Ribosomal RNAs:
Ribosomal RNAs:
Introduction:
They are also called structural RNAs for they act as structural components of Ribosome organelle. The ribosome in its entirety is constructed on ribosomal RNA as a scaffold on which riboproteins are sequentially built to produce a highly dynamic structure, which has astounding abilities to function as translation machine. It is deemed as the finest molecular machine.
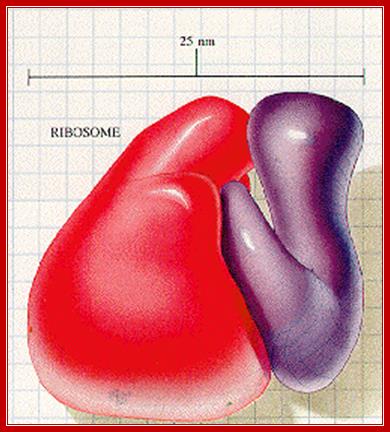
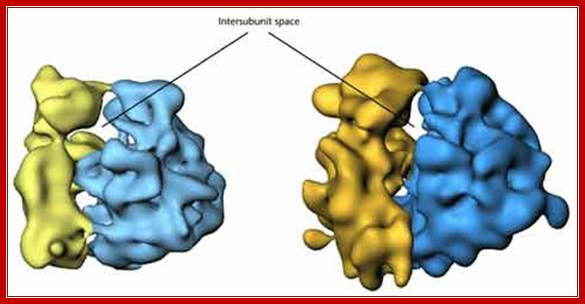
An excellent over view of ribosomal subunits hugging to each other: it is like mother sitting and hugging her child. liberary-online.blogspot.com ;www.keywordhut.com/ZWlmMw/
Based on the system and the size of the organelles, two types of ribosomes can be distinguished i.e. 70S ribosomes and 80S ribosomes; S’ stands for Svedberg’s units. Prokaryotic bacteria and Archaeal cells contain 70S ribosomes, nearly 20,000 (30-35000) per cell, and surprisingly eukaryotic organisms also contain 70s like ribosomes in mitochondria and plastids, for these organelles are considered as symbionts of bacterial origin. The organelle genomic RNA when compared to that of bacterial ribosomal RNAs, shows certain similarities in their sequence and structural organization. Ribosomes are the most dynamic cellular machine with 2.5 MD mass with RNA accounting 2/3rds of the ribosomes.
www.slideplayer.com/slide
https://www.studyblue.com;www.slideplayer.com/slide;
http://www.biochem.umd.edu/
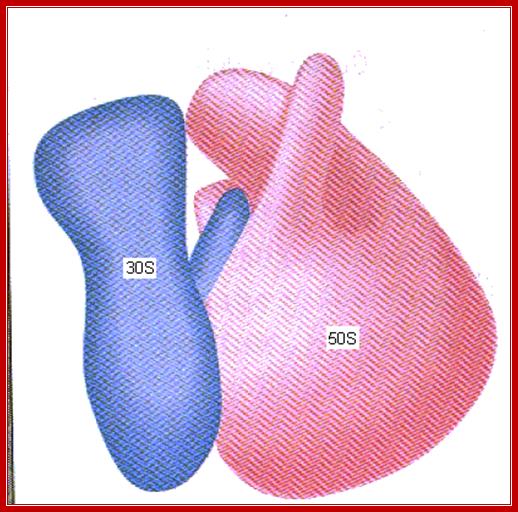
Eukaryotes have 80s type of ribosomes, ten to twenty million per cell, are found in cytoplasm. Each ribosome is actually made up of two subunits 60s and 40s and they can be separated into individual smaller subunits by lowering Mg2+ concentration and they can be sedimented into individual components.
When purified ribosomes from both sources are subjected to Phenol-chloroform extraction, one can obtain both RNAs and Riboproteins, which can be further purified to homogeneity; the following are the components of ribosomes.
