Processing of tRNA precursors:
Processing of tRNAs in Bacterial Cells:
- In bacterial systems rRNA operons contain tRNA genes intercalated in spacer regions of rRNA genes.
- But in sup B & E clusters, the rRNA operon contains several tRNA genes of different types are strung together with no rRNA segments.
- The Tyr-T operon contains tRNA genes as well as a gene for protein-P.
- Each of these is transcribed from a single promoter and the transcript is precursor RNA, often polycistronic, which is processed later.
- As described earlier, tRNA segments in the precursors are cut at 5’ end by RNase-p, while the 3’ is processed by RNase D.
- The released tRNA segments are further folded in 3-D structures and they are modified at specific bases as well.
- Approximate number of tRNAs per cell is about >20000.
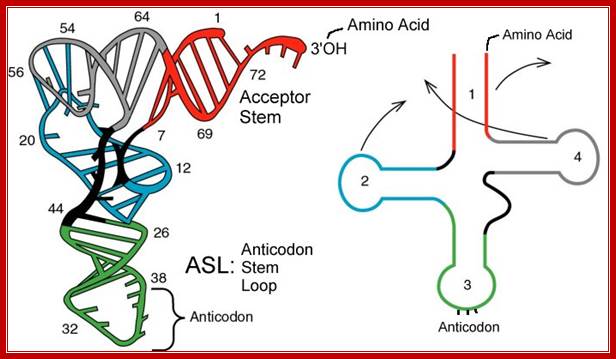
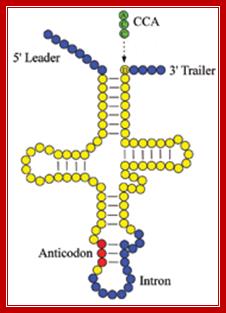
Folded Secondory structure of tRNA; http://www.bio.miami.edu/
Types of tRNA genes:
|
||||||||||||||||||||||
|
Types of tRNA genes |
Number |
|
Intron containing tRNA |
+/_ 671 |
|
Split tRNAs |
12 |
|
Initiator Met tRNAs |
93 |
|
AUA coding Ile-tRNAs |
81 |
|
Pyrolysine-tRNA |
4 |
|
Selenocsyteine tRNA |
8 |
|
Other tRNAs |
2872 |
This database contains total 3,741 sequences and secondary structures of various types of tRNA genes (details are shown on the right table), and 783 sequences and secondary structures of tRNA introns. If you use this database in your work you might want to cite: ;The above data is collected from Archaeal and primitive eukaryotes; required one to one tRNAs for decoding 63 codon but manage with 21 to 24 tRNAs by using wobble mechanism to properly base pair between codon and anticodon nucleotides. The minor AUA-decoding isoleucine tRNA in H. marismortui and other archaeal species is most likely derived from a CAU anticodon-containing tRNA; it is the rarest of the rare anticodon. It is similar to lysine, but with an added pyrroline ring linked to the end of the lysine side chain. It forms part of an unusual genetic code in these organisms, and is considered the 22nd proteinogenic amino acid. http://splits.iab.keio.ac.jp/
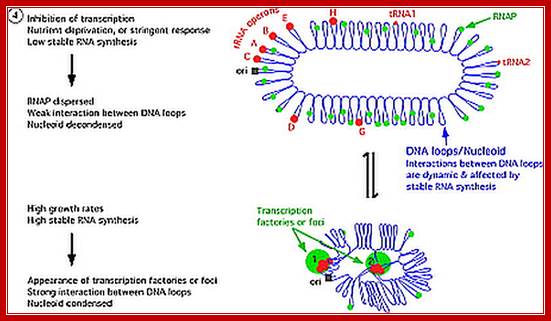
E.coli genome is in condensed state with many loops, specific loops containing ceretain genes are engaged in transcriptional activity, transcriptional loci or called transcriptional factories are shown in green. Model linking stable RNA synthesis, RNAP distribution and the dynamic structure of the nucleoid: The E. coli chromosome is represented as blue lines folded in loops, the ori of replication as a black square, the seven rRNA operons as large red circles with letters, and two representative tRNA operons as small red circles. The RNAP molecules are represented as small green circles. For simplicity, only two putative transcription factories/foci, which make the nucleoid more compact by pulling different stable RNA operons into proximity, are indicated here (bottom part of the diagram, large green circles labeled 1 and 2) [adapted from the study (Cabrera and Jin, 2003)].

