Transfer RNA (tRNA):
Salient features:
Cytoplasm contains the third kind of RNA, which is smaller, structurally compact and performs transfer of amino acids from cytoplasmic pool on to mRNA-ribosomal complex, so it is called tRNA. This RNA is as important as mRNA; mRNA has coded information and tRNA has decoding information; CODON<->ANTICODON.
- Its function is extraordinarily precise and perfect, and it is involved in what is called in molecular jargon, ‘second genetic code or directory’ and another term used is ‘second half of the genetic code’, which actually reads and deciphers codons with absolute accuracy of amino acids to proteins. As mRNA carries genetic code, the tRNAs carry the second code called anticodons and perform decoding of the information; without tRNA’s anticodon, mRNA codons ‘USE-LESS”.
The number of tRNA per bacterial cell E. coli K38 can be about 64,000 to 375,000 which is equivalent to four times the number of ribosomes. The number of genes in E.coli K12 is between 86; in other species it ranges from 86 and 99 (contain 45-52 different species) The number in eukaryotic genes that code for tRNAs is 497 but the number in the can be five millions or more. Plastids and mitochondria have their own tRNA genes and tRNAs, most of them are coded for by their respective organelle genomes. Mitochondria has 22 tRNA genes and plastids contain 30 genes.
- As they carry amino acids, specificity determines the function of tRNAs.
At least 20 different kinds of tRNAs for 20 different amino acids are expected to be present in every cell, that is what one finds is true. The 21st a.a is selenocysteine tRNA; it uses UGA codon for decoding. As codons exhibit third base degeneracy so also tRNAs exhibit first base degeneracy.
- The quantity of each kind of tRNAs is determined by the kind of proteins produced by the cell and the requirement of the cell type. In addition, each of tRNAs do contain iso-accepting species, which suggests multiple numbers of isoforms for each kind of tRNAs, which is determined by the kind of amino acid it carries.
tRNAs in Bacterial Cells:
- In bacterial systems rRNA operons contain tRNA genes intercalated in spacer regions of rRNA genes.
- But in sup B & E clusters, the rRNA operon contains several tRNA genes of different types are strung together with no rRNA segments.
- The Tyr-T operon contains tRNA genes as well as a gene for protein-P.
- Each of these is transcribed from a single promoter and the transcript is precursor RNA, often polycistronic, which is processed later.
- As described earlier, tRNA segments in the precursors are cut at 5’ end by RNase-p, while the 3’ is processed by RNase D.
- The released tRNA segments are further folded in 3-D structures and they are chemically modified at specific bases as well.
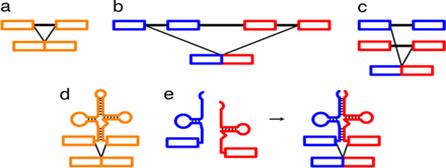
Folded Secondary structure of tRNA; http://www.bio.miami.edu/
Types of tRNA genes:
|
||||||||||||||||||||||
|
Types of tRNA genes |
Number |
|
Intron containing tRNA |
+/_ 671 |
|
Split tRNAs |
12 |
|
Initiator Met tRNAs |
93 |
|
AUA coding Ile-tRNAs |
81 |
|
Pyrolysine-tRNA |
4 |
|
Selenocsyteine tRNA |
8 |
|
Other tRNAs |
2872 |
This database contains total 3,741 sequences and secondary structures of various types of tRNA genes (details are shown on the right table), and 783 sequences and secondary structures of tRNA introns. If you use this database in your work you might want to cite: ;The above data is collected from Archaeal and primitive eukaryotes; required one to one tRNAs for decoding 63 codon but manage with 21 to 24 tRNAs by using wobble mechanism to properly base pair between codon and anticodon nucleotides. The minor AUA-decoding isoleucine tRNA in H. marismortui and other archaeal species is most likely derived from a CAU anticodon-containing tRNA; it is the rarest of the rare anticodon. It is similar to lysine, but with an added pyrroline ring linked to the end of the lysine side chain. It forms part of an unusual genetic code in these organisms, and is considered the 22nd proteinogenic amino acid. http://splits.iab.keio.ac.jp/
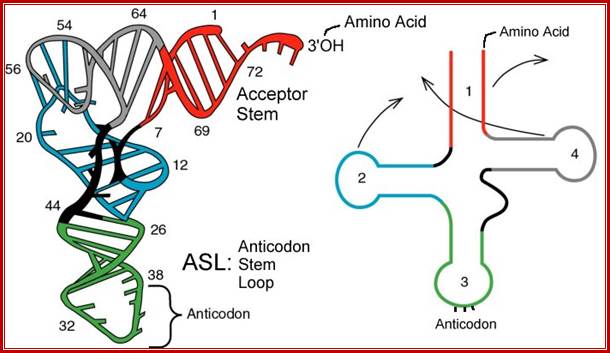
E.coli genome is in condensed state with many loops, specific loops containing ceretain genes are engaged in transcriptional activity, transcriptional loci or called transcriptional factories are shown in green. Model linking stable RNA synthesis, RNAP distribution and the dynamic structure of the nucleoid: The E. coli chromosome is represented as blue lines folded in loops, the ori of replication as a black square, the seven rRNA operons as large red circles with letters, and two representative tRNA operons as small red circles. The RNAP molecules are represented as small green circles. For simplicity, only two putative transcription factories/foci, which make the nucleoid more compact by pulling different stable RNA operons into proximity, are indicated here (bottom part of the diagram, large green circles labeled 1 and 2) [adapted from the study by (Cabrera and Jin, 2003)].

E.coli genetic map with positions of rRNA oeprons; http://www.nibb.ac.jp/
tRNA sup B-E Operon in prokaryotes:
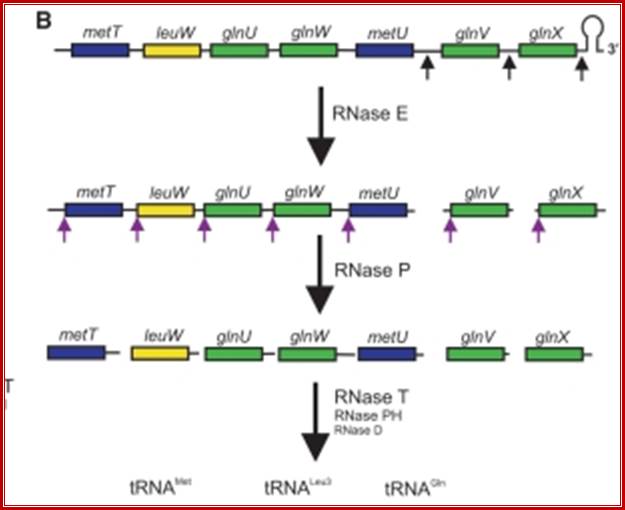
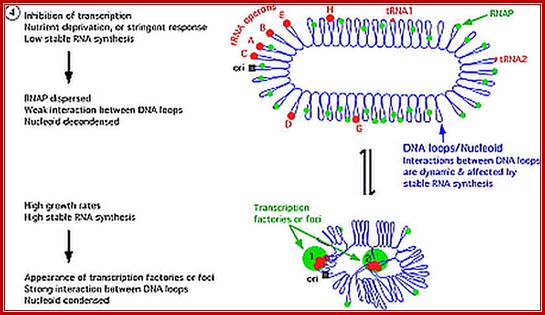
Seven tRNA encoding polycistronic RNA is stepwisely processed, by using Rnase E, Rnase P and Rnase T/pH/D; Noboru Nakajima, Haruo Ozeki etal; http://www.kiveand.com/
Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E.
The first step in the current model for the processing and maturation of mono- and polycistronic tRNA precursors in Escherichia coli involves initial cleavages by RNase E 1–3 nt downstream of each chromosomally encoded CCA determinant. Subsequently, each mature 5′ terminus is generated by single RNase P cleavage, while the 3′ terminus undergoes exonucleolytic processing by a combination of 3′ → 5′ exonucleases. Here we describe for the first time a previously unidentified pathway for the maturation of tRNAs in polycistronic operons (valV valW and leuQ leuP leuV) where the processing of the primary transcripts is independent of RNase E. Rather, RNase P cleavages separate the individual tRNA precursors with the concomitant formation of their mature 5′ termini. Furthermore, both polynucleotide phosphorylase (PNPase) and RNase II are required for the removal of the 3′ Rho-dependent terminator sequences. The 3’ end is cleaved first by endonucleases, and then trimmed by exonuclease like RNaseD. Our data indicate that RNase P substrate recognition is more complex than previously envisioned. Bijoy K. Mohanty and Sidney R. Kushner
5’pppGC—met—-leu—gln—gln—met---gln---glu---AUG-T/t
In E.coli Ribosomal RNA genes are organized into seven operons, called rrns and ribosomal protein genes into six operons. Some of the tRNAs are organized into polycistronic operons as shown below, eg B.subtilis. First interspace region is cut by RNaseE at 3’ region of each tRNA gene; they are cut by RNaseP at the 5’end of each tRNA’s segments. All stable tRNAs have to be processed at 3’ end before they amino acylated. Majority of tRNAs are processed at 3’ end by a set of exonucleases and exonucleases. Some of the defective tRNAs get poly Adenylated for degradation.