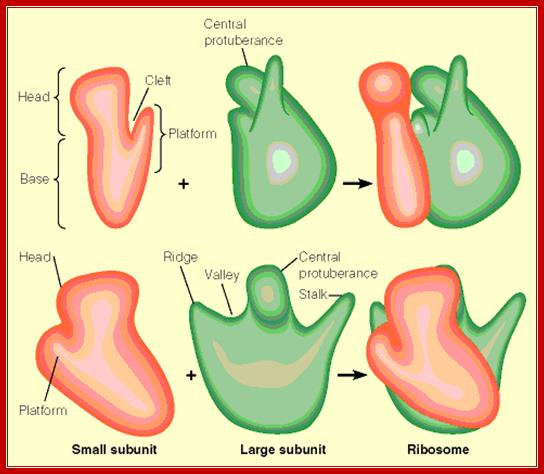
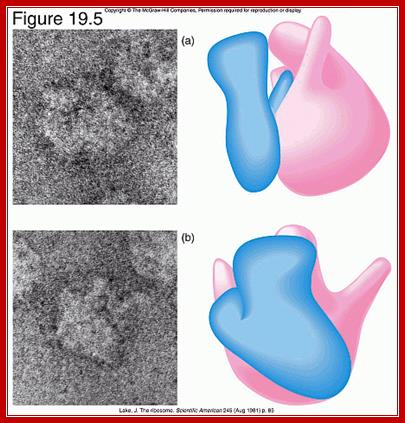
Front and side views of ribosomal subunits; M. Oaks, A. Scheirman,
http://www.pearsonhighered.com
Components of 70S Ribosomes:
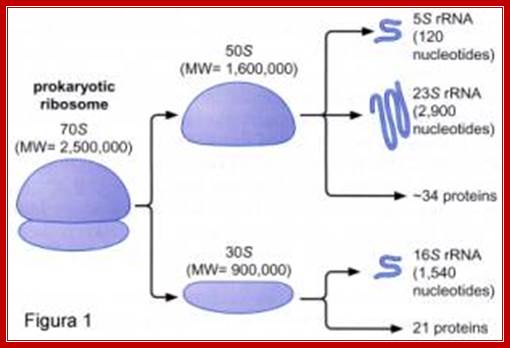
http://www.giv.no
Components of 80s ribosomes
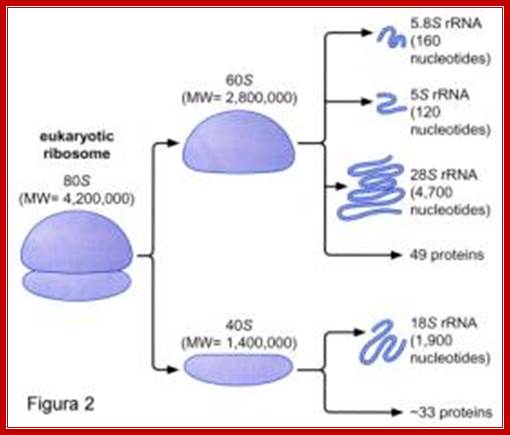
Spezielle Genetik und Molekularbiologie SS 2006 ; http://slideplayer.com/
|
70s |
Size and ntds |
No.Proteins |
No. sites modified |
|
50s |
5s-120 |
|
23 |
|
|
23s-2900 |
34 |
|
|
30s |
16s-1540 |
21 |
11 |
|
|
|
|
|
|
80s-ribosomes |
Size and ntds |
No.Proteins |
No. sites modified |
|
40s |
18s-1843-1900 |
33 |
43-44 2’O CH3 +~U |
|
60s |
28s-4710-4599 |
40 |
74,2’O-CH3, + ~U |
|
5.8 |
160 |
|
nil |
|
5s |
120 |
|
|
Organelle Ribosomes:
Chloroplasts and mitochondria are semiautonomous structures. They have their own heritable genetic material i.e. DNA; they have ribosomes and enzymes for performing specific functions. Their genomes codes for 13 and ~120proteins (mit and Cp), two rRNAs and twenty two tRNA and rest of the required proteins are coded for by the nuclear genome. Both chloroplast and mitochondrial ribosomes contain ~70s ribosomes similar to that of bacterial cells, there are some significant differences in their structural proteins and sizes.
Differences between chloroplast ribosomes left and bacterial ribosomes right; Manjuli R. Sharma et al; http://www.pnas.org/
Ribosomal RNA and Riboproteins:

Schematic rRNA 2° structures of a) E. coli LSU, b) E. coli SSU, c) S. cerevisiae LSU, and d) S. cerevisiae SSU; These 2° structures are derived from 3D structures, and include non-canonical base pairs. The domain colors in the LSU are, Domain 0, orange; I, purple; II, blue; III, magenta; IV, yellow; V, pink; VI, green, 5.8S, brown, 5S, light green. The domain colors in the SSU are, 5′, blue; C, brown; 3′M, pink; and 3′m green. Fully detailed 2° structures of rRNAs, including base pairs and additional information, from E. coli, T. thermophilus, H. marismortui, S. cerevisiae, D. melanogaster, and H. sapiens are available ; http://journals.plos.org at http://apollo.chemistry.gatech.edu/RibosomeGallery.
- Secondary structures of each of the rRNAs have been determined by their sequence analysis. The 16s rRNA and 23s rRNA, each of them, show four domains and each of them are distinguished by their binding to specific riboproteins.
- The 16s RNA’s domain I starts from 5’ end progresses into domain II, III and IV in an order. At the 3’ end of the IV th domain of the 16s rRNA, it has a small segment with a sequence that binds to the 5’ end of non-coding Shine-Delgarno sequence of mRNAs which is in the leader sequence of mRNAs. The sequence is 3’ AUUCCUCCACUAG—5’.
