Processing of Eukaryotic pre-rRNAs in:
AND Histone pre mRNA
Both in prokaryotes and eukaryotes rRNA transcripts are produced as long precursor, which are further processed into functional segments, by cutting and base modifications.
· In prokaryotes the transcripts, depending upon the operons, contain tRNA segments also within rRNA precursors.
· In such cases 5’ end of the tRNA is generated by RNase-P. This is a complex of nearly 300 ntd long RNA and associated with 3 proteins.
· The RNA by itself by folding into 3-D structure can perform cleavage, but presence of protein enhances RNAs function as an enzyme; so this RNA is named as Ribozyme. It is the first non-protein molecule to be considered or confirmed as an enzyme. Till then it is believed that only proteins are capable performing enzyme activity.
· Rnase III and RNase D (proteins) perform further processing.
· Before the cuttings and trimmings, the rRNA gets methylated at various sites; methylation is used for sequence specificity. Methylation provides site identity for other processes.
· A similar situation also prevails in EK ribosomal transcripts; the difference is EK transcripts, though longer, they don’t have tRNA segments tucked with in the transcript.
· Each of the tRNAs, rRNAs, SnRNAs, and 5s rRNA genes are all transcribed as independent entities by different RNAP enzymes.
· Pre ribosomal RNAs are processed similar to that of PKs with a difference.
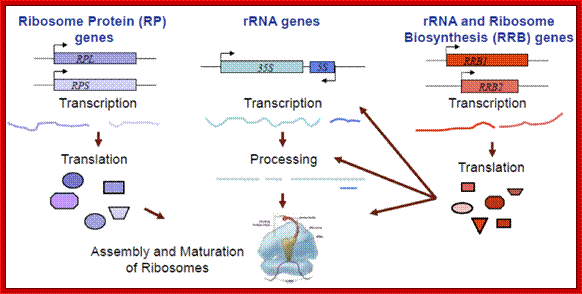
Ribosome biogenesis requires the coordinated regulation of three extensive gene networks, including 137 cytoplasmic RP genes, 150 rRNA genes and some 200 RRB genes. It is one of the largest regulon; http://mmcalear.faculty.wesleyan.edu/

The ribosome synthesis pathway in eukaryotes. The initial stages of ribosome synthesis take place in the nucleolus. The first step is the association of newly synthesized 35S rRNA with 40S processing proteins and 40S ribosomal proteins which form a complex with the future 18S rRNA sequence even before the transcript is completed (the co-transcriptional assembly stage). After completion of rRNA transcription, the 35S rRNA and its associated proteins form a 90S pre-ribosome particle which contains numerous 40S processing factors and 40S ribosomal proteins but very few 60S processing proteins or 60S ribosomal proteins. RNA cleavage releases U3 snoRNP and separates the 90S particle into 40S and 60S pre-ribosome particles. The latter recruits 60S processing proteins and 60S ribosomal proteins, and the separate pre-ribosome complexes are exported out of the nucleolus into the nucleus and cytoplasm. Most of our knowledge about this pathway has been compiled from studies in budding yeast; see text for further details. http://genomebiology.com/
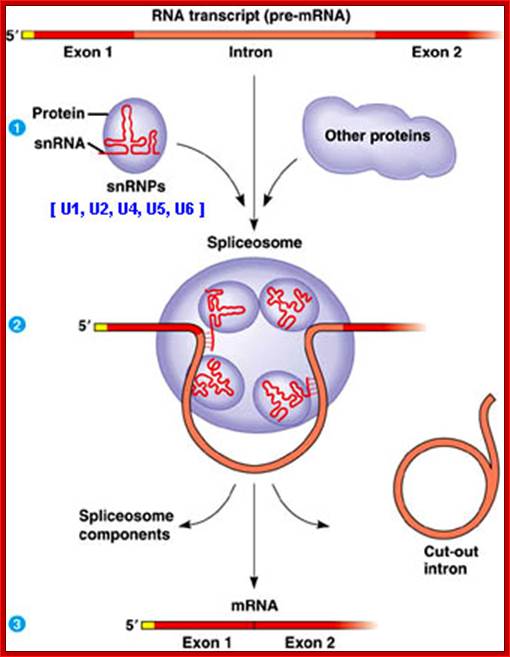
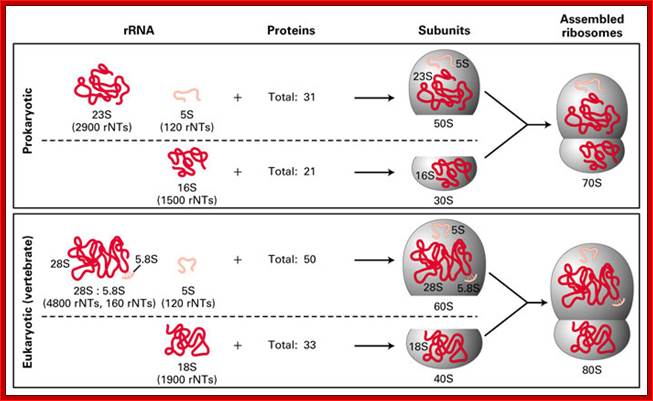
