences called leader or 5’UTR. There are introns in 5’ UTRs (5’UIs)
(35%), in humans which increase the expression.
- Many mRNAs contain uORFs in 5’UTR regions ex. GCN4 mRNA (there are four such uORF and the fourth one is the real ORF.
- Similarly most of the mRNAs at the 3’ end after TER codons, contain another 3’UTR trailer sequence, which vary in length and sequence.
- Secondary structure in 5’ leader and 3’ terminal regions play important roles in the regulation of mRNA activity, stability or and its ability and efficiency to translate. The said 3’ sequences are used for making the mRNA inactive to be stored for a long time. Unfertilized eggs, developing oocytes and plant seeds do store mRNA as informosomes.
- The initiator codon should be in proper context for proper initiation. In eukaryotes, ACC AUG G or A / G CC AUG G (A/GCCACCAUGG) sequence is the most important sequence context, without which translation initiation fails, even if initiates, it will be terminated shortly for it encounters a terminator codon. It is this sequence called Kozak sequence that acts at the time translation initiation; it is considered as the start of ORF . The small ribosomal subunit binds to this sequence covering Kozak, AUG and G.
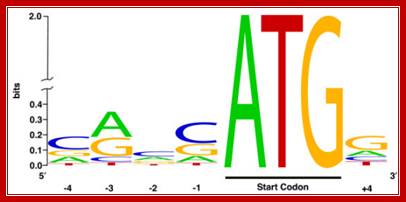
Kozak sequence is a consensus sequence (A/G)CCATGG) found adjoining the initiator codon AUG in eukaryotic mRNA, Harhay GP, et al ;http://openi.nlm.nih.gov/

Consensus RBS Sequences. The +1 A is the first base of the AUG initiator codon (shaded) responsible for binding of fMet-tRNAfMet. The underline indicates the ribosomal binding site sequence, which is required for efficient translation; http://themedicalbiochemistrypage.org/
- In addition to initiator sequences, positioning the Kozak sequence in right context, the succeeding sequences have to be in the order and should end in a terminator codon, it is only then such sequence is called open reading frame.
- The 5’ end the mRNA is added with 7’ methyl Guanine covalently to the first nucleotide, invariably to ‘A’ by an unusual 5’ P to 5’P linkage (7’CH3 G5’-P-P-P5’A---), called ‘cap-O’. Subsequent addition of more methyl groups to 7’A or to 2’ OH groups of ribose sugars in succeeding nucleotides make them to be called, cap-1, cap-2 etc.
- Only eukaryotic mRNAs contain cap structures and few non coding RNAs such as Sn RNAs, sno RNAs and few viral RNA genomes have such structures.
- Caps increase the efficiency of translational initiation of mRNA.
- Cells do contain specific CAP binding proteins.
- At the 3’ end, mRNAs are added with a 200-to 250-nucleotide long poly (A) tail. However EK mitochondrial mRNAs ex. From yeast and Neurospora, the poly (A) tail is just ~50 ntds long.
- A poly-A signal with AAAUAA and G/U rich sequence is used for cleaving the extended 3’ end of mRNA at 28-30 ntds from the signal, then poly-As are added to the cut end.
Histone mRNAs lack Poly-A tails, but contain stem loop structure at 3’ end. Some nc RNA like Lnc and Xist RNA too contain poly-A tail. Some histone mRNAs synthesized in non-cell cycle dependent stages contain poly-A tails. But yeast histone mRNA are poly-Adenylated?
Only capped mRNAs with poly-A tail and spliced ones are transported out of nuclei. But histone mRNA does not contain Poly-A tail, but the length varies. Its 3’ end is also processed to make it functional.
- Stored mRNAs contain very short poly-As and they are silent. They are activated by specific stimuli and poly-Adenylation restore their translation activity. Many such RNAs are found in oocytes and also in neuronal cells. In neuronal cell such mRNA are involved in learning?
- Many ncRNAs such as tRNA, rRNA, snRNA and snoRNAs polyadenylated for degradation. This is done in the nucleus by an enzyme complex called TRAMP.
· Synthesis and translation are separated in “time and space”. It takes nearly 30 to 45 minutes for the functional mRNAs to appear in cytoplasm after its synthesis.