- Similarly there are specific ribo-protein binding sites in each of the domains, ex. s4 & s20 bind to domain I, s8, s7 & s15 bind to domain II, s7, s9, s13 and s19 bind to domain III; thus each of the binding domain can be identified.

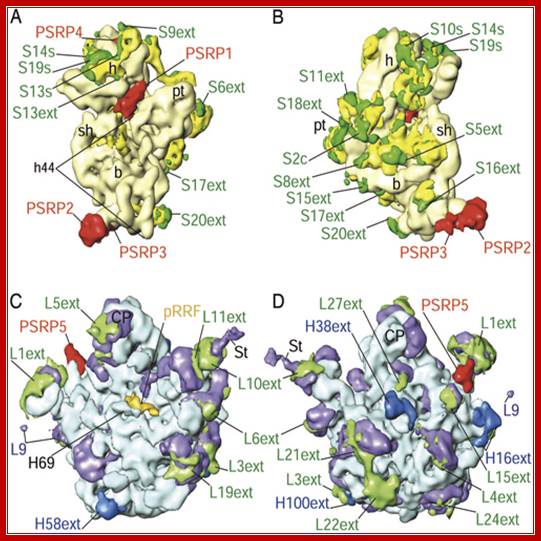
Ribosomal subunit 3D models and rRNA in 3d organization; https://www.cancerwatch.org
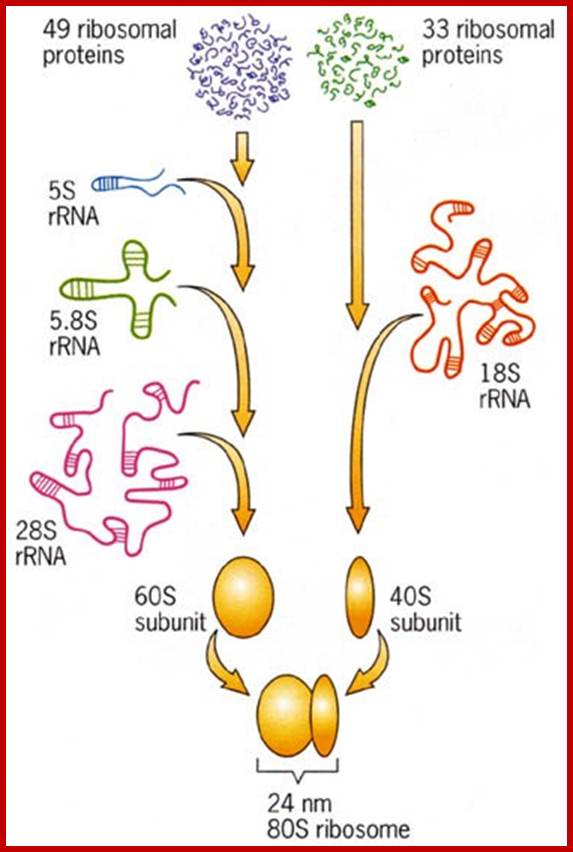
Eukaryotic mammalian rRNA and proteins assemble into 80S ribosomes; http://www.biologyexams4u.com; www.suggest-keywords.com
https://cnx.org/contents; http://micro.magnet.fsu.edu/
- Binding of tRNA and other factors to specific regions have been discerned by a variety of techniques such as electron microscopy, immuno labeling, neutron scattering techniques and others. Even eukaryotic subunit rRNAs show such domains identified by their ability to bind to certain riboproteins and other translational factors. Even the binding of RNA species such as tRNA has been established.
Some commonality of the sequence and secondary structures can be observed when one compares the 5’end of the 23s RNA of prokaryotes with that of 5’ ends of eukaryotic 5.8sRNA. A secondary structure of rRNAs provides space for the binding of ribosomal proteins to suggest overall structural features of ribosomes.
- It can be discerned that rRNAs from eukaryotes also show similar structural features. Prof. Venkataram (Hyderabad), popularly called Venki, has done invaluable work in solving Ribosome structure, using crystallography and X-ray diffraction studies for which he was awarded Nobel Prize with other two scholars in 2009. He deserves ‘Bharat Ratna’ from India too?
Riboproteins (Prokaryotic):
- Riboproteins, isolated from small and large subunits, have been numbered as small and large riboproteins. They are numbered as s1 to s21 and L1 to L31 respectively. The numbering is based on the mobility of each of the riboprotein subunits on a 2-D polyacrylamide gel. The s1 is the largest protein found at the left top most corner of the gel and s21 is the smallest, found at the right bottom most corner of the gel.
- Each of the riboproteins from both 70s and 80s ribosomes have been purified and antibodies have been obtained and many proteins have been studied using X-ray diffraction, immuno-diagramming, neutron scattering, electron microscopy, NMR and cross linking to other riboproteins
- In prokaryotes s20 and L26 are common to both ribosomal subunits and located at the interface of the subunits. L7 and L12 are found 4 copies each. Their N- terminal is acetylated and forms stable complex with L10.