B subtilis, rrnB/trnB operon, P and T promoter and terminator; at the 3’ end of rRNA segments one finds 21 tRNAs clusters, and transcribed by the RNAP using its bacterial promoter.
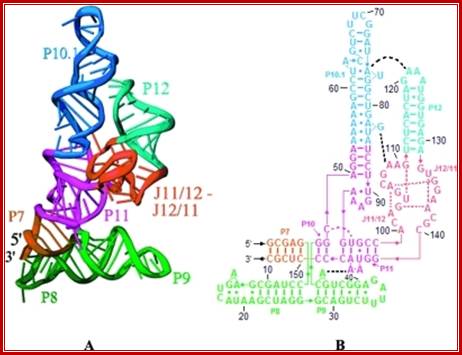
The 3-D model shows the RNA structural form of RNase-P, b.subtilis; Tertiary structure (A) and secondary structure (B) Liang R, Kierzek E, Kierzek R, Turner DH - Biochemistry (2010);http://openi.nlm.nih.gov/
Processing of tRNAs:
RNase E initiates processing by endonuclease downstream of the CCA determinant of each tRNA within the operon leading to the formation of immature tRNAs, which are further processed by RNase P to generate mature 5′ termini. The mature 3′ ends result primarily from the activity of RNase T, but RNase PH, RNase D, RNase BN and RNase II can substitute in the absence of RNase T. Many tRNAs that are processed lack 3’ terminal CCA, in such cases, nucleotidyl transferase adds CCA3’ nucleotides.
The tRNA genes are in the arrangement- 5'-leader- tRNAArgCCG -57 base pairs- tRNAHisGUG -20 base pairs- tRNALeuCAG -42 base pairs- tRNAProUGG -3'. Coordinate expression of the component tRNAs in vivo and the absence of intercistronic promoters indicated that all four tDNAs reside in the same operon.
These tRNA sequences are preceded by a single promoter region where a “Pribnow box” sequence is present seven base pairs upstream from the transcription start site. The spacer regions separating the seven tRNA sequences are different from each other both in size and in nucleotide sequence (Organization and structure of an E. coli tRNA operon containing seven tRNA genes; Noboru Nakajima, Haruo Ozeki and Yoshiro Shimura)
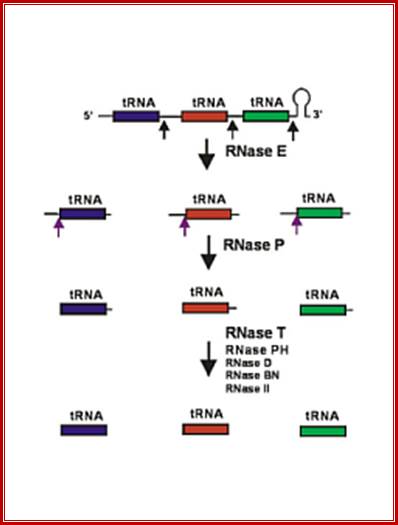
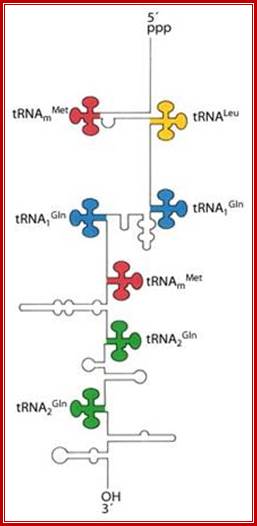
Structure of an Escherichia coli tRNA operon containing linked genes for arginine, histidine, leucine, and proline tRNAs; General model for tRNA processing in E. coli. In the first step the endoribonuclease RNase E cleaves in the intercistronic regions of polycistronic tRNA precursors to generate pre-tRNAs that have a small number of extra nucleotides at both their 5' and 3' ends. The mature 5' termini are generated by cleavage with the ribozyme, RNase P, while the mature 3' end arises from the action of a series of 3' � 5' exonucleases. The most important of these enzymes are RNase T and RNase PH. Model is based on data from Li and Deutscher (Li and Deutscher, 2002) and Ow and Kushner (Ow and Kushner, 2002). http://www.genetics.uga.edu/;
RNase P:
(1) RNAse P is composed of a 375 nt RNA and a 20 kDa protein.
(2)The catalytic activity is in the RNA. The protein is thought to aid in the reaction, but is not required for catalysis. “All enzymes are not proteins”.
(3) This was discovered Sidney Altman and awarded Nobel prize for this work.

Clustered tRNA genes, their polycistronic transcripts undergo processing; i.e. cutting and trimming, 5’ end is generated by RNaseP, and 3’ ends by a number of 3” exonucleases.
General mode of tRNA processing in E.coli;
RNase E (endonuclease first cleaves in the intercistronic regions which generates pre-tRNAs containing few nts at both ends. RNase P generates mature 5’ end and the 3’ end is generated by a series of exonucleases- RNaseT and RNasePH.
Ow and Kushner 2002;http://www.genetics.uga.edu/

A cluster of tRNA genes- When transcribed the polycistronic tRNA assumes structural features as shown in the diagram
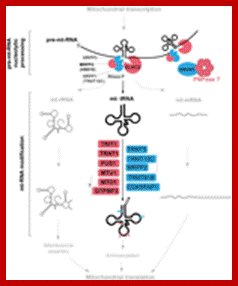
Mitochondrial tRNA maturation and factors involved. Key protein factors involved in post-transcriptional nucleolytic processing and in chemical modifications of mitochondrial tRNAs. Factors associated with human disease are indicated in red; other factors characterized thus far are in blue. Note: PNPase can be either directly or indirectly involved in mt-tRNA processing (red question mark). The bacterial homologs of MTO1 and GTPBP3 form a complex, however, the interaction between the human proteins has not been shown (question mark). http://journal.frontiersin.org
In some specialized cells, few species of tRNAs are found in abundance than the others. This is determined by the protein requirement.
- Primarily each tRNA is a short polynucleotide chain of 72 to 97 ntds. Most of them are synthesized as long precursor RNAs, which are then processed and bases are specifically modified. In prokaryotes tRNAs synthesized as part of rRNA transcript and in some it is synthesized as polycistronic transcript.
The 3-D folding is unique to each tRNA. Many of the cross linking are non-Watson–Crick base pairing, and in some 2’OH groups of ribose are also involved in cross-linking and some modified.
- Sequencing of all 20 or more different types of tRNAs, from more than several thousands of species, has been done. Even some of the tRNA genes have been synthesized in the Lab e.g. Hargobind Khorana has synthesized Ala tRNA in the lab.