http://www.suggest-keywords.com/;http://www.slideserve.com/
tRNA sup B-E Operon in prokaryotes:
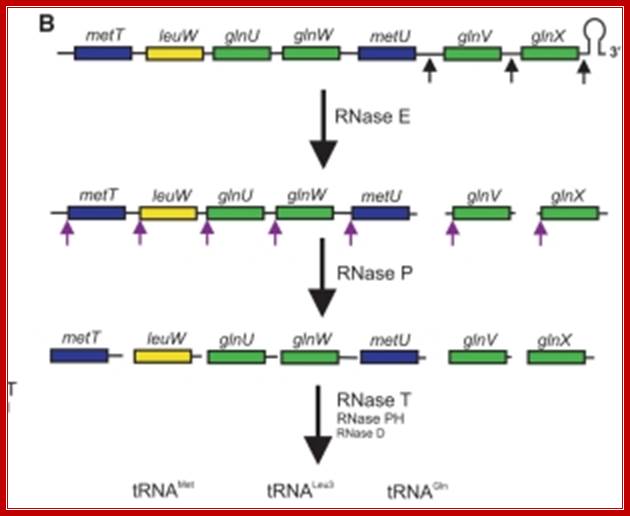
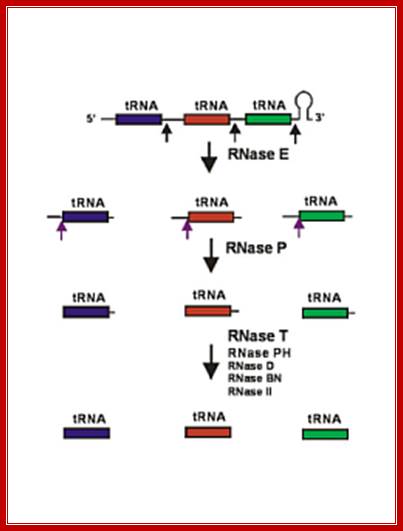
Seven tRNA encoding polycistronic RNA is stepwisely processed, by using Rnase E, Rnase P and Rnase T/pH/D; Noboru Nakajima, Haruo Ozeki etal
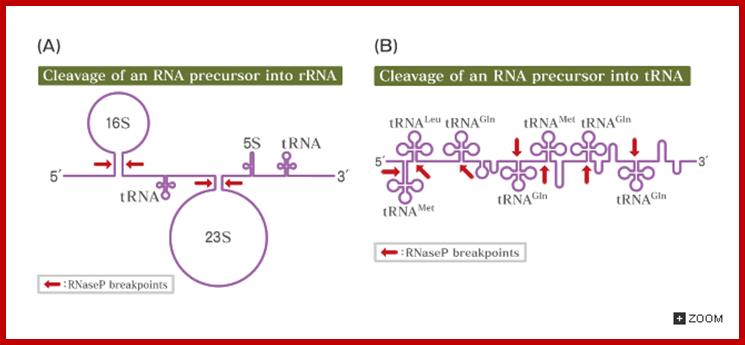
Cleavage of pre RNA and pre tRNA transcripts; http://csls-text.c.u-tokyo.ac.jp/
Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E.
The first step in the current model for the processing and maturation of mono- and polycistronic tRNA precursors in Escherichia coli involves initial cleavages by RNase E 1–3 nt downstream of each chromosomally encoded CCA determinant. Subsequently, each mature 5′ terminus is generated by single RNase P cleavage, while the 3′ terminus undergoes exonucleolytic processing by a combination of 3′ → 5′ exonucleases. Here we describe for the first time a previously unidentified pathway for the maturation of tRNAs in polycistronic operons (valV valW and leuQ leuP leuV) where the processing of the primary transcripts is independent of RNase E. Rather, RNase P cleavages separate the individual tRNA precursors with the concomitant formation of their mature 5′ termini. Furthermore, both polynucleotide phosphorylase (PNPase) and RNase II are required for the removal of the 3′ Rho-dependent terminator sequences. The 3’ end is cleaved first by endonucleases, and then trimmed by exonuclease like RNaseD. Our data indicate that RNase P substrate recognition is more complex than previously envisioned. Bijoy K. Mohanty and Sidney R. Kushner
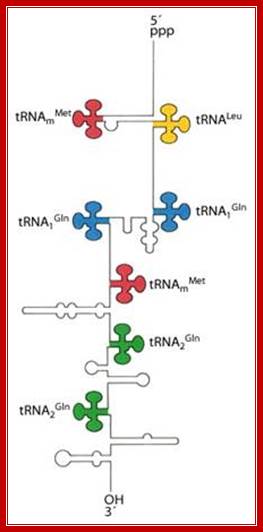
5’pppGC—met—-leu—gln—gln—met---gln---glu---AUG-T/t
In E.coli Ribosomal RNA genes are organized into seven operons, called rrns and ribosomal protein genes into six operons. Some of the tRNAs are organized into polycistronic operons as shown below, eg B.subtilis. First interspace region is cut by RNaseE at 3’ region of each tRNA gene; they are cut by RNaseP at the 5’end of each tRNA’s segments. All stable tRNAs have to be processed at 3’ end before they amino acylated. Majority of tRNAs are processed at 3’ end by a set of exonucleases and exonucleases. Some of the defective tRNAs get poly Adenylated for degradation.

B subtilis, rrnB/trnB operon, P and T promoter and terminator; at the 3’ end of rRNA segments one finds 21 tRNAs clusters, and transcribed by the RNAP using its bacterial promoter.
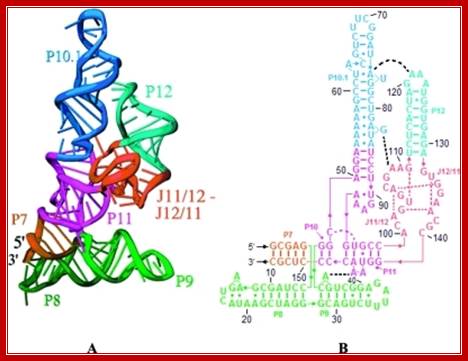
The 3-D model shows the RNA structural form of RNase-P, b.subtilis; Tertiary structure (A) and secondary structure (B) Liang R, Kierzek E, Kierzek R, Turner DH - Biochemistry (2010);http://openi.nlm.nih.gov/
Processing of tRNAs:
RNase E initiates processing by endonuclease downstream of the CCA determinant of each tRNA within the operon leading to the formation of immature tRNAs, which are further processed by RNase P to generate mature 5′ termini. The mature 3′ ends result primarily from the activity of RNase T, but RNase PH, RNase D, RNase BN and RNase II can substitute in the absence of RNase T. Many tRNAs that are processed lack 3’ terminal CCA, in such cases, nucleotidyl transferase adds CCA3’ nucleotides.