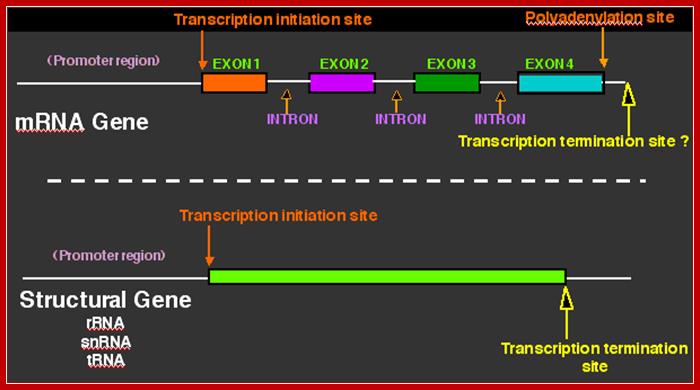
· All mRNAs are synthesized as long precursor RNAs called heterogeneous nuclear RNAs (hnRNAs), which are also called pre-mRNAs. Because of the heterogeneity in size and characters of the mRNAs produced, they are also called hnRNA.
· The hn-RNAs, in most of the cases, longer than their counter part cytoplasmic functional mRNAs.
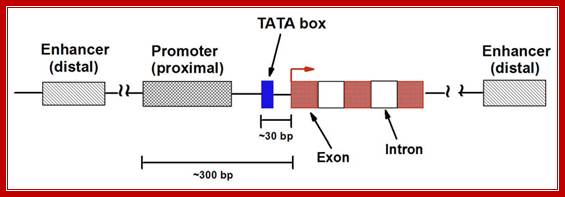
· They (hnRNAs) contain several segments of non-coding sequences, called Introns or intervening or interspersed sequences among coding regions called Exons.
· More than ~1000 intron less mRNAs are found in human genome transcripts, ex. Histone mRNAs, IFN alpha and IFN-beta mRNAs and many more lack any introns.
· The number of introns and the size of each introns and the position of introns vary from one species of mRNA to the other.
· Introns are removed and specific Exons are joined in a sequence by a process called splicing.
· Only spliced mRNAs are transported out of the nucleus.
· The same mRNA produced in different tissues can be spliced in different ways, by what is called alternative splicing to produce different polypeptides in stage specific or tissue specific or cell type manner, depending on the needs.
· Alternative polyadenylation-In some species a single polycistronic Pre mRNAs contains polyadenylation sites at different positions, thus it produces different mRNAs but its 5’ region remains the same.
· Some mRNAs are edited in tissue specific manner by specific enzymes or by Editosomes.
· Many systems perform Trans-splicing resulting in all mRNAs having the same leader sequence. Here leader sequences and mRNA or pre-mRNAs are synthesized separately, and then they are processed to produce Trans-spliced mRNAs.
· Majority of the mRNAs exhibit rapid turnover, yet their half-life is more than 10 to 20 minutes; some have long half-lives up to 120 days.
· Some mRNAs associated with a variety of mRNPs remain untranslated for quite a period of time, such mRNAs are called informosomes or stored mRNAs. They are activated differentially, when required, with specific signals.
· mRNP code is not unique to eukaryotic mRNAs, it is also prevalent in bacterial mRNAs.
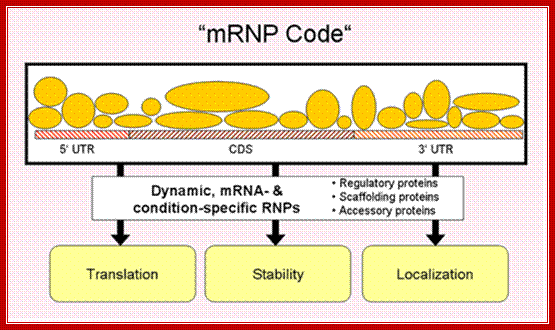
Different proteins assemble on a given mRNA to form a ribonucleoprotein complex (mRNP), the composition of which changes dynamically, depending on the cellular context. The combinatorial control of associated regulatory, scaffolding and accessory proteins ultimately determines fate of the mRNA ("mRNP code"); http://surrey.ac.uk/ ,k.a.gerber
· In eukaryotes (Eks) certain species of mRNAs are positioned in specific locations inside the cell, ex. positioning of mRNAs in developing segments of Drosophila larvae, mRNAs for egg polarity fixation, and Actins in developing Myeloblast cells. Ex. Oskar and Nano (Posterior), Bicoid (anterior); Staufen a dsRNA binding protein is involved in immobilization of Bicoid RNA at anterior end, Gurken (anterior-dorsal) mRNAs.
· Such localization is made possible by ‘zip code’ or ‘RNA transport signal sequences (21ntds)’ to which specific proteins bind and then they are transported on microtubule tracts using Kinesin or Dyneins motor proteins. In some their poly-(A) tails are actually held by cross-linking to Actin filament network. Microtrabacular network of Actin like filaments of small sizes may have a role? Actin mediated motor proteins are Myosins.
· Some mRNAs contain quadruplex RNA structure in 5’UTR.
· Some eukaryotic mRNAs are also known to contain several types of repeat in the untranslated regions, including short interspersed elements (SINEs) such as Alu elements, long interspersed elements (LINEs), Minisatellite and microsatellites. In human mRNAs, repeats elements are found in about 12% of 5' UTRs and 36% of 3' UTRs. A lower repeat abundance is observed in other taxa, including other mammals.