- Amino acid sequence of most of the riboproteins has been determined, where the number of amino acids ranges from 46 (of smallest subunit at right bottom most in the gel) to 557 in the largest subunit s1 (Left top most in the gel).
- Among the largest L1 is 45 KD and L34 is 25KD? There is sequence homology among the protein subunits, but in general, they are rich in Lysine and Arginine, both are basic amino acids.
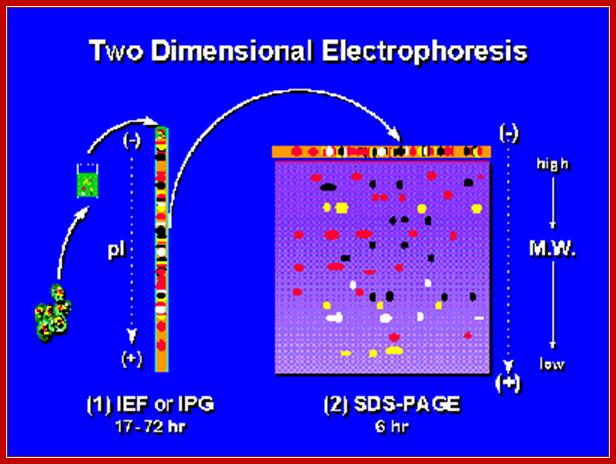
Two-dimensional Electrophoresis; In first dimension Isoelectric focusing or immobilized pH gradient Gel (IPG); followed by SDS-PAGE at 90o to that of first (2nd dimension). http://en.wikibooks.org/

Gel Electrophoresis of Ribosomal Proteins;
Ribosomes contains mainly basic proteins (proteins that are positively charged at neutral pH). In addition the ribosome also contains some neutral or acidic proteins. The proteins can be extracted from the ribosome and separated by two-dimensional gel electrophoresis. These 2D-techniques usually separate proteins according to charge in the first dimension and according to size or charge (at a different pH) in the second dimension.Nobelprize.org

Two-dimensional polyacrylamide gel analysis of ribosomal proteins from wild-type and L27-lacking E. coli. Proteins were extracted from 10A 260 units of purified 70 S ribosomes from strains LG90 (left) and IW326 (right) and separated by acidic two-dimensional PAGE Iwona K. Wower‡, ‡ and Robert A. Zimmermann§
- Structural motifs of riboproteins in general show 3 b sheets and 2 a helical strands and resemble that of Sn RNPS, rho, and eIf-4B. Such motifs are called RRMs (rRNA Recognition Motifs).
- Most of the riboprotein genes have been identified and their operons have been established. Each operon consists of several cistrons controlled by a single promoter element with -10 and -35 sequences.
- In prokaryotes the riboprotein genes have been organized into seven clusters called rRNA operons. Some of these operons not only contain riboprotein genes but also contain some other genes like RNAP subunits and some translational factors. Their synthesis is regulated at translational level, which depends upon the availability and the abundance of rRNA segments. The mRNA is translated similar to other polycistronic mRNAs.
- If the concentration of rRNA falls short, the translation of ribo- protein mRNAs is inhibited by their own proteins by binding to specific regions of the mRNAs thus prevent translation, which is called autogenous regulation.
- Subunits of riboproteins act as self-assembling systems with rRNA providing a template or threading for sequential assembly. As rRNA is synthesized (synthesized as a precursor RNA), they are methylated and pseudo-uridinylated at specific sites, which acts as recognition points for processing of RNA and also for assembly of proteins.
- Assembly of riboprotein with rRNA is hierarchical, in the sense, some proteins bind first to certain RNA sequence derived secondary structures, then another set of proteins bind either by RNA-protein interaction or protein-protein interaction or by both.
- There is a gradual build up of the assembly line, which ultimately produces a highly organized and a dynamic structure, where the position of each protein is fixed with respect to each RNA fold or sequence.
- Though it exhibits a specific 3-D shape, it is not a rigid structure but it always undergoes conformational changes according to the binding of other translational components or regulatory factors. Structurally, as well as functionally, ribosomes are flexible and highly dynamic.
- In the overall organization of ribosomal structure, some regions don’t have any RNA but only proteins. The whole structure appears to be porous; through which some of RNA folds protrude to the surface.
Assembly:
- Assembly or association of riboproteins with rRNA is sequential and stepwise. Methylation of 2’OH of ribose sugars at adenine and guanine nucleotides at specific position is critical, and such methylations are performed by specific methylases and they use sequence driven secondary structures as motifs for identification of sites.