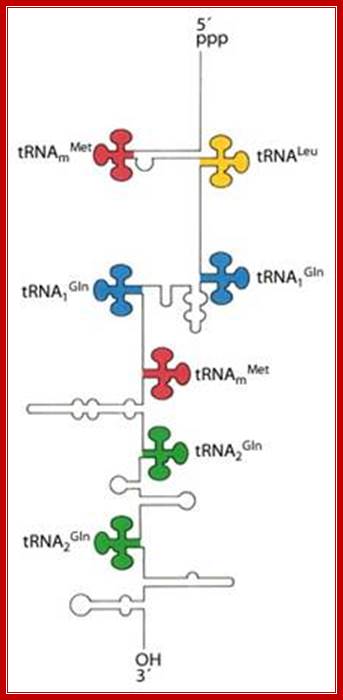
The tRNA genes are in the arrangement- 5'-leader- tRNAArgCCG -57 base pairs- tRNAHisGUG -20 base pairs- tRNALeuCAG -42 base pairs- tRNAProUGG -3'. Coordinate expression of the component tRNAs in vivo and the absence of intercistronic promoters indicated that all four tDNAs reside in the same operon.
These tRNA sequences are preceded by a single promoter region where a “Pribnow box” sequence is present seven base pairs upstream from the transcription start site. The spacer regions separating the seven tRNA sequences are different from each other both in size and in nucleotide sequence (Organization and structure of an E. coli tRNA operon containing seven tRNA genes; Noboru Nakajima, Haruo Ozeki and Yoshiro Shimura)
Structure of an Escherichia coli tRNA operon containing linked genes for arginine, histidine, leucine, and proline tRNAs; Processing starts with endoribonuclease RNase; Sequence specific cleavage by RNases P, F, D; (1) RNase P is an endonuclease that cleaves the precursor to generate the 5' end of the mature tRNA.(2) RNase F is an endonuclease that cleaves the precursor 3 nucleotides past the 3' end of the mature tRNA.(3) RNase D is an exonuclease that trims in a 3' to 5' direction to generate the 3' end or the mature tRNA. https://www.nobelprize.org; ttp://www.genetics.uga.edu/;
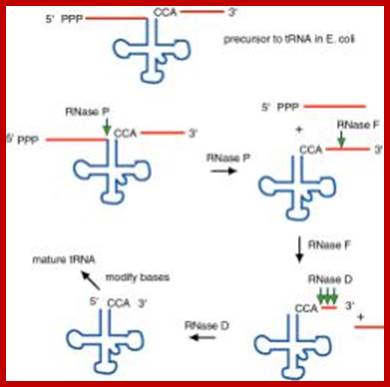
The ends of tRNA in E. coli are produced by the action of three nucleases that cleave the precursor to tRNA. A schematic of the pre-tRNA is shown at the top, with RNA extending from the 5’ and 3’ ends of the RNA that will become the mature tRNA (shown as a cloverleaf). The site of cleavage is indicated by the short vertical arrows above the lines denoting RNA, and they are labeled with the name of the enzyme cutting at that site. The enzymes catalyzing each reaction are listed above or adjacent to the reaction arrows. The catalytic activity of RNase P is in the RNA component
RNAse P is composed of a 375 nt RNA and a 20kDa protein. (2) The catalytic activity is in the RNA. The protein is thought to aid in the reaction, but is not required for catalysis. All enzymes are not proteins!. (3) This was one of the first instances discovered of catalytic RNA, and Sidney Altman shared the Nobel Prize for this. http://www.bx.psu.edu/ http://www.genetics.uga.edu/;
Eukaryotic tRNA Processing:
In prokaryotes tRNA genes are either found within rRNA operons or some are organized into exclusive tRNA operons. But in eukaryotes majority of the tRNA genes are localized in certain regions of chromosomes as clusters. The number and kind of tRNA genes found in each of the loci vary and such loci can be identified by tRNA specific RNA or DNA probes. Each kind of tRNA genes are found in multiple copies which can range from 50 to 200 each. In human chromosomes some tRNAs are located in chromosome 6 and other chromosomes. And each of the genes have their own specific promoters, mostly in the coding regions and they are transcribed by RNA polymerase III assisted by its TFs. The precursor tRNAs are then processed by RNase-P and other enzymes such as RNase III, RNase E, RNase D and few more. RNase-P cleaves and generates 5’end of tRNA and RNase E and D generate 3’ end. In some the 3’ end lacks CCA, but the CCA is added by another set of enzymes. In eukaryotes tRNA do contain introns of different sizes, which are found at the 3’ end of anticodon loop. Introns are spliced out by two modes.