· Processed mRNAs are bound by specific mRNPs and transported out of the nucleus.
· For transportation, mRNAs must have a cap structure, with out which they cannot be transported out of the nucleus. In addition specific proteins are required for transportation; otherwise they remain within the nucleus.
7’CH3GpppA-5’UTR—A/GCC.AUG.G-//--UAA.3’UTR-(A)n 3’

mRNA processing, 5’ capping; https://www.boundless.com
At 5’UTR region some mRNAs contains stem loop structures that have regulatory functions. Some of the mRNAs contain internal ribose entry elements called IREs and Iron response stem loop structures, also called IREs.
The 3’UTR contain many sequence elements such as Antisense RNA binding sequences, regulatory stem loops like IRE-BP, ARE-AUUUA sequences, mRNA localization sequences called Zip Code, EDEN-embryonic deadenylation elements, DICE-Differential control elements, CPE-cytoplasmic adenylation elements, mi/siRNA binding elements, SECIS element-selenocysteine binding sequence, CRE-cysteine rich stability element, CITE-3’cap independent translation enhancer element, and SL- stem loop element.
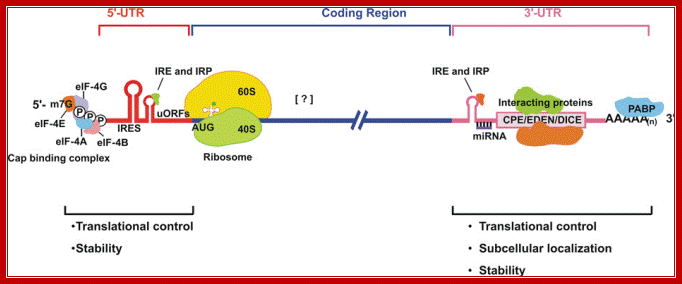
Structural organization of eukaryotic mRNA and the different points of possible regulation of translation through various trans-acting factors;5′-m7G, cap structure; eIF, eukaryotic initiation factor; CPE, cytoplasm polyadenylation element; EDEN, embryonic deadenylation signal; DICE, differential control element; PABP, poly(A)-binding protein; the possible sites of interaction of transacting factors (yet many unknown) in the coding sequence. Regions of mRNA involved in subcellular localization and stability are also indicated. ; Sangeeta Chaterjee and Jayant K.Pal; http://www.biolcell.org
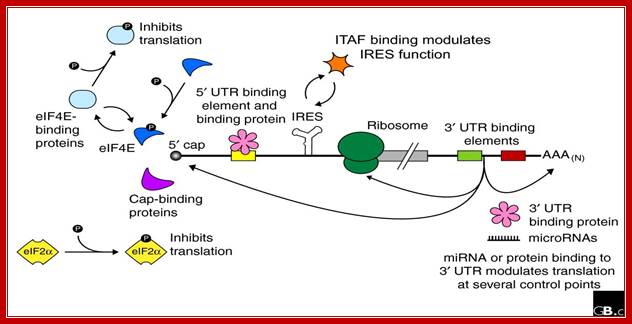
Baker and Coller; http://genomebiology.com
Regulation of eukaryotic mRNA translation occurs at numerous control points. Recognition of 3' UTR sequence or structural elements (green and red boxes) by RNA-binding proteins leads to either activation or repression of translation, often through alteration of the 3' poly(A) tail or through interactions with proteins that bind at the 5' terminal cap structure (that is, the initiation factor eIF4E or cap-binding proteins). Repression of translation by miRNAs can occur through inhibition of translation initiation or elongation, and may also lead to changes in the status of the mRNA 3' poly-(A) tail. Elements found within the mRNA 5' UTR (yellow box) can bind regulatory proteins that repress translation by inhibiting 48S ribosome scanning. Global regulation of mRNA translation is commonly achieved through modification of the translational apparatus (that is, by phosphorylation of the translation initiation factors eIF2α and eIF4E) and the ribosome itself, or modulation of protein partner binding affinities (such as the phosphorylation of the eIF4E-binding proteins). Translation can be initiated independent of the mRNA 5' cap through a structured internal ribosome entry site (IRES) in the 5' UTR whose efficiency in initiating translation is, in turn, modulated by trans-acting factors (ITAFs).