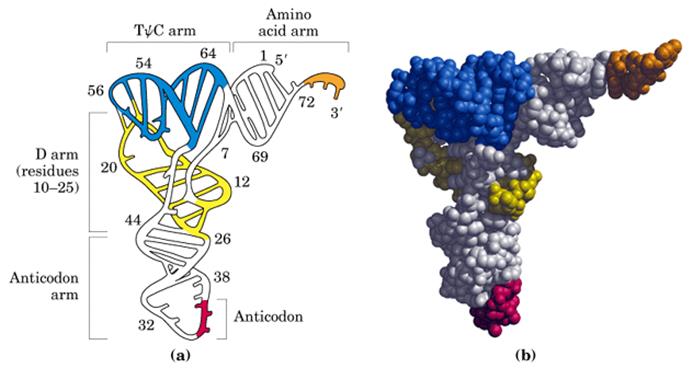
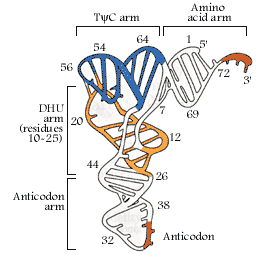
The diagram is Holley’s Clover Leaf model, just a secondary structure. Most of the amino-acyl tRNAs will have G at the 5’ end and it is always base paired with C and it is phosphorylated, but in initiator tRNAs the 5’ nucleotide is not G but A, thus the 5’ end remains unpaired’ http://biology.kenyon.edu/

Secondary structural model

tRNA 2-D model; http://mamit-trna.u-strasbg.fr/
The above diagram represents a 3-D model derived from the secondary structural model that is shown before. The first tRNA model was proposed by Holley, which was called as ‘cloverleaf’ model for its secondary structural appearance resembled trifoliate leaves of Trifolium repens or what is called clover leaf.
- Whether it is a secondary structure or tertiary structure, when viewed one can discern at least four distinct domains or structural motifs, and they are invariably found in all species of tRNAs.
The domains are, from 3’ end (starts with 3’ACC) of the polynucleotide chain: 1) amino acid acceptor end, 2) amino acid acceptor stem, 3) TψCG stem, 4) TUCG loop, 5) an extra arm found in between TψCG stem, 6) anticodon stem, 7) anticodon loop, 8) DHU stem, 9) DHU loop ending in 5’G (in E.coli), which is paired with C in the amino acid acceptor stem.
- The amino acid acceptor end found at 3’, has a consensus sequence 3’ACC--, that leads to a 7 base pair stem with G: U, U: A pairings. It is to the 3’Adenine’s ribose-OH group amino acids are covalently added with carbonyl bonds. In many tRNAs 2’O is added with amino acyl group. Type I tRNAs have ACC ends as they are synthesized and Type II tRNAs don’t have when they are synthesized, but added later. The amino acid acceptor sequences may be involved in binding to a.a. tRNA synthase.
The TψCG loop is 7 bp long and consists of 3-5 bp stem. The TψCG is an invariant sequence. This region might be involved in binding of tRNA to ribosomal surface either at A or at P sites.
3-D model of tRNA; http://www.nobelprize.org
- The anticodon loop again contains seven bases with 4-5 bp long stem. Anticodons that are geometrically and stearically projected like three fingers, whose sequence differ from one species of tRNA to the other. However, more than one combination of triplet sequences as anticodons can code for the same amino acid. But very often in tRNAs, the three nucleotides, as anticodons are bracketed on either side by a modified purine (like Isopentinyl Adenine) towards 3’ side and a uracil (or pseudouridine) on 5’ side. The anticodon loop with its specific sequences may have a role in binding to a.a. tRNA synthase. Anticodons are responsible for decoding the information present in mRNAs. This is the only RNA called noncoding RNA but contain codon-decoding sequences, yet can we call it as non-coding RNA?
In the upstream of anticodon stem and DHU stem sequence in DHU loop Di Hydroxy Uridine (DHU) is invariant, and the loop consists of 8-12 variable nucleotides. The DHU nucleotide is invariant. The variable sequence is specific-to-specific tRNAs. The DHU stem is 3 to 4 base pair long. The DHU loop and its inner angle may provide specificity in tRNA and enzyme recognition?
- Another additional structural feature that is found in tRNAs is an extra arm of variable length, found in between TψC stem and anticodon stem. The length varies from 2 to 21 ntds, based on the length they are classified into class I and class II tRNAs. Class I contain 3-5 ntds long arm and class-II have 13 to 21 ntds long arms.
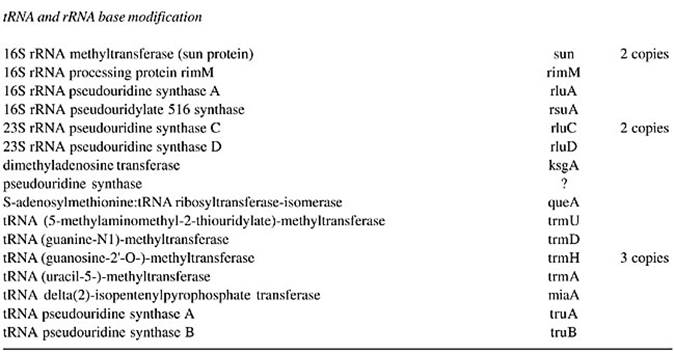
Bases in all tRNAs, in specific positions are modified which happens after synthesis. Each species of tRNA is modified specifically to make it distinctly different.