http://slideplayer.com
Ribosomes, tiny organelles composed of approximately 60 percent ribosomal RNA (rRNA) and 40 percent protein. However, though they are generally described as organelles, it is important to note that ribosomes are not bound by a membrane and are much smaller than other organelles. Some cell types may hold a few million ribosomes, but several thousand is more typical. The organelles require the use of an electron microscope to be visually detected; http://micro.magnet.fsu.edu/
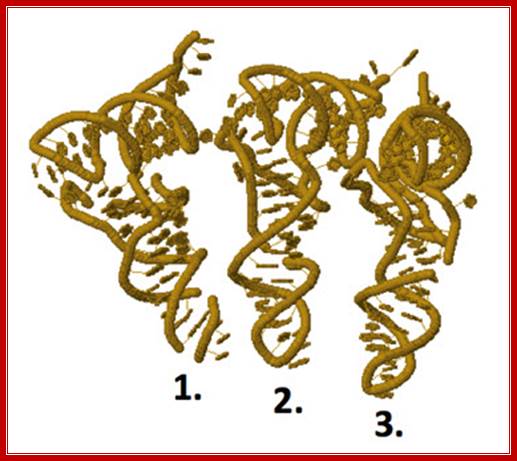
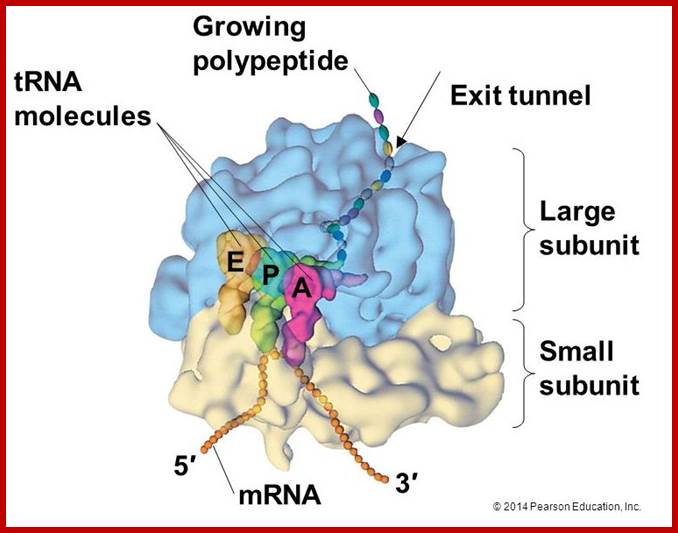
Mloecular odel of tRNA bond to different ribosomal sites;E-site tRNA (1), P-site tRNA (2), A-site tRNA (3); http://www.chegg.com/

The ribbon diagram shows the positioning of tRNA on large ribosomal surface; A, amino acyl tRNA site, P peptidyl RNA site and E Exit site. http://liberary-online.blogspot.com
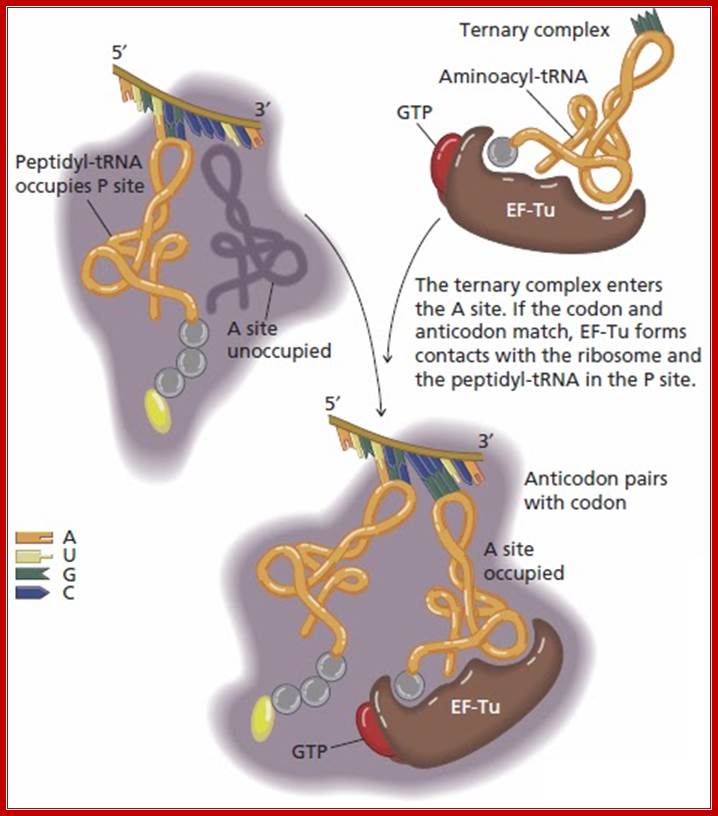
Laurence Moran; http://sandwalk.blogspot.in/
Assembly of small ribosome subunits:
16sRNA + 16 s riboproteins à 21 s particles (can assemble at 20^oC),
21s particles + 6s riboproteins >à 26 s particles,
26 s particles ----> 30 s particles.