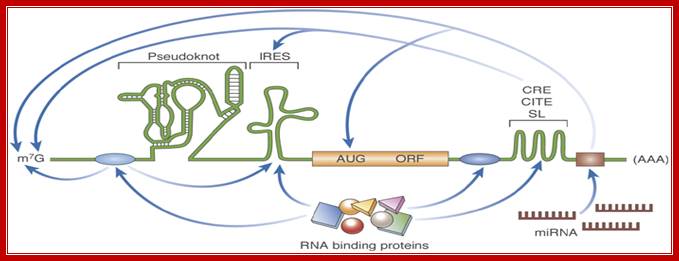
RNA structure: new messages in translation, replication and disease; mRNA with many 5’ and 3’ regulatory elements- %’ UTR may contain pseudo knot, IREs and 3’ may contain Zip code, CBE,CITE, mi antisense sequence. Lisa Roberts, Martin Holcik; http://embor.embopress.org/
Schematic illustration of discreet RNA regulatory elements: The translation of an mRNA is regulated by diverse mechanisms that involve both structural and non-structural RNA elements, as well as interactions with RNA-binding proteins. IREs found in the 5' UTR promote cap-independent translation. Pseudoknots can be located in the 5' UTR, the 3' UTR, or the coding region, and their localization influences their effect on translation; for example, initiation, frameshifting and termination. 3' UTR structural elements such as C rich stability enhancer elements CREs, 3’Cap independent translational enhancer element-CITEs and Stem loop SLs often function through long-range RNA interactions. The miRNA target sites are located in the 3' UTR. The canonical mRNA features of the Cap (m7G) and poly-(A) are targets of several regulatory interactions. The RNA-binding protein-binding sites are shown as blue ovals, and the miRNA-binding site is shown as a brown rectangle. AUG, initiation codon; CITE, cap-independent translational enhancer; IRES, internal ribosome entry site; m7G, 7-methyl-guanosine; miRNA, microRNA; mRNA, messenger RNA; ORF, open-reading frame; SL, stem loop; UTR, untranslated region.
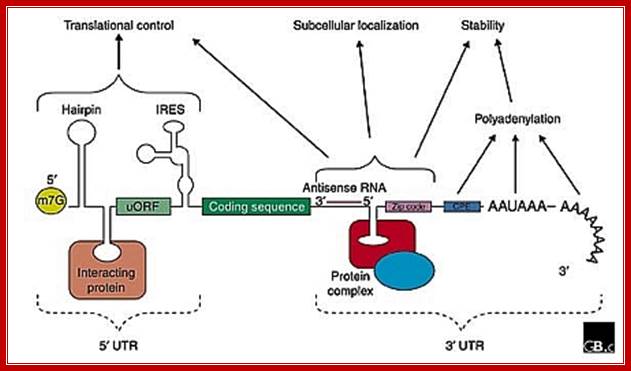
The generic structure of a eukaryotic mRNA, illustrating some post-transcriptional regulatory elements that affect gene expression. Abbreviations (from 5' to 3'): UTR, untranslated region; m7G, 7-methyl-guanosine cap; hairpin, hairpin-like secondary structures; uORF, upstream open reading frame; IRES (iron response), internal ribosome entry site; CPE, cytoplasmic polyadenylation element; Zip lock, antisense elements, Iron response elements 3’IRES, ARE elements, AAUAAA, polyadenylation signal.; Mignone er al;http://genomebiology.com/; http://openi.nlm.nih.gov/
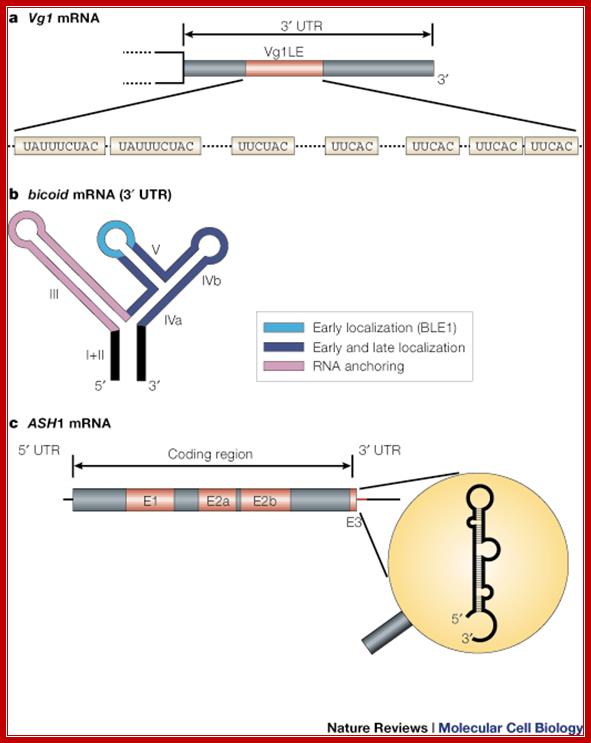
Types of mRNA zip codes; mRNA localization; message on the move; Ralf-Peter Jansen; http://www.nature.com/
a | The Vg1 3' UTR contains a 340-nucleotide zip code that is sufficient for mRNA localization, termed the Vg1LE, or Vg1 localization element. The functional units in this type of element are short repetitive sequences (either UAUUUCUAC or UUCAC)59, 104. b | The bicoid localization element (BLE) in the bicoid 3' UTR is an example of a zip code with a modular architecture. bicoid mRNA undergoes several sequential transport steps, each involving different, partially overlapping, regions in the highly structured 3' UTR. (Light blue, early localization; dark blue, early and late localization; purple, mRNA anchoring.) c | ASH1 mRNA is an example of a zip code element that lies in the coding region (E1, E2a, E2b) and in the 3' UTR (E3). The E3 element also represents an example of a structure-based zip code, because the displayed stem loop structure but not the primary sequence is important for the function of the zip code
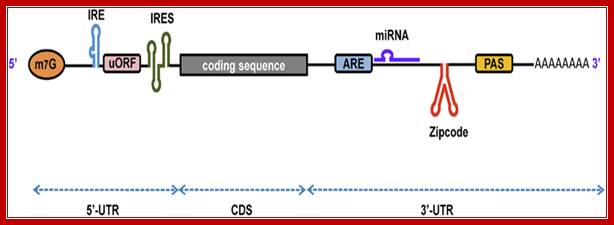
Firoz Ahmed et al; http://journal.frontiersin.org/