tRNA base modifications; https://www.rpi.edu/dept/bcbp/molbiochem
- Specific enzymes perform modifications. The Uridine can be modified into pseudouridine, dihydroxy Uridine, ribothymidine and thio Uridine; cytosine can be added with methyl group at 3’ or 5’ sites. Hydroxyl group can be added at 5’ position of cytosine; adenine in most of the case is modified into Isopentinyl-Adenine at specific positions, or Inosine; even guanines are modified at N’7 as Methyl guanine. More than 90 different kinds of modifications at different positions have been found among various members of tRNAs, yet tRNAs are structurally and functionally stable and specific. What function the modified bases perform is not fully elucidated.
Their half-life can be anywhere between 100 to 150 days or more; tRNAs are the most stable form among all species of RNAs.
- Structurally tRNAs have 3-D conformations, which make them very compact and almost crystalline. The 3-D folding is due to cross-linking between G:G, G:U and A:G pairing which are non-Watson-Crick base pairing. Unusual hydrogen bonding in cross-linking may involve 2’OH group of ribose or even phosphate groups.
The vertical length of tRNAs is ~60Å and horizontal tail is also of the same length. The diagonal distance between the 3’ tail and the anticodon loop is ~76Å. At the inverted outer L corner, D-loop and T-loops are located. The D-stem and anticodon stem are at the inner angle of the inverted L region. The acceptor stem is in line with T-loop and T-stem.
- Among various species of tRNAs, there are few special species called Initiator tRNAs and the rest of them are referred to as elongation aminoacyl tRNAs (a.a. tRNA).
Structural Features of tRNAs (General):
|
Motif from 3’ end |
Features |
|
1 st -Amino acid acceptor terminal |
It has 3’ACC sequence in all tRNAs, in some they are present when they are synthesized, in some they are added later. |
|
2 nd- Amino acid accepter stem |
It is 7 bp long, contain 5G: C and 2A: U base pairings. |
|
3 rd -TψC G stem |
3 to 5 bp long |
|
4 th –TψCG loop |
7nucleotides,TψC invariable sequences |
|
5 th- Extra arm |
Located between TψC and anticodon stem, the arm length can range from 3 to 21 ntds. |
|
6 th- Anti codon loop |
7 ntds long, 3 ntds act as anticodons, anticodon nucleotides are often bracketed by modified ψ at 5’ side and modified A (like IPA) on 3’ side, and triplets are projected as rigid structures. |
|
7 th -Anticodon stem |
Consists 4-5 bp |
|
8 th -DHψ loop |
Consists of 8-12 variable bases, Di-Hydroxy Uridine is the only invariable nucleotide |
|
9 th -DHψ stem |
Is 3-4 bp long |
|
10 th – 5’ G |
Extension of the DHψ stem ends in 5’ G inmost of amino acyl tRNAs and pairs with C. |
Initiator tRNAs:
In bacteria and eukaryotic cells, there are specific tRNA which initiate translation and they lay first amino acid on translational machinery, so they are called initiator tRNA and no other tRNA can perform this critical step or function i.e. initiating protein synthesis. Rarely mutant initiator tRNA initiate translation from any codons (surprising).
- Though initiator tRNAs are typically similar to other a.a. tRNAs, they are different in terms of length, sequence and also in structure in subtle ways.
The initiator tRNA in bacteria binds to formylated CH3-CO~ methionine at its 3’ end, whereas eukaryotic initiator tRNA takes on an unmodified methionine. Such structural features are recognized by their specific amino-acyl tRNA synthetases.
- During translational initiation, activated initiation factors recognize initiator tRNAs only and not any other tRNA though some of them carry methionine (they are called amino acyl tRNAs), that means the initiator-tRNA has unique structure that can only be recognized by the translation initiation factors.
Prokaryotes and eukaryotes have different initiating factors.
- The f-met tRNA in bacterial systems consists of 76 nucleotides. The amino acid acceptor 3’ACC- stem has 6 G: C pairs.
The TψC stem has four G: C pairs and one A:U ; It has a seven nucleotide long loop, no TψC sequence. It has a short arm of 5 nucleotides.
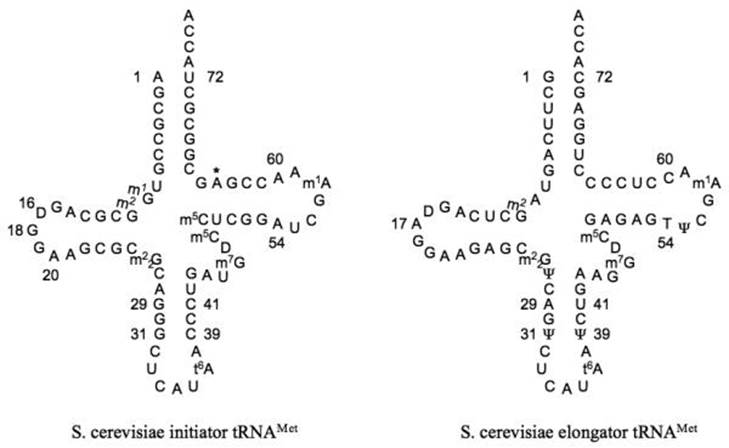
Cloverleaf diagram of S. cerevisiae initiator (left) and elongator (right) methionyl tRNAs. Identity elements in the sequence are indicated by numbers. The position 64 Oribosylphosphate modification is indicated by an asterisk. There is some confusion in the literature and databases about the identity of base 64 in the elongator tRNA. In most cases, including all elongator tRNA genes in the S. cerevisiae genome, this base is a C. However, in a few cases it is reported as a U. It is possible that this is the result of deamination of C in a fraction of the tRNAs; in this case, sequencing of the corresponding cDNA would give a U at this position; ; http://rnajournal.cshlp.org/

Chemistry of life; http://chemistryolife.blogspot.in/
- There are seven nucleotides long anticodon loop with 4 G: C pairs and one A: U base pairs in the stem.
The DHψ stem has 4 G: C and 7-9 ntd long loop. At the 5’ end it has C or A instead of G as in the case of bacterial cells, and it cannot pair with its opposite nucleotide for it is Adenine, so it remains unpaired.
- In eukaryotes, especially in yeasts, the initiator tRNAs have an unusual structural modification at 64 th position where the 2’OH of ribose residue is phosphorylated.
Initiator tRNA features-prokaryote and Eukaryote:
|
Motif |
Features-PK (E.coli) |
Features-EK (hu) |
|
3’ end |
CCA3’ |
CCA3’ |
|
1st- amino acyl acceptor |
CCA3’ to which methionine covalently is added, then it is modified |
CCA3’; methionine is covalently added |
|
2nd-aa acceptor stem- |
6G:C , 1G:Apairs |
6G:C, 1A:U |
|
3rd TψCG stem |
4G:C and 1A:U |
3G:C,2A:U |
|
4th-TψCG loop |
7ntds, TψC, |
7ntds,o TψC |
|
5th extra arm |
5ntds or more |
5tds or more |
|
6th Anticodon stem |
4G:C and 1A:U |
4G:C, 1A:U |
|
7th Anticodon loop |
7 ntds, 3anticodon bracketed by U and IPA |
7ntds, 3’IPA, |
|
8th DHU stem |
4G:C |
3G:C,1G:U, DHU |
|
8th DHU loop |
7-11 ntds, DHU- consensus, |
No DHU |
|
5’ end |
C, cannot pair with A |
5’A pairs with U |
|
In yeast initiator tRNA, 64th ntd is phosphorylated |
|
|
Amino Acyl-tRNAs:
Each of the twenty or more different species of tRNAs carry 20 different amino acids, one species to one amino acid basis.
- Recently Seleno-cysteine, another amino acid has been added to the list 20 amino acids; so also 21st tRNA.
When a tRNA is loaded with an amino acid, it is called aminoacyl tRNA.
- There are twenty distinct tRNAs for twenty different amino acid residues.
There can be more than one form of tRNAs called isoforms, for the same amino acid, as in the case of degenerate codons which code for the same amino acid but with different codon sequences mostly in the third base.
- Each species of tRNA has distinctly different anticodons and each recognize specific codons.
Each kind of tRNAs has their own specific nucleotide sequence with specific base modifications, which determines its structural and functional characteristics.
- Each tRNAs, in spite of having structural and functional distinct characters, amino acid is added specifically to the same 3’ A end of ACC sequence. And all different tRNAs have the same 3’ACC sequence to which different, but specific amino acids are covalently added.
In spite of having same 3’ end sequence of ACC-, it is the subtle, but individualistic structural features that distinguish one tRNA from the other.
Loading of specific amino acids on to the CCA at the 3’end of each of the tRNA is somewhat puzzling, for the 3’ends of all tRNAs have the same sequence. Then how different tRNAs are loaded with specific amino acids. This is achieved by specific amino acyl- tRNA synthetases.
As there are twenty one different tRNAs with isoforms for twenty different amino acid residues, there are twenty one different amino acyl-tRNA synthetases which are species specific.
|
Residue |
PDB |
Full Name of Nucleoside |
Standard |
|
10 |
2MG |
N2-Methylguanosine |
M2G |
|
16 & 17 |
H2U |
Dihydrouridine |
D or hU |
|
26 |
M2G |
N2, N2-Dimethylguanosine |
M22G |
|
32 |
OMC |
2'-0-Methylcytidine |
M2'C |
|
34 |
OMG |
2'-0-Methylguanosine |
M2'G |
|
37 |
YG |
Wybutosine* |
Y-base |
|
39 & 55 |
PSU |
Pseudouridine |
"Psi" |
|
40 & 49 |
5MC |
5-Methylcytidine |
M5C |
|
46 |
7MG |
7-Methyguanosine |
M7G |
|
54 |
5MU |
5-Methyluridine |
T |
|
58 |
1MA |
1-Methyadenosine |
M1A |
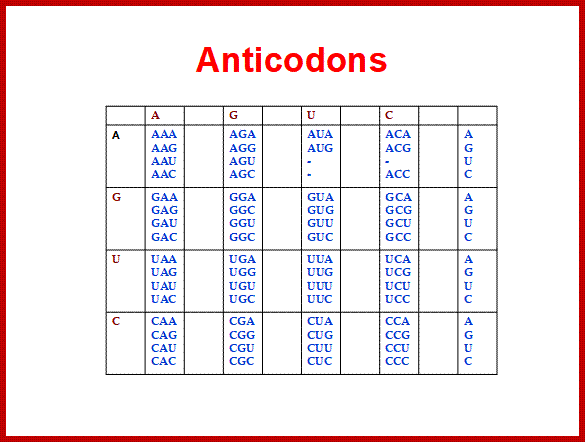
Codon and Anticodon directory:

http://www.cpalms.org
mallonsnyderlaw.com
tRNA, the Adaptor Hypothesis and the Wobble Hypothesis
This structure was first recognized by Robert Holley (right - image from Nobel web site) in 1965. He determined the nucleotide sequence of the yeast tRNAAla which is 76 nts in length. He shared the 1968 Nobel Prize in Physiology /Medicine with Marshall Nirenberg and Hargobind Khorana for tRNA synthesis and processing. Hargobind Khorana not only synthesized ala.tRNA but also generated codon directory using synthetic codon sequences for different anticodons.
Many tRNA genes (in both eukaryotes and prokaryotes) are synthesized as larger precursor molecules. The mature final forms of these RNA molecules are obtained by processing reactions that involve both the removal of nucleotides and in some instances, by the addition of nucleotides and modifications of bases and ribose sugars. The important feature is modification of bases at specific positions.
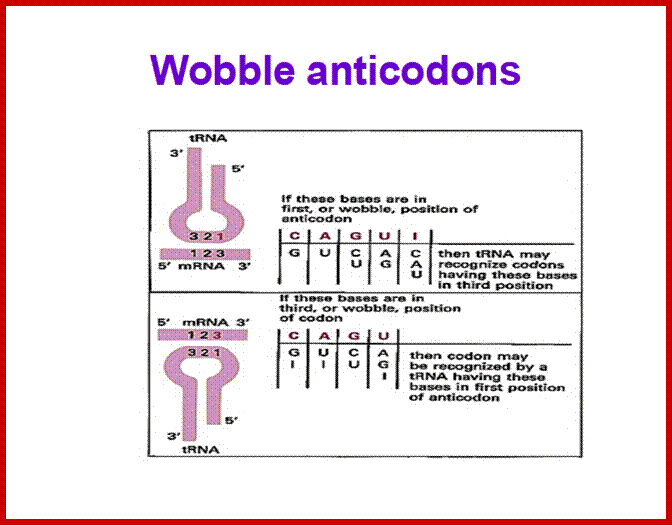
If tRNAs use the said anticodon sequences against 61 codons, leaving apart terminator codons, there should be at least 61 different tRNAs. Actually it is not the case. tRNAs use what is called wobble mechanism; a single tRNA can read more than one codon including degenerate codons. For example 1st base of the anticodon from 5’ end can wobble with more than one base in the 3rd base of the codon in mRNA. The diagram below shows which base wobbles with which base. The 3rd base degeneracy of codons in mRNA and the 1st base of the anticodon wobble base pairing provide great leeway for the translation process without compromising with meaning of the codon for a specific amino acid. N most the cases the first letters of the codons are important.
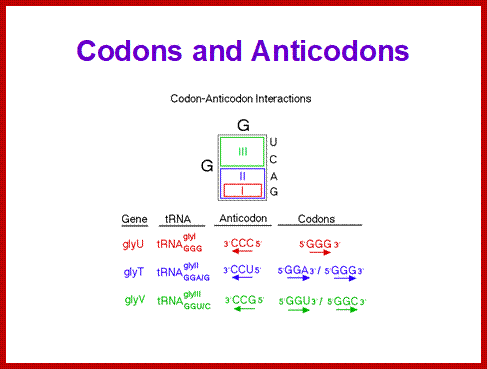
5” anticodon G a base pair with U or C of the codon at 3’end, similarly C of anticodon base pair with G, A with U and U with AG and I with U, C or A. This accommodate of different tRNA can base pair with third 3’ codon nucleotide. Another feature is different or non identical tRNAs carry the same amino acids is called isoaccepting tRNAs www.bio.miami.edu
http://wpinfotech.com/
https://www.suggest-keywords.com
• Anti-Codon 5’ = 3’ Codon : wobble base pairing ; base pairing between 5’ 1st base pairing with mRNA 3rd base,
tRNA 1st letter =3rd letter mRNA

Translation of proteins; http://themedicalbiochemistrypage.org/
C = G, A = U, U = A
U = G, G = C, G = U
I = U, I = C, I = C

www.keyword-suggestions.com
Extended anticodon: S.H Kim proposed the efficiency of the anticodon recognition is determined between anticodon loop and the proximal anticodon stem sequence:
|
Ü |
A |
U |
G |
C |
|
VII |
Ü |
Ü |
PY/PU |
Ü |
|
VI |
GC/GC |
Ü |
PY/PU |
GC/CG |
|
V |
PU/PY |
Ü |
Ü |
Ü |
|
IV |
GC |
GC/AG |
Ü |
CG |
|
III |
AU/UA |
AU/CG |
Ü |
CG |
|
II |
A |
A |
A/U |
A/C |
|
I |
Me2 16A |
16A |
M2A/G |
Am2A/m6A |

http://faculty.samford.edu/
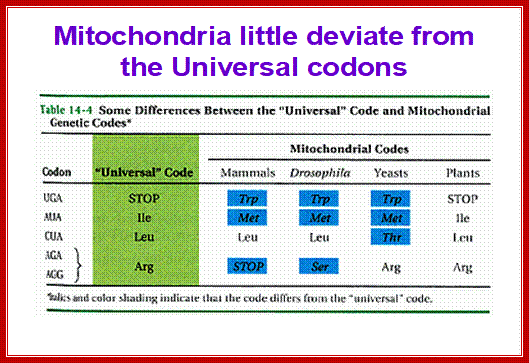

The ancestor mitochondria contain a tRNALeu with a UAG anticodon that pairs with CUN codons. Both the CUN codons and tRNALeuUAG were lost during evolution. A duplicated copy of tRNAHis then evolved to decode CUN. Although this new tRNA is no longer recognized by histidyl-tRNA synthetase (HisRS), it becomes a substrate for the coevolved threonyl-tRNA synthetase (MST1). With the emergence of the orthogonal MST1-tRNAThrUAG pair, CUN codons reappeared in the mitochondrial genome to complete the codon reassignment event25. This naturally evolved system could serve as a model for sense codon recoding.; Patrick O'Donoghue;;http://www.nature.com/
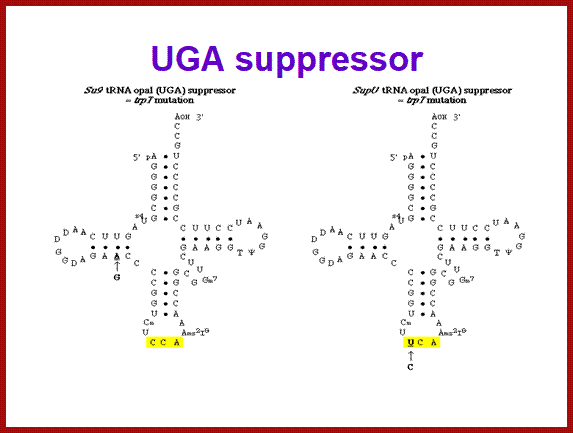
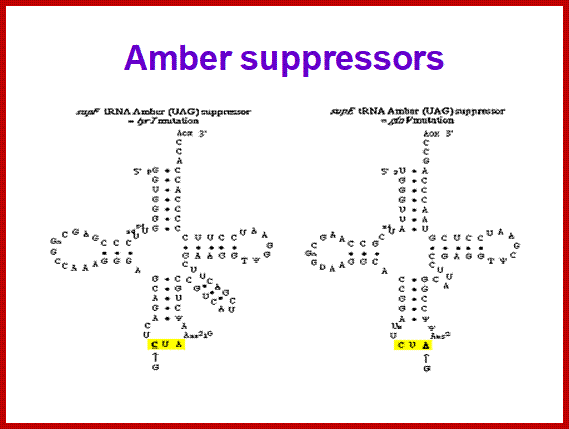
SCUB synonymous codonb usage Bas occurs across different species or with in the genome of the same species, example Bombax mori. Some of the mutated tRNA with their changes in anticodon sequences acts as suppressors of mutation such as UAG amber suppressors, UAA suppressors, UGA suppressors. Even other mutated 3rd base codons are often read without much problem. Even some mutated codons are read correctly by similar mutated anticodon of tRNAs. If the amino acid incorporated happens to be in the middle and compacted region (not active site or substrate binding site) of the mRNA the reading frame does not affect the structure and function of the protein.
http://faculty.samford.edu/
http://www.sci.sdsu.edu/