Assembly of Large Ribosome subunits:
23SRNA + 5sRNA -à 33 s particle,
33 s [articles -à 41 s particles,
41 s particles -à 50s particles
During dissociation also, certain subunits dissociates fast, even at the earliest steps of preparation; they are called split proteins. Such proteins are found both in small and large subunits. Even during assembly, certain proteins associate at 0^oC, this is because great affinity of some proteins to certain RNA sequence. Cold sensitive mutants block such assembly; they are called Subunit Assembly Defective mutants (SAD mutants). Proteins, which associate, first are hard to disassociate and they are called core groups, and proteins, which assemble last, are the first to dissociate. The following figure depicts sequential steps in the assembly.
Assembly of small ribosome subunits:
16sRNA + 16 s riboproteins à 21 s particles (can assemble at 20^oC),
21s particles + 6s riboproteins >à 26 s particles,
26 s particles ----> 30 s particles.
Assembly of Large Ribosome subunits:
23SRNA + 5sRNA -à 33 s particle,
33 s [articles -à 41 s particles,
41 s particles -à 50s particles
rRNA 5’-------------------------------------------------------------------3’
I I I I
1st level I s4 I I s8
2nd level s15 I s20 s7
3rd level s17 s13
4th level s16
5th level s12 s9 s19
6th level s18 s5
Assembly sequence:
30s = 17.5sRNAàs4,s8,s15-às1,s5,s7,s13--->s2,s3,s6,s9,s10
s17, s20 s16, s21 s11, s12, s14, s18/19
50s= 25sRNA--->L1,4,5,8,9,10---->L3,7,11,14-->L2, 6,12,10,28,31,32,
13,17,18,20, 15, 19, 23
21,21,22,23,
24,25,27,29,
30, 33.
30s [16s RNA] O^oC 40^oC O^oC
+[ s21 proteins]--------------------> 21s--------------->26s------------->30s
50s [23sRNA] o^oC 44^oC O^oC 50^oC
+5sRNA+34L] ---------------->33s---------------->41s------------->48s----------->50s
Proteins]
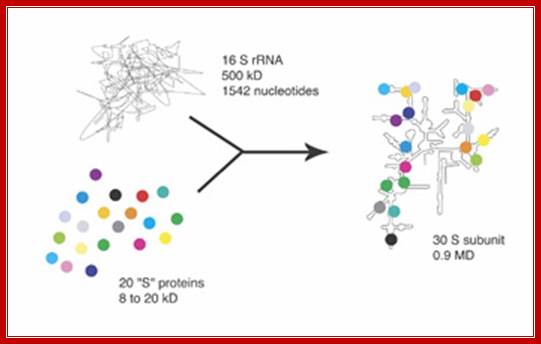
James R. Williamson et al ; http://www.scripps.edu/
Assembly reaction of the 30S ribosomal subunit. A total of 20 proteins (represented as circles) bind to the 16S ribosomal RNA (shown as a line). In the absence of the proteins, the RNA is only partly folded (left), but it becomes highly ordered as it folds during assembly. The complex reaction shown here occurs in a stepwise manner to ensure proper assembly, but the details of this process remain to be elucidated.Williamson
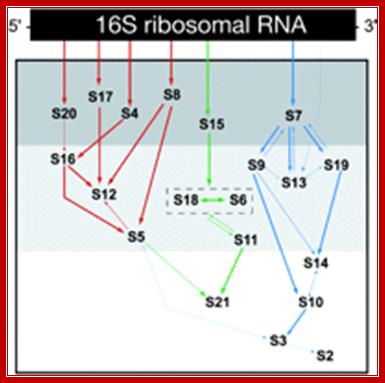
Ribosome Biogenesis and the Translation Process in Escherichia coli;

Assembly of 30s ribosomal protein on 16srRNA; JOEL F. GRONDEK and GLORIA M. CULVER; http://rnajournal.cshlp.org/
Studies of Escherichia coli 30S ribosomal subunit assembly have revealed a hierarchical and cooperative association of ribosomal proteins with 16S ribosomal RNA; these results have been used to compile an in vitro 30S subunit assembly map. In single protein addition and omission studies, ribosomal protein S13 was shown to be dependent on the prior association of ribosomal protein S20 for binding to the ribonucleoprotein particle. While the overwhelming majority of interactions revealed in the assembly map are consistent with additional data, the dependency of S13 on S20 is not. Structural studies position S13 in the head of the 30S subunit > 100 Å away from S20, which resides near the bottom of the body of the 30S subunit. All of the proteins that reside in the head of the 30S subunit, except S13, have been shown to be part of the S7 assembly branch, that is, they all depend on S7 for association with the assembling 30S subunit. Given these observations, the assembly requirements for S13 were investigated using base-specific chemical footprinting and primer extension analysis. These studies reveal that S13 can bind to 16S rRNA in the presence of S7, but not S20. Additionally, interaction between S13 and other members of the S7 assembly branch have been observed. These results link S13 to the 3′ major domain family of proteins, and the S7 assembly branch, placing S13 in a new location in the 30S subunit assembly map where its position is in accordance with much biochemical and structural data. JOEL F. GRONDEK and GLORIA M. CULVER
- As the 5’ end of the precursor rRNA emerges during its synthesis, s4, 8 and 15 bind to this region tightly. Then s17 and s7 join directly on to the RNA, later other proteins join by protein-protein or protein-RNA interactions.