A schematic representation of eukaryotic mRNA with functional elements. UTR, untranslated region; CDS, coding sequence; m7G, 7-methyl-guanosine cap; IRE, iron-responsive element; uORF, upstream open reading frame; IRES, internal ribosome entry site; ARE, AU-rich element; PAS, poly(A) signal, Firoz Ahmed1, Vagner A. Benedito et al
- In developing Oocyte of Xenopus and such cells many mRNAs required remain dormant and stored. When activated they are translated. Such mRNAs at 3’UTR certain GU rich sequences called EDEN Embryonic Deadenylation Elements which on binding to EDEN-BP 2.87 kDa (oligomer 27a.a) removes 3’ poly-A chain using PARN Poly-A ribonuclease and render the mRNA inactive.
- Zip code- this region has specific sequences to which certain small molecular weight RNAs binds that lead to inactivation of mRNAs or it is get degraded.
- In developing Oocyte of Xenopus, a large number of mRNAs produced are processed and transported into cytoplasm where they remain untranslated and stored. Such mRNAs covered with a variety of mRNPs, such threads are called informosomes. The kinds of proteins bind to mRNA are species specific, some are cell specific and the rest of them are general.
- Some of the mRNAs stored as informosomes have shortened poly-(A) tail (20Å). Fertilization provides signals, which make several of the mRNAs, specifically get activated. At this point of time hitherto inactive mRNAs are added with poly-(A)s and such mRNAs are translated. The CPSF and PAP factors activate the process.
- Even dry seeds, with 6-8% of water, having inactive embryos; on imbibing water to full capacity (80-90%), several of the stored informosomal mRNAs become active and translate efficiently. This increase in translation is not accompanied with new synthesis of mRNAs. But it is due to activation of stored mRNAs.
- In the dry seeds of Arabidopsis thaliana, more than 12,000 stored mRNA species were detected, including all ontological categories. A large number of them are produced in response to ABA. With imbibition of water PAPs become active and add Poly-A s to 3’end and thus activate stored mRNA into active mRNAs. Majority of them have intact poly-A chains.
Even tissues, which are kept in dark and etiolated, when, exposed to light or hormones or both, mRNAs are activated and then they are translated. Such mRNA under conditions such as mentioned above or cells remain in stationary phase dictated by cell cycle events loose most of the poly-A tail. Such dormant mRNAs at 3’ UTR contain CPE cytoplasmic poly adenylation elements bound by CPEB which in turn bound by Maskin proteins. The Maskin in turn binds to TF4E thus the mRNAs are in circular state and rendered inactive under dark or unfavorable conditions.

Luc Paillard H. Beverley Osborne
Two alternative models of EDEN/EDEN-BP mechanism: A poly(A)+ transcript is translated. The EDEN sequence present in the 3′ UTR of a target mRNA binds EDEN-BP. According to a first model (A), EDEN-BP stimulates deadenylation. The resulting transcript is translationally repressed as a consequence of its poly(A)– status. According to another model (B), EDEN-BP represses translation. The translationally repressed mRNA would then be deadenylated. Whatever the correct model, the presence of an EDEN sequence in the 3′ UTR with bound EDEN-BP, leads to deadenylation and translational arrest. ORF, open reading frame.