Top sketch shows the clustering of tRNA genes in plant Arabidopsis thaliana. Below clusters of tRNA genes, each of the have their own promoter elements. http://www.genetics.uga.edu/;
http://biosiva.50webs.org/
http://slideplayer.com
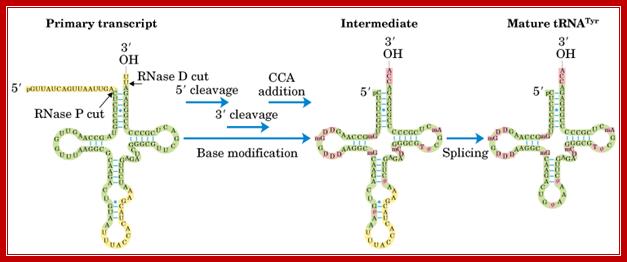
Processing of t-RNA:
Both prokaryotic and eukaryotic pre t-RNA undergo post transcriptional modification. The steps for post transcriptional modification of pre-t-RNA are as follows:
i) First, the flanking regions of the 3’-OH and 5’ phosphate ends are cleaved by the endonuclease action of RNase D and RNase P respectively.
ii) Then, the introns in the anticodon loop space, spliced out by splicing reaction.
iii) Tri nucleotide CCA is added to the 3’-end to give 32’-OH ACC terminus. This reaction is catalyzed by t-RNA specific nucleotidyl transferase. It is unique reaction because this enzyme catalyzes transfer of 3 nucleoside phosphodiester bond formation in one step.
iv) Finally, the t-RNA undergoes base modifications to give mature t-RNA. http://biosiva.50webs.org/
This diagram illustrates the trimming of precursor tRNA from the 5’ and 3’ ends; note the 5’ end is generated by RNase-P. The 3’ is trimmed by RNase-D, and then CCA is added nucleotide by nucleotide. Yeast tRNA splicing involves cutting and rejoining. The intron in yeast tRNAPhe base pairs with the anticodon to change the structure of the anticodon arm. Pairing between an excluded base in the stem and the intron loop in the precursor may be required for splicing. Splicing of tRNA depends principally on recognition of a common secondary structure in tRNA rather than a common sequence of the intron. http://genes.atspace.org/
- In eukaryotes tRNAs are organized as individual genes, where each gene has its own promoter element, but interestingly most of the genes are clustered in certain regions of the chromosomes.
- Each locus may contain 50 to 60 tRNA genes of different kinds, spanning about 50kbp area. Such loci are found distributed among all chromosomes. In many case they are clustered very near to telomeres. However the total number of tRNA genes found can vary from 250 in yeast to 2000 in human cells. In Drosophila there are 50 to 60 sites of 50kbp each and each contain about 18 –20 tRNA genes. In Xenopus, a 3-kbp site containing 7 (Phe, Tyr, Met, Asn Ala Leu and Lys) genes, are repeated 150 times. In this region one finds 5srRNA genes too located in chromosomes 13-18. In rats one cluster containing 3-4 tRNA genes, spanning about 2kbp is repeated 10 times. In Arabidopsis there are 124 tRNA genes.
- Moreover DNA polymerase III enzymes transcribe each of the tRNA genes. And they use internal promoters, i.e. promoter elements located with in the coding regions.
- The total number of tRNAs found in a eukaryotic cells is about 2 million (all species put together).
- Most of the precursor tRNAs are cut and processed by RNase-P from 5’ end and by RNase E/F and RNase D from 3’ end.
- Some processed tRNAs don’t contain the 3’ACC called type II tRNAs, where 3’ACC are added nucleotide by nucleotide by nucleotidyl transferase.
- Those tRNAs containing 3’ACC at the time of synthesis are called type-I tRNAs.
- An enzyme, RNase II degrades all types of tRNAs.
- The terminal 3’ A is constantly subjected turnover and tRNA nucleotidyl transferase is required for the repair all the time.
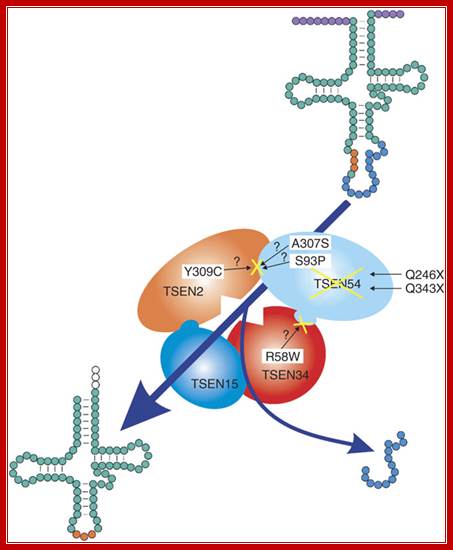
tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia;The tetrameric enzyme complex consists of the two catalytic subunits, TSEN2 and TSEN34, and two structural subunits, TSEN15 and TSEN54. Identified mutations in the PCH families are indicated by the corresponding amino acid changes. Top right, unprocessed tRNA with intron (blue) and anticodon loop (orange). After processing by the tRNA endonuclease, the intron is removed, resulting in the mature tRNA, bottom left. The processing of 5' leader and 3' trailer (purple) is performed by other enzymes; Birgit S Budde and etal (~20) http://www.nature.com/
Mario Morl and Anita Marchfelder; http://gcat.davidson.edu/
Modifications:
|
PDB Abbreviations for Modified Nucleosides in tRNAPhe |
|||
|
Residue |
PDB |
Full Name of Nucleoside |
Standard |
|
10 |
2MG |
N2-Methylguanosine |
M2G |
|
16 & 17 |
H2U |
Dihydrouridine |
D or hU |
|
26 |
M2G |
N2, N2-Dimethylguanosine |
M22G |
|
32 |
OMC |
2'-0-Methylcytidine |
M2'C |
|
34 |
OMG |
2'-0-Methylguanosine |
M2'G |
|
37 |
YG |
Wybutosine* |
Y-base |
|
39 & 55 |
PSU |
Pseudouridine |
"Psi" |
|
40 & 49 |
5MC |
5-Methylcytidine |
M5C |
|
46 |
7MG |
7-Methyguanosine |
M7G |
|
54 |
5MU |
5-Methyluridine |
T |
|
58 |
1MA |
1-Methyadenosine |
M1A |

a | Transcription
of a tRNA gene by RNA polymerase III (Pol III) generates a primary transcript,
which in some cases contains an intron, with 5' and 3' extensions. After 5' and
3' processing, base modifications (red circles) and CCA-nucleotide addition at
the 3' end, the resulting tRNAs can follow different export routes.