- Proteins s1, 3, 4, 5, 9, 12, 18 and the 3’ end of 16s RNA are involved in mRNA binding. Peptidyl transferase function at P-site involve proteins L-2, 11, 15,16,18,23, and 27 in association with 23s RNA.
- About 40 ntds long region of 16S RNA is located in the platform of 30s ribosomal subunit. Peptidyl transferase occupies valley in the ribosome. Two L7 and two L12 together act as GTPase.
Role of rRNA in protein synthesis (Prokaryotic):
- In the molecular organization of ribosomes, both RNA and proteins are ordered and occupy certain specific invariant positions and perform specific function. As a 3-D structure, it goes through several conformation changes with each binding events and catalysis.
- The 3’ terminus 16s RNA of 30s ribosome directly interacts with 5’ end Shine-Delgarno sequence of mRNA and facilitates initial binding of 30s ribosomal surface so as to bring the first codon exactly to P site.
- Specific regions of 16s RNA interact with tRNA for the binding at P and A sites. 23 S RNA interacts with CCA terminus of Peptidyl tRNA. It is envisioned that there is RNA-RNA interaction and protein–protein interaction as well as protein-RNA interaction, thus both subunits are held together while they perform functions.
- There must be some proteins, which perhaps act as motor proteins in moving ribosomes on single stranded mRNA in GTP/ATP dependent manner, similar to helicase.
- Cleavage of 3’ region of 16S RNA by E3 Colicin abolishes initiation of translation for this region binds to Shine Delgarno sequence found in PK mRNAs.
- Methylation of Adenine at 6th position and at 3’ end (di methylation to Adenine) of 16s RNA, facilitates dimrization of 30s and 50s units. If methylases are absent dimrization fails.
- Sensitivity to Kusugamycin depends upon methylation or absence of methylation at the above-mentioned sites. Mutation at these sites abolishes sensitivity to the drug, but methylation at these sites makes it Kusugamycin resistant. Kusugamycin blocks initiation of translation by releasing F-met tRNA from ribosomal surface.
- Mutation in the of 3’ end of 16s RNA suppress terminator codon function. A mutation in rRNA can lead to frame shift function, because recognition between mRNA and rRNA doesn’t take place. A region at 1400th ntds in 30s, is directly involved in the binding of peptidyl tRNA at P-site.
- Both 16s and 23s RNAs are involved in organizing A and P site. The CCA-end of tRNA at P- site protects 23SRNA from RNase digestion. The 23s rRNA is involved in organizing the exit site E, found in 50s subunit.
- The 23s RNA is involved in catalyzing peptide bond formation; hitherto it is believed that an enzyme called di peptidyl transferase found in larger ribosomal subunit is involved in peptide bond formation.
Structural Features (Prokaryotic):
Structurally prokaryotic ribosome has 200 x 220 A^o dimension and the size of eukaryotic ribosome is slightly larger.
- For initiating translation process, to begin with the small subunit should be separated from the large subunit; it is at later part of initiation both subunits join later.
The larger subunit looks like a cup shaped palm having a central protuberance curved inwards, a blunt thumb like structure and a last finger like structure projecting outwards.
- The extended finger acts like a stalk and contains L7/L12 with GTPase activity.
The central protuberance contains 5s RNA.
- The valley is located between the blunt thumb like projection and central protuberance. The ridge or thumb like portion contains L1/9.
Actually the valley provides peptidyl transferase activity. The large subunit has a narrow tunnel like region, which extends from peptidyl assembly site to exterior, through which nascent polypeptide chain is threaded through with NH3+ end ahead. The length of the tunnel can hold about 20 to 30 amino acid long polypeptide chain and has the diameter to accommodate the chain.
It is at the posterior end, where polypeptide chain exits, contains a site for the binding of large ribosome to endoplasmic reticular membrane.