http://www.sci.sdsu.edu/
The structure of each kind of amino acyl-tRNA synthetases are designed to accept only one specific amino acid and only one specific tRNA. This discerning action of identifying specific amino acid and specific tRNA is often called second coding.
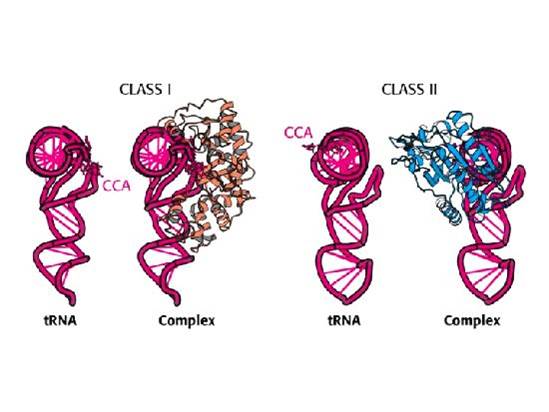
Aminoacyl-tRNA synthetase classes of E. coli / Structures; Class 1 - links to 2' hydroxyl. Most are monomeric; Class 2 - links to 3' hydroxyl (except Phe-tRNA). Most are dimeric; Translation outline;http://oregonstate.edu/
|
The Two Classes of Aminoacyl-tRNA Synthetases |
|
|
Class I |
Class II |
|
Arg |
Ala |
|
Cys |
Asn |
|
Gln |
Asp |
|
Glu |
Gly |
|
Ile |
His |
|
Leu |
Lys |
|
Met |
Phe |
|
Trp |
Pro |
|
Tyr |
Ser |
|
Val |
Thr |
Table showing two classes of a.a tRNA synthetases;

http://biochem4.okstate.edu/
Amino acyl synthase in action:
https://www.slideshare.net

Copyright © Thomson;http://journals.prous.com/
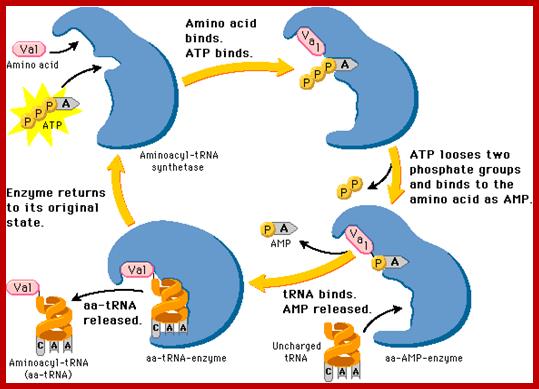
Aminoacyl tRNA synthetase enzyme gets activated with binding of ATP Actually ATP binding induces conformational change so it accepts correct Amino acid. Te binding of a.a leads to the hydrolysis of P~P leaving mono phosphor ATP.
Basically, first the binding of ATP to the enzyme induces conformational change, then amino acid binds (specifically) thus active amino acid AMP complex. The amino acid binds to its specific site in such a way the carboxy terminal of amino acid is bound to tRNA. One of the OH group of the 3’end of the tRNA generates O=C~O bond.. One of the important structural features that determine the specificity between a.a and tRNA is the DHU loop and the anticodon nucleotides. As the tRNA and amino acyl-AMP in place the enzyme brings about covalent bond formation between the 2’ or 3’ OH of the tRNA and carbonyl end of the amino acid. This makes the enzyme to undergo conformational change resulting in the release of charged aa-tRNA.
A very fascinating feature of the structure of some of the translation and chain termination factors do have structural features similar to tRNA. Activated EF-Tu binds to amino acyl-tRNA. During amino acylation the enzyme binds and the amino acid gets adenylated (activated) and binds to its specific binding site, then specific tRNA binds (they can be simultaneous also). EF-Tu and EF-G are structurally mimic each other and chain releasing factors and ribosome recycling factors mimic EF-G.
Transfer of aminoacyl group to tRNA adenine –OH group is very critical; it is at this stage proof reading is done to ensure specific tRNA is loaded with specific amino acid. The important domain responsible for this activity is one acceptor stem with its end and the anti-codon loop. It is the enzyme that ultimately decides specificity.
Processing pre-tRNAs:
Most of the tRNA genes are found in clusters, but each of them have their own promoter elements and they are transcribed by RNA pol-III independent of each other. The precursor is slightly longer than the processed one. Here in eukaryotes also RNase-P is responsible for cleaving and generating the 5’ end. Mitochondria too contain RNase P called MRNP. Similarly most of the eukaryotic cells contain RNase P. Some of the are associated with specific proteins and they have specific functions. RNase F and endonuclease cuts at 3’end, then RNase D trims nucleotide by nucleotide till reaches the 3’ACC. These enzymes, events produce functional tRNAs. Many of the pre tRNAs lack 5’CAA3’ ends, in which case specific enzymes add CCA in sequence to generate the 3’ terminal A with 3’ OH group.