Intron-containing and intron-free tRNAs (aminoacylated or not) are exported
either by the exportin-t/Los1-dependent pathway, or by other less well
characterized routes. After export to the cytoplasm and dissociation of the
tRNA cargo from its receptor, intron-containing tRNA is spliced by the
tRNA-splicing machinery, which is bound to the outer mitochondrial membrane in
yeast. Mature tRNA is then channelled to the translational machinery by release
factors such as Cex1 and eEF1![]() .
.
b | For microRNA (miRNA), only the Pol
II-dependent generation of the primary mRNA (pri-mRNAs) is depicted. miRNAs can
be encoded by an miRNA gene situated in the intron of a coding gene (left
branch) or from a separate miRNA gene (right branch). In both cases, the
primary transcript is cleaved by the Drosha–DGCR8 complex to generate a
pre-miRNA with a stem–loop structure. Moreover, a spliced mRNA can be generated
concomitantly with the excision of an intronic miRNA. The pre-miRNA with its
typical ![]() 2-nucleotide overhang at its 3' end is specifically recognized by
exportin-5 and is transported to the cytoplasm, where it dissociates from its
receptor after RanGTP hydrolysis. Following release from exportin-5, Dicer
further cleaves the pre-miRNA to finally generate a single-stranded miRNA
species, which assembles into the RNA-induced silencing complex (RISC).
Highlighted in blue is the RNA region that corresponds to the mature effecter
miRNA. Alwin Köhler & Ed Hurt;http://www.nature.com/
2-nucleotide overhang at its 3' end is specifically recognized by
exportin-5 and is transported to the cytoplasm, where it dissociates from its
receptor after RanGTP hydrolysis. Following release from exportin-5, Dicer
further cleaves the pre-miRNA to finally generate a single-stranded miRNA
species, which assembles into the RNA-induced silencing complex (RISC).
Highlighted in blue is the RNA region that corresponds to the mature effecter
miRNA. Alwin Köhler & Ed Hurt;http://www.nature.com/
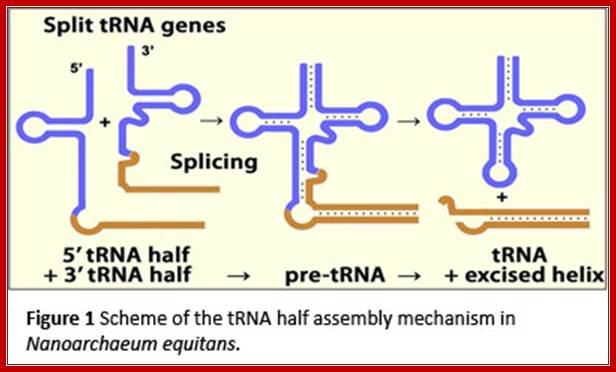
The two tRNA segments are coded in two different loci, the transcripts get associated and processed to produce functional tRNA; http://www.staff.uni-marburg.de/
“sRNA-guided 2'-O-methylation in Archaea:
Authors have developed a heterologous, in vitro modification system
using Methanocaldococcus jannaschii proteins and pre-tRNATrp
from H. volcanii. Using this system, authors have proven that 2'-O-methylations
at positions 34 and 39 of the pre-tRNA occur through guide-target pairing in trans,
as opposed to cis. The intron as part of the pre-tRNA or in free linear
or circular form, produced during splicing reaction can act as guide in these in
vitro reactions. Using the same system we have also shown that methylation
of the two nucleotides, guided by the two sites of a single box C/D RNA occur
sequentially. Box C'/D' RNP-guided U39 methylation first requires a box C/D
RNP-guided methylation of C34. We also show that dynamic guide-target
interactions contribute to this sequential modification. Based on these and
earlier results (retention of circularized introns in vivo) we propose
an in vivo scenario, where the intron splicing and RNA methylation
events occur in concert. In addition, we have also constructed an H.
volcanii strain that has deletion of intron in its pre-tRNATrp
gene. As expected, this strain lacked 2'-O-methylations at positions 34
and 39 of its tRNATrp. Surprisingly, the strain showed no detectable
phenotype, in spite of deletion within a single-copy essential gene in the
genome”.