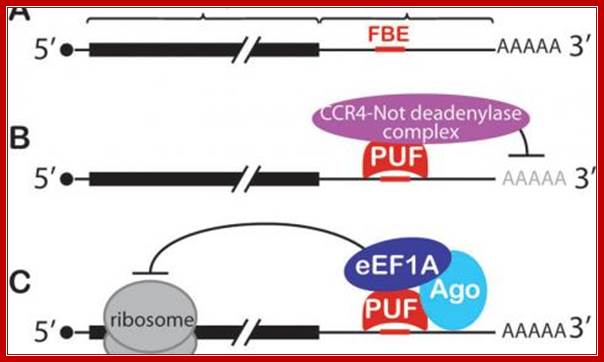
Conserved PUF mechanisms of mRNA control. A: FBF binding elements (FBEs) were among the first PUF regulatory elements identified; most are located in the mRNA's 3' untranslated region (3'UTR). B: PUF (Pumilo and FBF (fem-3 binding factor) proteins recruit deadenylase to repress target mRNAs. C: Both nematode FBF and human PUM2 form a ternary complex with an Argonaute (Ago) and translation elongation factor (eEF1A) to repress translation; Judith kimble; http://www.hhmi.org/

Translational regulation of the transferrin receptor and ferritin production; Production of the transferrin receptor (TfR) and ferritin is regulated at the level of mRNA by iron regulatory proteins (IRPs), which bind to iron response elements (IREs) on the 3'- and 5'- untranslated regions of their respective mRNAs1. a | In iron deficiency, the IRPs bind to the IREs, protecting the TfR mRNA from nuclease digestion and preventing the synthesis of ferritin. b | When iron is abundant, the modified IRP no longer binds to the IREs — in IRP1 the IRE binding site is blocked by a 4Fe–4S cluster (green rectangle), whereas in IRP2 the protein is targeted for destruction in the proteasome — allowing TfR mRNA to be destroyed and allowing the expression of ferritin; Luigi Zecca, Moussa B. H. et al; Nature Reviews; http://www.nature.com/
Some of these mRNA also contain EDEN (Embryonic Deadenylation Element), which are bound by EDEN binding proteins. EDEN-BP binding sequence is discerned as UGUCCUUUUAUAUGUAA or UR repeats and a single 36 ntd region -UAUAUGUAUGUGUUGUUUUAUGUGUGUGUGUGUGCU.

Evolutionary conservation of deadenylation by CELF1 protein and GU-rich sequences. (a). InXenopus and Drosophila eggs, after fertilization, EDEN-BP (CELF1 homologue) bound to EDEN-containing maternal mRNAs, causing deadenylation and subsequent translational activation.(b). In mammalian cells, CELF1 binds to GREs within the 3' UTR of specific transcripts and promotes their deadenylation (by deadenylases) and subsequent decay by the Exosome; ( (CUGBP and embryonically lethal abnormal vision-type RNA binding protein 3-like factor 1 ); Daniel Beisang et al; http://www.intechopen.com/