RNA genes found in clusters is subjected to cleavage; http://csls-text3.c.u-tokyo.ac.jp/
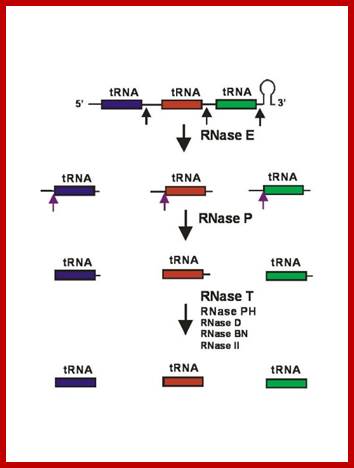
Processing of polycistronic tRNA transcripts; Kushner: http://www.genetics.uga.edu/
tRNAprecursor; http://bioinfosu.okstate.edu/
https://www.slideshare.net
Number of genes in eukaryotes such as yeast, worm, fruit fly, chicken, rat and mouse etc is about 170 to 570, and isoacceptors is about 41 to 55. Sea anemone is expected to contain 17,000 genes; 17-36% are pseudo genes. Interestingly tRNAs having the same anticodon but differ in its internal sequences, called isodecoders, they are about ~274. The tRNA genes are organized in clusters with one promoter with specific promoter elements and transcribed by RNAP III. In between each of the tRNAs a spacer of various length is present. Processing of these tRNA individually takes place by 5’PNase and 3’ RNase D and RNase E. But individual tRNA genes are transcribed by RNAP III with specific TFs
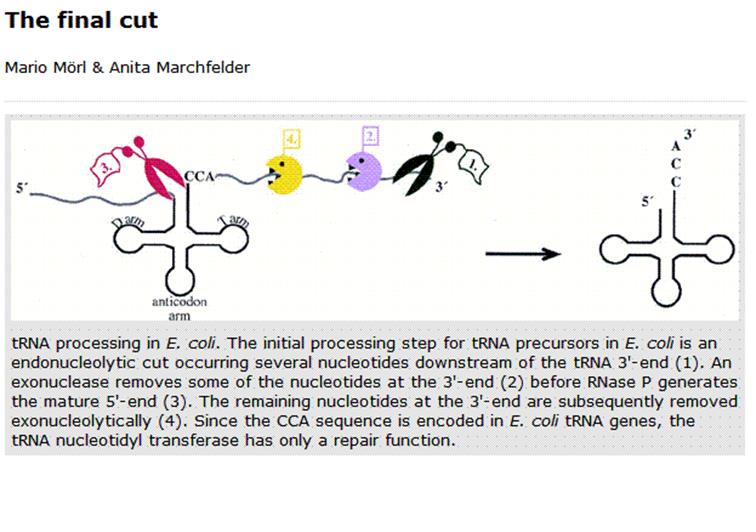 Mario Mori & Anita Marchfelter; ;http://gcat.davidson.edu/
Mario Mori & Anita Marchfelter; ;http://gcat.davidson.edu/
As the tRNA is transcribed it assumes secondary structure which facilitates the enzymes to process to generate 3-D tRNA. While the transcript is produced certain bases at specific positions are modified, similar to that of rRNA methylation and pseudouridinylation.
Modified nucleosides in eukaryotic tRNAs and chemical structures. A: Clover-leaf structure of eukaryotic tRNA. Each circle represents a nucleotide, numbered from 5'- to 3'- end. Modified nucleosides found at different positions are shown. B: Chemical structures of some modified nucleosides.
Transfer RNA (tRNA) is the adapter molecule mainly responsible for decoding mRNA into the corresponding peptide sequence. tRNA molecules are generally 75-87 nucleotides long and form clover-leaf shaped structures through base pairing in the acceptor stem; D-stem, TΨC stem and anticodon stem (Figure 1A). Modified tRNA nucleosides are found universally in living organisms. Some are conserved across all domains of life (e.g. Ψ, D, m1G, m7G, Cm, Um and Gm), indicating an evolutionary ancient enzyme
According to the RNA modification database -
Nearly 107 different modified nucleosides were found in RNA as on 2008. Among these, 92 are present on tRNA molecules. All modified nucleosides are derivatives of the four normal nucleosides: adenosine, guanosine, uracil and cytosine. The modifications vary from a simple methylation on the ribose or base moiety to complicated side chain modifications in different positions of the purine/pyrimidine rings. ring,http://library.med.utah.edu/RNAmods/ webcite, ,
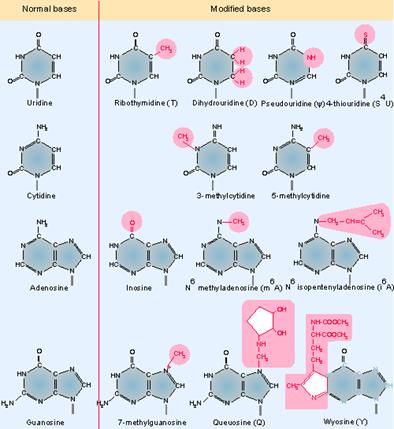
Genes IV;
http://genes.atspace.org/
The above figure shows some of the more common modified bases. Modifications of pyrimidines (C and U) are less complex than those of purines (A and G). In addition to the modifications of the bases themselves, methylation at the 2′-O position of the ribose ring also occurs. Transfer RNA is unique among nucleic acids in its content of "unusual" bases. An unusual base is any purine or pyrimidine ring except the usual A, G, C, and U from which all RNAs are synthesized. All other bases are produced by modification of one of the four bases after it has been incorporated into the polyribonucleotide chain. There are >50 different types of modified bases in tRNA.
For the 21 known modified nucleosides mentioned above, authors used loss-of functional studies and identified five genes responsible for four specific modified nucleosides, m1G (AtTRM10), m2G (AtTRM11), m7G (AtTRM82) and ncm5U (AtKTI12 and AtELP1). Modified nucleosides participate in fine-tuning the activity of tRNAs during translation. For example, defects of certain tRNA modifications result in decreased translation efficiency and increased translational error.

Scheme of RNase P and RNase Z endonucleolytic processing of a tRNA precursor (left) and crystal structure of RNase P in complex with tRNA;http://www.ibvf.csic.es/
Intron Splicing- 1st type:
Some of the pre-tRNA produced, especially in some eukaryotes, have intron just towards the 3’ end of the anticodon region. This intron containing tRNA do not function. The length of the intron varies from 12-23 ntds or more. There are two sets of enzymes, which perform processing.
One type of enzymes cut the intron region which is normally base paired with anticodon stem region at its 3’ end generating 5’P and 3’ OH at the other end. The RNA ligase performs the ligation reaction.
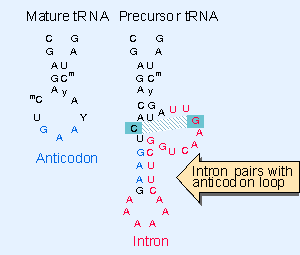
Introns in yeast tRNA-f pairs with the anticodon to change the structure of the anticodon arm. Pairing between an excluded bas in the stem and intron loop in the precursor may be required for splicing; Genes IV; http://genes.atspace.org/