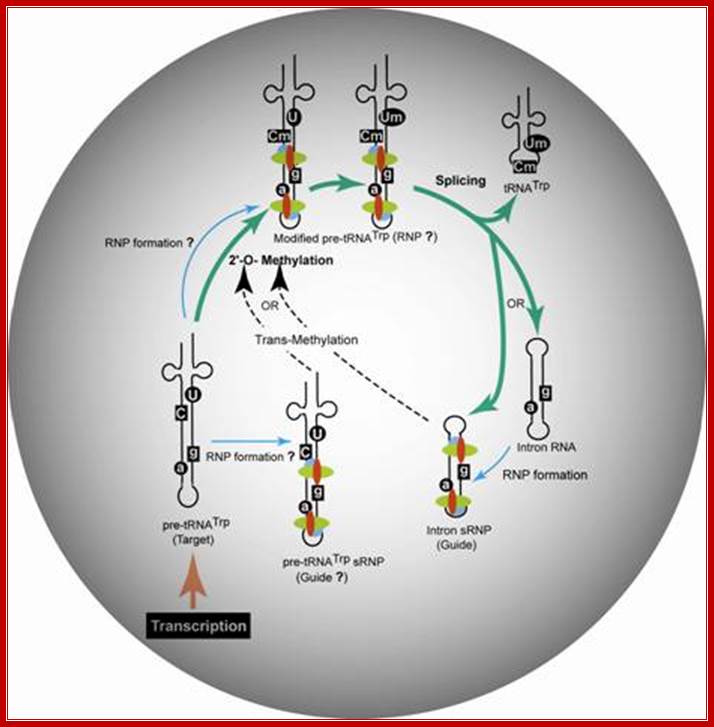
Proposed model for the trans-2'-O-methylation of H. volcanii pre-tRNATrp in vivo. Thick arrows indicate the pre-tRNATrp processing pathway proceeding from transcription of the pre-tRNA through nucleotide methylation and splicing to the production of tRNATrp and excised intron. RNP assembly is denoted by thin arrows, and nucleotide modification guided intermolecularly by the intron-encoded box C/D RNP is denoted by dashed arrows.;Gupta Lab; http://www.siumed.edu/
“Proposed model for the trans-2'-O-methylation of H. volcanii pre-tRNATrp in vivo. Thick arrows indicate the pre-tRNATrp processing pathway proceeding from transcription of the pre-tRNA through nucleotide methylation and splicing to the production of tRNATrp and excised intron. RNP assembly is denoted by thin arrows, and nucleotide modification guided intermolecularly by the intron-encoded box C/D RNP is denoted by dashed arrows”.
“Pseudouridine formation in tRNAs of Archaea
Pseudouridine
(ψ) is almost universally present at position 55 of tRNAs in all three
domains of life. Bacterial TruB protein and its homolog Pus4 in yeast convert U
at this position to ψ. This reaction is catalyzed in Archaea by the Pus10
protein, which is not a member of TruB/Pus4 family of pseudouridine synthases.
Pus10 homologs are found in Archaea and higher eukaryotes, but not in Bacteria
and yeast. This coincides with the presence of ψ54 in most tRNAs of
Archaea and a few tRNAs of higher eukaryotes and its absence in Bacteria and
yeast. Instead, most tRNAs of Bacteria and eukaryotes contain ribothymidine at
position 54. We have shown that archaeal Pus10 proteins can produce ψ54 in
addition to its tRNAs ψ55 synthase activity. The homology of eukaryotic
Pus10 with archaeal Pus10 suggests that the former may also have a tRNA
ψ54 synthase activity. Additionally, human Pus10 is involved in
TRAIL-induced apoptosis. This implies a dual functional role of Pus10 in higher
eukaryotes”.
Processing of tRNA precursor:
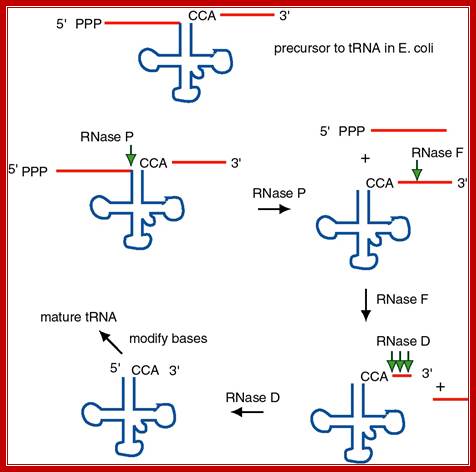
The ends of tRNA in E. coli are produced by the action of three nucleases that cleave the precursor to tRNA. A schematic of the pre-tRNA is shown at the top, with RNA extending from the 5� and 3� ends of the RNA that will become the mature tRNA (shown as a cloverleaf). The site of cleavage is indicated by the short vertical arrows above the lines denoting RNA, and they are labeled with the name of the enzyme cutting at that site. The enzymes catalyzing each reaction are listed above or adjacent to the reaction arrows.; Gene expression and protein synthesis;BMB 400; http://www.personal.psu.edu/
The catalytic activity of RNase P is in the RNA component.
(1) RNAse P is composed of a 375 nt RNA and a 20 kDa protein.
(2) The catalytic activity is in the RNA. The protein is thought to aid in the reaction, but is not required for catalysis. All enzymes are not proteins!
(3) This was one of the first instances discovered of catalytic RNA, and Sidney Altman shared the Nobel Prize for this
Molecular Basis of tRNA Processing Reactions-
Molecular Basis of tRNA Reactions; Michelle Mitchell and Hong Li; http://www.landesbioscience.com/
This chapter provides a review of recent advances in the molecular basis of transfer RNA (tRNA) processing events. tRNAs are transcribed into precursors that must undergo removal of 5′ and 3′ leader sequences, addition of a universally conserved CCA sequence, removal of introns and covalent modification of nucleotides (the latter are considered in other chapters). With the exception of the 5′ end processing that is carried out by a ribonucleoprotein particle, all processes are carried out by protein enzymes. These processing enzymes recognize features shared by all tRNA and facilitate cleavage or formation of phosphodiester bonds. Structural and complementary biochemical findings reveal an overriding theme that underscores enzyme modularization and local flexibility in tRNA.
Wobble base pairing uses tRNA 1st base of the anticodon (5>3’) and 3rd base of the codon (5>3), when base paired they are in antiparallel orientation. The first base and the third base while decoding mRNA codon they use what is famously called Wobble base pairing (F.Crick).