Binding of CPEB to UUUUAU sequence and regulating cytoplasmic polyadenylation @2001 Nature Publishing group, Mendez,R and Richeter, J.D, Nature Review Molecular Biology, The fertilized eggs contain all the required mRNAs for rapid cell division and also contain factors to prevent premature translation and remain dormant; Maternal mRNA and PolyA Tail in Oocytes; Ren-Jang Lin, Ph.D; http://www.nature.com/
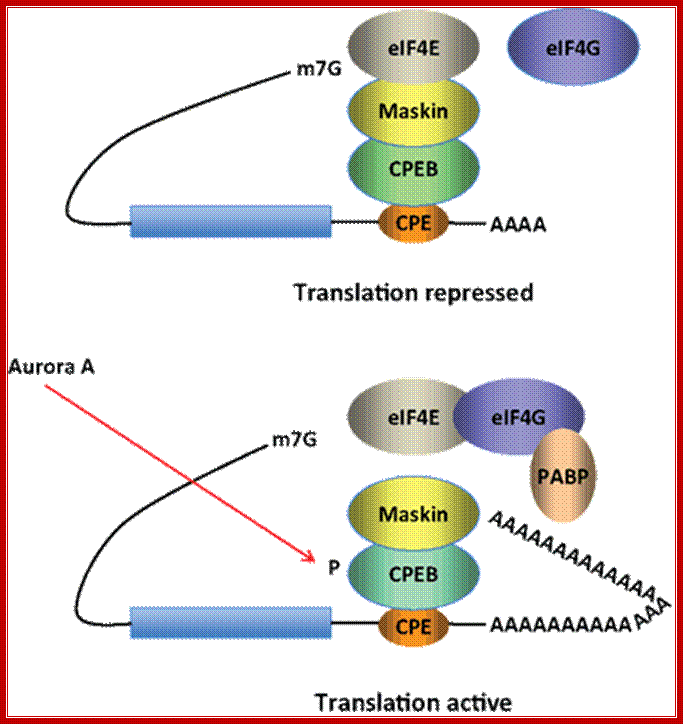
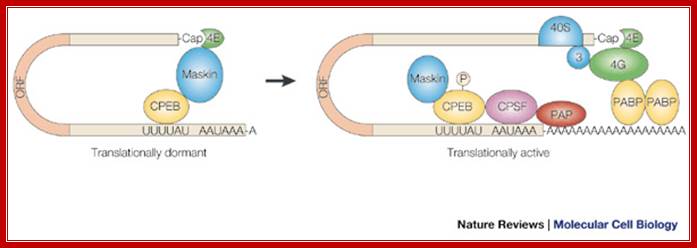
Model for CPEB activity. Some mRNAs contain a cytoplasmic polyadenylation element (CPE), which is bound by CPEB. CPEB also interacts with Maskin (or Neuroguidin, a functionally related protein), which in turn interacts with the cap (m7G) binding protein eIF4E. Such mRNAs are translationally inactive because Maskin inhibits the association of eIF4E and eIF4G, another initiation factor that helps recruit the 40S ribosomal subunit to the 5′ end of the mRNA. Cues such as NMDA receptor activation stimulate the kinase Aurora A, which phosphorylates CPEB, an event that causes poly(A) tail elongation. poly(A) binding protein (PABP) binds the newly elongated poly(A) tail and recruits eIF4G. The PABP-eIF4G dimer helps to displace Maskin from eIF4E, allowing eIF4G to bind eIF4E and initiate translation. R. Suzanne Zukin et al ;http://journal.frontiersin.org/
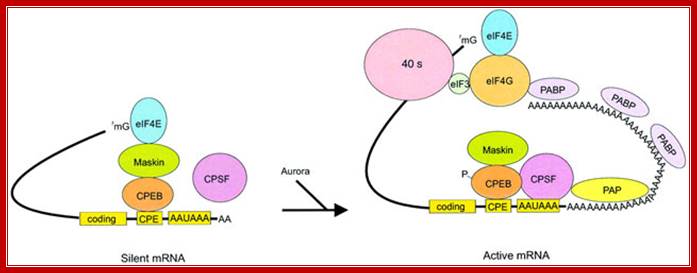
Model of polyadenylation-induced translation. Dormant CPE-containing mRNAs (e.g. cyclin B1) in immature oocytes are bound by CPEB, which in turn is bound to maskin, which in turn is bound to eIF4E, the cap-binding factor. The binding of maskin to eIF4E precludes the binding of eIF4G to eIF4E, thus inhibiting the formation of the initiation complex. The cleavage and polyadenylation specificity factor (CPSF) may or may not be loosely associated with the hexanucleotide AAUAAA at this time. Following progesterone stimulation, the kinase aurora is activated and phosphorylates CPEB Ser174, an event that causes CPEB to bind and recruit CPSF into an active cytoplasmic polyadenylation complex, presumably helping it to associate with the AAUAAA. CPSF recruits poly(A) polymerase (PAP) to the end of the mRNA, where it catalyzes poly(A) addition. The newly elongated poly(A) tail is then bound by poly(A)-binding protein (PABP), which in turn associates with eIF4G. eIF4G, when associated with PABP, then displaces maskin from, and binds to, eIF4E, thereby initiating translation. eIF4G, through eIF3, interacts with the 40S ribosomal subunit. Model of polyadenylation-induced translation.