http://www.pnas.org
Intron-Splicing is similar that of Cis-splicing
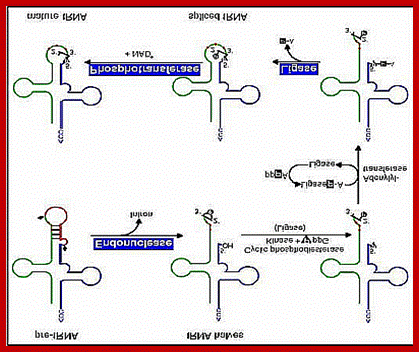
Intron Splicing-2nd type:
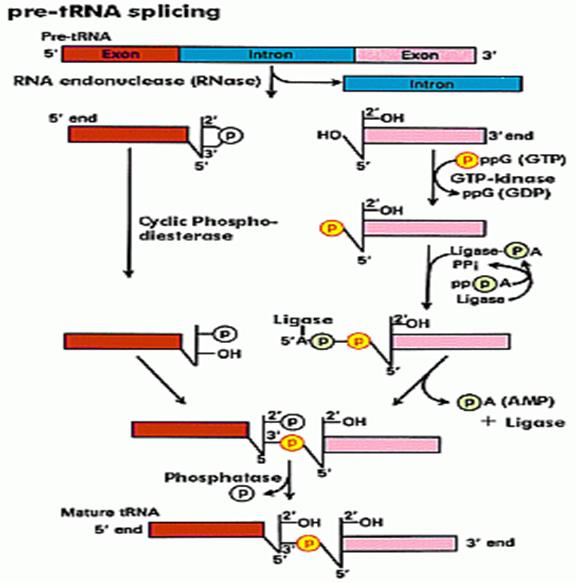
Douglas W. Smith; http://classes.biology.ucsd.edu/
Specific endonucleases recognize the 3’ and 5’ ends of the respectively and cut; and the intron is removed. The cut end, if you read from the 3’ end contain 5’OH group and the other end contains cyclic phosphate group. First, the 5’ OH group is added with a phosphate donated by the GTP kinase; at the same time the 3’ cyclic phosphate is cut to generate 3’ phosphate group. At this point activated RNA ligase binds to OH-phosphate group forming RNA ligase-P-P. Then the ligase ligates to bring about 5’-3’ ligation. One has to follow the positions of P and OH groups with respect to tRNA ends; Douglas W.smith.
RNA ligase:
In the second type, specific enzymes cut at the 3’ end of the intron producing a 5’OH group. And at the other end with it produces a unique 2’-3’cyclic phosphate group. The cyclic phosphate is cleaved to generate 2’P by a special enzyme called cyclic phosphatase. Then the 5’OH is P-lated by another phosphorylating enzyme. These modifications provide 5’’ to 3’ linkage groups, which are linked by a RNA ligase to join the 3’ OH ends with 5’ P-group, then at the P at the other site, is removed by another specific phosphatase.
Split tRNAs and trans-splicing
These are rare type of tRNA; their function is same, but they are derived from two different tRNA genes by recombination events.
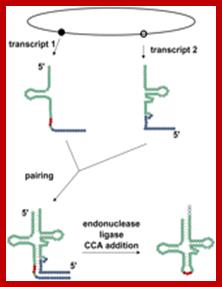
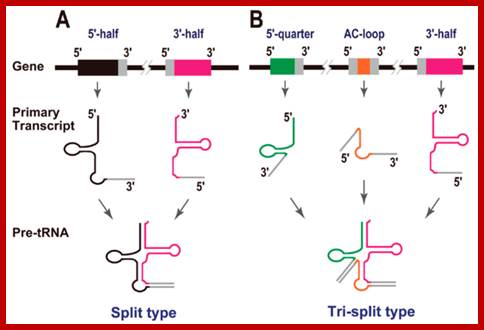
Trans splicing http://www.mdpi.com
Trans-splicing of tRNA reactions: Two fragment of tRNA segments coded for by two genes are ligated together to generate a tRNA with extended tail; it is then trimmed to generate functional tRNA.
Long distance-Trans splicing; Alissa M. Anderson; http://www.pnas.org.
RNA splicing pathways observed in eukaryotes (a–c) and engineered in yeast by (a) Canonical pre-mRNA cis-splicing yielding mRNA from pre-mRNA composed of two exons (orange boxes) and an intron (black line). (b) A transcription-induced chimera generated by read-through transcription from one gene (blue) into a downstream gene (red), followed by cis-splicing. (c) Trans-splicing of exons from two distinct pre-mRNAs (red and blue boxes). (d) Cis-splicing of a pre-mRNA embedded with a permuted tRNA. (e) Trans-splicing of two hybrid RNAs each composed of an mRNA fragment and a half tRNA. Note that in the tRNAs the aminoacyl acceptor stem is at the top and the anticodon loop is at the bottom (d and e);Di Segni et al. Alissa M. Anderson
In trypanosome brucei tRNAs coded for by nuclear genome are processed then the same are transported into mitochondria. It is rare case. Even 5s rRNA are also transported into mitochondria in large numbers more than ~1000. Once tRNAs are processed, one or two specific tRNAs, transported out into cytoplasm;.
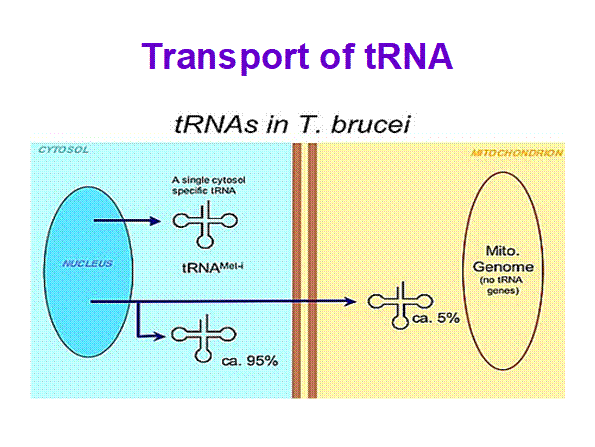
Mitochondrial Biogenesis in Trypanosoma brucei; Specific tRNAs are transported into mitochondria too; André Schneider; http://schneider.dcb.unibe.ch/
Mitochondrial protein import
The mitochondrial genome codes for only a small number of proteins so that almost all of the ~1000 proteins that constitute a mitochondrion are synthesized on cytosolic ribosomes and targeted to mitochondria post-translationally. Extensive studies during the last decades have identified four hetero-oligomeric protein complexes that catalyze mitochondrial protein import and whose core components are conserved among all eukaryotes. Two of these complexes are found in the outer membrane (the TOM complex and the SAM complex) and two in the inner membrane (the TIM22 complex and the TIM23 complex). A genome analysis revealed two aspects where the protein import pathway of T. brucei appears to be different and possibly simpler than in other eukaryotes. These are the apparent absence of a conventional TOM complex and the existence of only a single TIM complex. Thus, studying mitochondrial protein import in T. brucei is likely to provide more insight into the enigmatic evolutionary origin of mitochondrial protein import in general. André Schneider
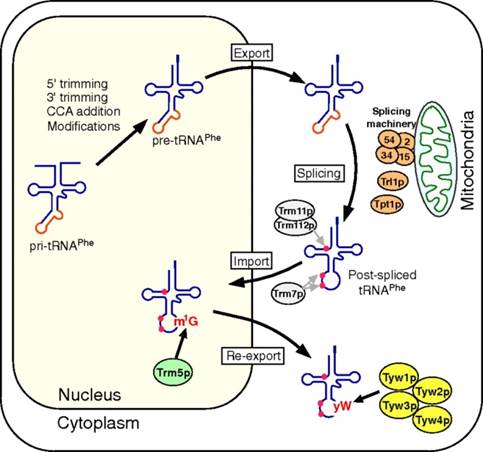
Takayuki Ohira http://www.pnas.org/
Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast; Schematic depiction of the tRNAPhe maturation pathway in S. cerevisiae. The primary transcript of tRNAPhe (pri-tRNAPhe) is end-matured by 5′/3′ trimming and the addition of CCA to form precursor tRNAPhe (pre-tRNAPhe). Ten positions are then modified in the nucleus, after which the pre-tRNAPhe is exported to the cytoplasm, where the intron is removed by the splicing machinery at the mitochondrial outer membrane. The spliced tRNAPhe is then modified by the cytoplasmic methyltransferases Trm11p/Trm112p and Trm7p, which make the m2G10 and Cm32/Gm34 modifications, respectively. The tRNAPhe is imported into the nucleus, where G37 is modified into m1G37 by Trm5p. The postspliced m1G37-bearing tRNAPhe is then reexported to the cytoplasm, where yW37 is generated through consecutive reactions catalyzed by Tyw1p, Tyw2p, Tyw3p, and Tyw4p; Takayuki Ohira,
Mitochondrial tRNA3’ end processing:
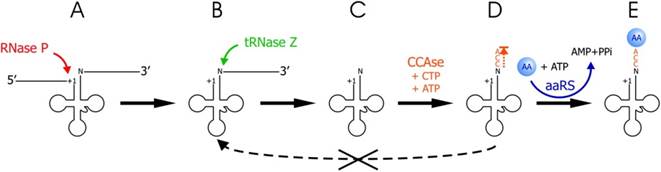
Copyright © 2014 Oxford University Press;http://nar.oxfordjournals.org/
The tRNA end processing pathway followed by aminoacylation. (A) tRNA is transcribed as a precursor, with a 5′ end leader and a 3′ end trailer. (B) RNase P has endonucleolytically cleaved the tRNA at +1. (C) tRNAse Z endonucleolytically cleaves the precursor on the 3′ side of the discriminator base (N; +73). (D) CCA-adding enzyme (CCAse) adds CCA to the 3′ end of the tRNA (N) produced by tRNase Z cleavage. (E) tRNA is charged with the cognate amino acid by a specific aminoacyl-tRNA synthetase (aaRS). Dashed line from CCA in (D) to tRNAse Z between (B) and (C) with an X through it indicates that 3′-CCA of mature tRNA is a tRNase Z anti-determinant. http://nar.oxfordjournals.org