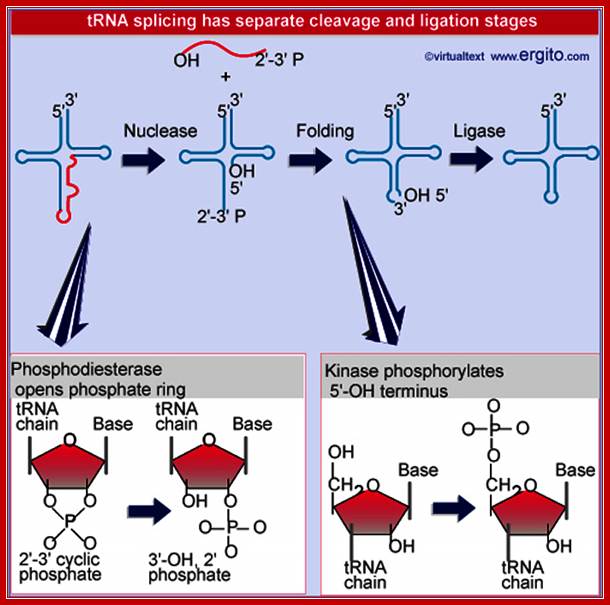
Nuclear splicing and RNA processing; Splicing of tRNA requires separate nuclease and ligase activities. The exon-intron boundaries are cleaved by the nuclease to generate 2¢F-3¢F cyclic phosphate and 5¢F OH termini. The cyclic phosphate is opened to generate 3¢F-OH and 2¢F phosphate groups. The 5¢F-OH is phosphorylated. After releasing the intron, the tRNA half molecules fold into a tRNA-like structure that now has a 3¢F-OH, 5¢F-P break. This is sealed by a ligase;; ttp://genes.atspace.org/
Processing of Introns in Eukaryotic tRNAs:
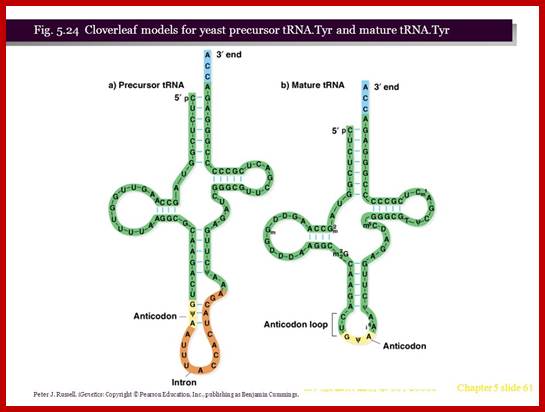
- Yeast and most eukaryotes contain an extra, but non-coding segment of nucleotides within tRNA; presence of which hinders the formation of 3-D structure of the tRNA and hinders its function as well. Such a segment is called tRNA-type introns.
- In most of the split or interrupted tRNAs, introns are found next to 3’ end of the anti codon structure.
- The size of the Intron varies from 14 to 60 ntds. In yeasts Intron containing tRNAs are found as subsets, ex. ser, leu, ile, trp, pro and lysine. tRNA-tyr contain 14 ntds long Intron, tRNA-phe has 17 ntd long, and tRNA-ile has 60 ntds long introns.
- In all 40 of the 400 tRNA genes in yeast cells contain introns.
- Enzymes responsible for splicing have been isolated and purified. They are found to be associated with nuclear membranes.
- One of the splicing enzymes is 90kd in size and has 5’-phosphokinase and ligase activities. Another splicing enzyme has endonuclease activity.
- Enzymes have universal features, for enzymes from yeast cells can act and process tRNAs of other systems including mammalian systems.
Yeasts, plants, mammals and other systems don’t possess any specific consensus sequence at splice junction sites. Instead they have well defined secondary and tertiary structural domains, which are very essential for splicing enzymes to recognize and act.
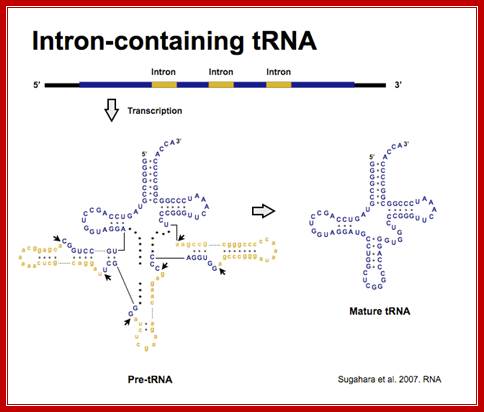
Sugahara J. et al. 2008. Mol Biol Evol.;http://splits.iab.keio.ac.jp/
- Interestingly, in all Intron containing tRNAs, the anti codon sequence is base paired with Intron sequence to form a distinct stem, and next to it is an Intron loop or a bubble.
- However mutation in these sequences of introns doesn’t affect splicing.
- The recognition structural feature is the secondary structure of D-arm, TUC arm and anticodon arm plus the secondary structural motif of the Intron.
- Splicing takes place in two different modes, one with ATP dependent manner and the other without ATP.
I type:
An endonuclease, using the secondary structural motif, binds and cuts at ends, producing, 2’-3’ cyclic phosphate at the 3’end. And Oh group at the 5’ end of the other segment. Then the phospho diesterase cleaves cyclic phosphate to produce 2’phosphate and 3’ OH group. A Kinase enzyme phosphorylates the 5’ OH group. The ligase, activated by ATP forms a covalent bond between 5’-P and 3’OH (a phosphodiester bond), thus seals the cut ends. A phosphotases removes the 2’ phosphate group.