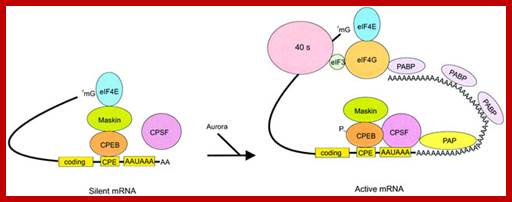
R. Suzanne Zukin et al;‘http://journal.frontiersin.org/
Model of polyadenylation‐induced translation. Dormant CPE‐containing mRNAs (e.g. cyclin B1) in immature oocytes are bound by CPEB, which in turn is bound to maskin, which in turn is bound to eIF4E, the cap‐binding factor. The binding of maskin to eIF4E precludes the binding of eIF4G to eIF4E, thus inhibiting the formation of the initiation complex. The cleavage and polyadenylation specificity factor (CPSF) may or may not be loosely associated with the hexanucleotide AAUAAA at this time. Following progesterone stimulation, the kinase aurora is activated and phosphorylates CPEB Ser174, an event that causes CPEB to bind and recruit CPSF into an active cytoplasmic polyadenylation complex, presumably helping it to associate with the AAUAAA. CPSF recruits poly(A) polymerase (PAP) to the end of the mRNA, where it catalyzes poly(A) addition. The newly elongated poly(A) tail is then bound by poly(A)‐binding protein (PABP), which in turn associates with eIF4G. eIF4G, when associated with PABP, then displaces maskin from, and binds to, eIF4E, thereby initiating translation. eIF4G, through eIF3, interacts with the 40S ribosomal subunit; http://emboj.embopress.org/
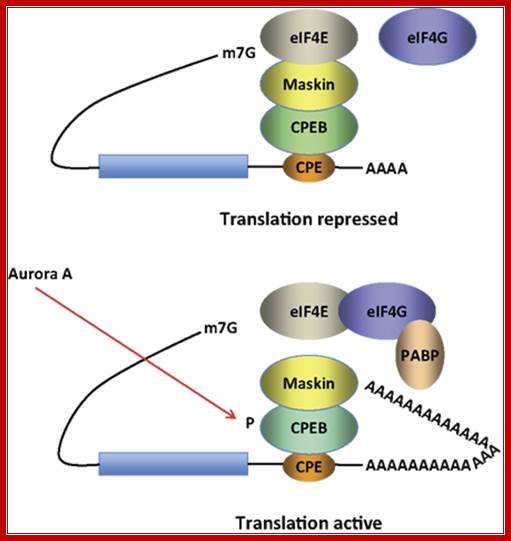
Model for CPEB activity: Some mRNAs contain a cytoplasmic polyadenylation element (CPE), which is bound by CPEB. CPEB also interacts with Maskin (or Neuroguidin, a functionally related protein), which in turn interacts with the cap (m7G) binding protein eIF4E. Such mRNAs are translationally inactive because Maskin inhibits the association of eIF4E and eIF4G, another initiation factor that helps recruit the 40S ribosomal subunit to the 5′ end of the mRNA. Cues such as NMDA receptor activation stimulate the kinase Aurora A, which phosphorylates CPEB, an event that causes poly(A) tail elongation. poly(A) binding protein (PABP) binds the newly elongated poly(A) tail and recruits eIF4G. The PABP-eIF4G dimer helps to displace Maskin from eIF4E, allowing eIF4G to bind eIF4E and initiate translation.
hnRNPs.
Most of the hnRNPs are heterogeneous in character and molecular weight. Based on their mobility in the gel and affinity to poly (A), poly (U), poly(C), poly (G) or poly (T) nucleotide chains, a large number of proteins, have been isolated and designated as hn RNP-A to hn-RNP-U and so on. Smallest of the proteins is A1, A2 and the highest Mol.wt proteins are U1, U2 proteins. The molecular mass of them ranges from 34 KD to 120kds. Most of the proteins have RNA binding motifs and also protein-protein binding motifs.
A list of hnRNPs proteins (not all):
A1A2, B1B2, C1C2, D1D2, E (4), F (2), G, H (2-3), I (2), J, K (3), L (2), M (4), N, P (2), Q (2), R (2), S2, T1, U1 and U2. Mol.wt of A to C range about 30-40kd, D to G 45-48kd, J, H, I 55-58kd, L, M, Q, P, N =68KD, T is95kd, R is 75kd, K is 66kd and U is about 116kd.
- Almost all hnRNPs are derived from normal and alternative splicing products. Among them, there are core proteins organized into 40s complexes, invariably they are made up of A1, A2, B1, B2, C1, C2 and the rest of them are grouped as D1 to U1 etc; the latter are called accessory proteins.
- The proteins have RNA binding motifs called RRN (RNP) motifs, made up of beta strands. Some of the sequences found in proteins are used for RNA binding contain RGG box, KH motifs, and SR motifs. Their versatile nature makes them ideal candidates for them to bind to hnRNA or pre-RNAs and prevents the RNAs from the formation of unwanted secondary structures.
- In addition, protein-RNA binding and protein-protein interactions bring pre-RNA into focus for modifications.
- Several of hnRNP proteins that bind to pre mRNA and also to processed mRNA have been identified and isolated. Core protein complexes of 200 Å size uniformly cover the pre-mRNA.
- As soon as an hnRNA is produced or being produced, it is covered with a variety of proteins, among them are SnRNA and snRNP complexes, for they are involved in splicing. Among them SM/SMN (severe motor neuron) proteins are important.
- Processed mRNAs are also bound by a variety of proteins called mRNPs, and some of them may be involved in transport of mRNAs out of the Nucleus. Inexplicably, some proteins move out of nucleus in association with mRNAs, and they are displaced with cytosolic mRNPs; and displaced nuclear mRNPs return to the nucleus, ex. Poly (A) binding protein PAB-II. Similarly a viral protein REV facilitates HIV RNA to move out of the nucleus; once mRNAs are transported out the nucleus the Rev Protein returns to the nucleus.