Protein exit tunnel in large ribosomal subunit; A. A. Bogdanov*, N. V. Sumbatyan et al;;http://protein.bio.msu.ru/
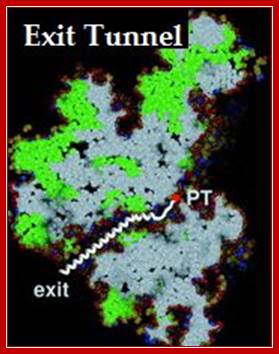
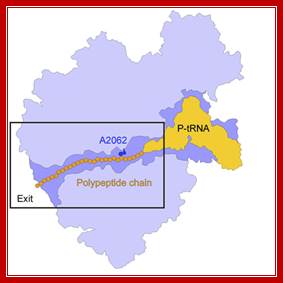
This diagram shows a tunnel through which the nascent polypeptide threads through as it is translated. Interplay between the Ribosomal Tunnel, Nascent Chain;Agata L. Starosta et al; http://www.cipsm.de/
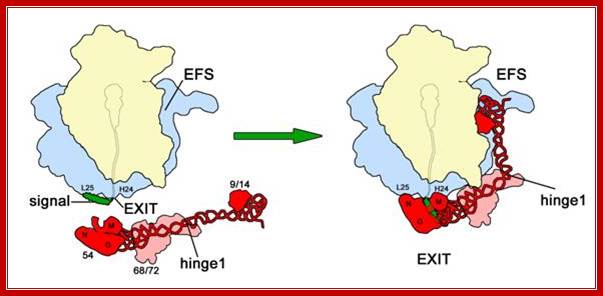
Signal sequence dependent SRP-Ribosome interaction; Binding/association of SRP protein at exit tunnel and guide the nascent protein into ER receptor and channel; http://edoc.hu-berlin.de/
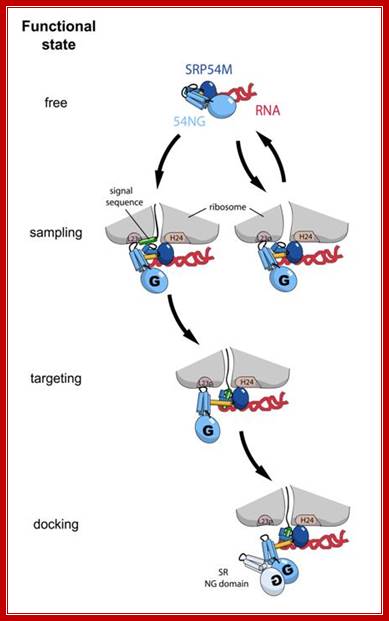
Model for the first steps of SRP cycle; SRP binds to protruding Hydrophopic signal sequences and guides towards ER channel; http://edoc.hu-berlin.de/
The small subunit is split in the top region into a platform and a head; the space between them is called cleft. The 3’ end of the 16sRNA is located in the platform. It is through cleft region the mRNA is threaded and both P-site and A-site are located on the surface of platform and at the base of the head.
Ribosomal site for the binding of mRNA to 16sRNA and the binding of initiation factors are located in the platform of 30s ribosome.
- Peptidyl tRNA and aminoacyl tRNA binding locations (P and A sites) are at the cleft region of 30S ribosome. The small ribosomal subunit has an additional site called A site to the right of A site, where the incoming a.a tRNAs are screened.
- The stalk (finger like) of 50s ribosome contain factor binding site and GTPase activity. Peptidyl transferase activity is located in the valley of large unit. Large subunit also has sites for the binding of peptidyl (P) and aminoacyl tRNAs (A) in line with small subunit, plus it has another site for the exit (E) of decoded tRNA.
The tunnel is 100-120 A^o long, 25A^o broad and can hold approximately 20-30 amino acid long polypeptide chain.
- ER Membrane binding domain in the large subunit is by the side of tunnel exit. Ribosome is the most dynamic structure which undergoes conformational changes at every step of chain initiation, elongation and termination during protein synthesis.
Ribosome Mediated Inhibitors of translation:
Kusugamycin: initiation (PK), displace F-met tRNA, mutants lack methylation of 16 s rRNA at the 3’end.
Streptomycin: initiation (PK), mutation in s12 of 30s ribosome causes resistance.
Kirromycin: elongation (PK), EF-Tu-GDP release is blocked by the antibiotic and no recycling.
Puromycin: elongation (PK), premature termination, because Puromycin has structure similar to tRNA configuration.
Erythromycin: peptidyl transfer (PK), blocks peptide bond formation, mutation in 23sRNA results in resistance.
Chloramphenicol: peptidyl transfer (PK), blocks peptidyl bond formation,
Cycloheximide: translocation (EK), inhibits peptidyl transferase on 60s subunit.
Fusidic acid: translocation (PK), EF-G-GDP cannot be released, no recycle.
Thiostrepton: translocation (PK) binds to 23sRNA and inhibits GTPase activity.

http://oregonstate.edu, Courtesy of M. Oaks, A. Scheirman, T. Atha, G. Shankweiler, and J. A. Lake, The Ribosome, W. E. Hill, A. Dahlberg, R. A. Garret, P. B. Moore, D. Schlessinger, and J. R. Warner, eds.,p. 181 (New York:American Society of Microbiology Press, 1990).