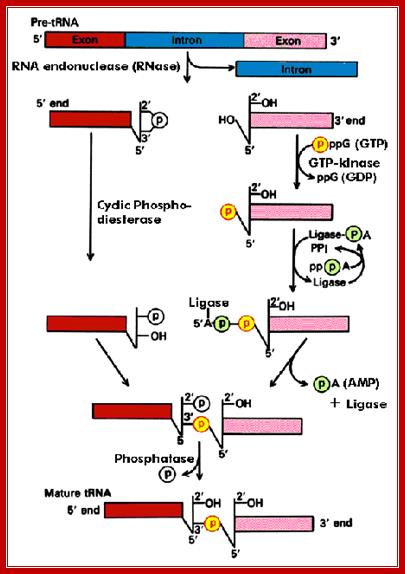
The diagram illustrates the steps involved in processing of tRNA introns; this diagram shows one mode of splicing. Molecular Biology BIMM100http://classes.biology.ucsd.edu/
II-Type:
Enzyme complexes identify the site and bind to it and cleave at both ends of the Intron, generating 3’ OH at the 3’ end of the anticodon and 5’ phosphate group at the at the other end. Such ends are recognized by Ligases and seal them in a typical way. Here it appears that the enzyme performs both cleavage and ligation similar to Topoisomerase-I or certain gp involved in the synthesis of (+) strand during replication of M13 and OX174 phage DNA.
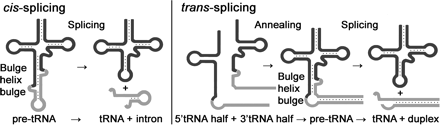
Trans splicing of tRNA derived from different segments:
The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge–helix–bulge motifs of joined tRNA halves.
Schematic representation of cis- and trans-splicing of N. equitans tRNAs. During conventional tRNA cis-splicing, the archaeal splicing machinery recognizes and cleaves a BHB motif in the pre-tRNA, leading to the excision of the intron. The initial step of the proposed trans-splicing mechanism requires the annealing of the primary transcripts of a 5′ tRNA-half gene and a 3′ tRNA-half gene. The intervening reverse complementary sequences (gray) form a duplex that facilitates the joining and folding of the tRNA body (black). Noncanonical BHB motifs are accommodated at the junctions suitable for recognition by the splicing endonuclease. http://www.pnas.org/
Split tRNAs and trans-splicing- Overview of SPLITSdb:
These are rare type of tRNA; their function is same, but they are derived from two different tRNA genes by recombination events.
The purpose of this database is to store and retrieve complete set of tRNA sequences in all archaeal species and primitive eukaryotic species to promote the next study of tRNA evolution and tRNA processing. This database contains total 3,741 sequences and secondary structures of various types of tRNA genes (details are shown on the right table), and 783 sequences and secondary structures of tRNA introns. If you use this database in your work you might want to cite: Sugahara J. et al. 2008. Mol Biol Evol.
|
Types of tRNA genes |
Num |
|
Intron-containing tRNA |
671 |
|
Split tRNA |
12 |
|
Initiator Met-tRNA |
93 |
|
AUA decoding Ile-tRNA |
81 |
|
Pyrrolisine-tRNA |
4 |
|
Selenocysteine-tRNA |
8 |
|
Other tRNA |
2872 |
http://splits.iab.keio.ac.jp/
In every living organism, tRNAs are known as a essential molecules to decode initiator codon and 61 sense codons during the translation mechanism and are encoded in the genome sequence. Although, some of these essential tRNA genes are still missing in archaeal species, this is inspite of the use of tRNA predicting softwares. The difficulty of predicting complete set of tRNA genes in a single species has been reaffirmed by the discovery of several types of unique tRNA genes in archaea and in primitive eukaryotes. For example, Archaeal parasite Nanoarchaeum equitans possess 'Split tRNAs', which the 5' and 3' halves are encoded on the separate genes and many Crenarchaeota possesses 'Intron-containing tRNAs'. Here, we have predicted and collected all the above types of tRNAs and further covered selenocysteine tRNAs and pyrrolysine tRNAs which are known to share major stop codons. Further, initiator Methionyl-tRNA, elongator Methionyl-tRNA and isoleucyl-tRNA were distinguished in SPLITSdb for easy interpretation of tRNAs with same CAU anticodon.
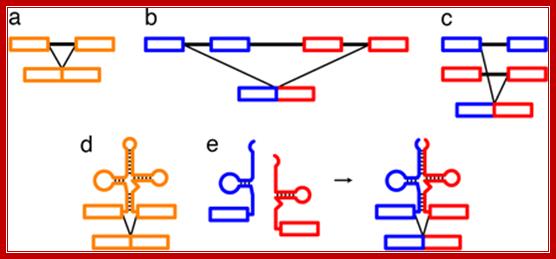
Long Distance splicing; http://www.pnas.org/
RNA splicing pathways observed in eukaryotes (a–c) and engineered in yeast by Di Segni et al. (5) (d and e). (a) Canonical pre-mRNA cis-splicing yielding mRNA from pre-mRNA composed of two exons (orange boxes) and an intron (black line). (b) A transcription-induced chimera generated by read-through transcription from one gene (blue) into a downstream gene (red), followed by cis-splicing. (c) Trans-splicing of exons from two distinct pre-mRNAs (red and blue boxes). (d) Cis-splicing of a pre-mRNA embedded with a permuted tRNA. (e) Trans-splicing of two hybrid RNAs each composed of an mRNA fragment and a half tRNA. Note that in the tRNAs the aminoacyl acceptor stem is at the top and the anticodon loop is at the bottom (d and e).