Ribosomal RNA, or rRNA forms the backbone of each ribosomal subunit. These rRNAs are first transcribed as a long molecule, and then cleaved to form smaller 28S and 18S units. Each of these is packaged with proteins to form the large and small ribosomal subunits. Proper conformation of the mRNA is necessary to give each ribosome its proper shape. Ribosomal RNA is often the target for antibacterial drugs, as the prokaryotic and eukaryotic rRNAs are usually sufficiently different to allow specific targeting of only the prokaryotic molecule. Some RNA molecules can also be catalytic. In eukaryotes, small nuclear ribonuclear proteins (snRNPs) remove the introns from newly transcribed mRNA molecules. These snRNPs are a mix of RNA molecules and proteins. Any mistakes in snRNA function will have cascading effects as they are necessary to produce every other RNA molecule (and therefore protein) within the cell. http://compbio.pbworks.com/ and (http://fig.cox.miami.edu/
The above three figues show the transcription, processing and assembly of ribosomes.
Number of rRNA and tRNA Genes:
|
Species |
16s/23s/5s or18s/28s-5.8s s rRNA genes |
5s rRNA genes |
tRNA genes |
|
E.coli |
7 |
- |
60 |
|
S.cerviciae |
140 |
140 |
250-400 |
|
Dictyostelium discoideum |
180 |
180 |
>850 |
|
D.melanogaster: XY: XX: |
150 250 |
150 150 |
850 850 |
|
X.laevis |
450-600 |
24000 |
1150 |
|
Homo sapiens |
280 |
2000 |
>2000 |
|
|
|
|
|
Size of Precursor and Processed rRNAs:
|
Species |
Precursor size (s value) |
Precursor size (in ntds) |
23s/28s final in ntds |
16s/18s final in ntds |
Percentage of precursor |
|
E.coli |
30s |
5.6 to 6 kb |
2914-3100 |
1500 |
80% |
|
Yeast37s |
37s |
7.2-8.95 kb |
3750-3800 |
1700-2000 |
80% |
|
Dictyostelium |
37s |
7.4 kb |
4100 |
1800-2000 |
|
|
Drosophila |
34 s |
7-7.4 kb |
4100 |
1800 |
78% |
|
Xenopus |
40 s |
7.875 kb |
4475-4500 |
1900-1925 |
79% |
|
Gallus domesticus |
45 s |
11.25 kb |
4625 |
1800 |
57% |
|
Mus musculus |
45 s |
12.4-13kb |
4712-5100 |
1900-1958 |
52% |
|
Homo sapiens |
45 s |
13.7kb |
5100 |
1900 |
|
|
Plants (general) |
|
7.9kb |
3700 |
1700 |
71% |
Eukaryotic ribosomal RNA genes are transcribed in the nucleolus. Each of the ribosomal rRNA genes are organized into tandem repeats of 50 to 100. When the nuclear membrane reassembles after telophase rRNA genes open out into histone free DNA loops. Each of these rRNA genes are interspersed with non coding and non-transcribing spacer regions. Ribosomal RNA genes have promoter elements described elsewhere. However the non-transcribing spacers do contain regulatory elements including enhancer regions.
The transcribed rRNA is larger than their products; it 45s rRNA; in yeasts it of 35s size. This precursor contains, from the 5’ end, 18s RNA , 5.8sRNA and 28sRNA segments separated by internal transcribed spacer regions (ITS). On either side of this block the precursor also contains external transcribed spacers at both 5’nds (ETS).
Processing of rRNAs is complex for some of the bases are modified, some 2’OH group of riboses are added with methyl groups and some Uridines are converted to pseudouridine. All the said modifications are site specific but driven by sequences.
While such modifications are taking place the rRNA is cut and trimmed by host endo and exo enzymes.
Processing in eukaryotic rRNAs requires U3 Sn RNA and a host of sno-RNAs (small molecular weight nucleolar RNAs), which are in real terms smaller in length, ranging from 87 to 275 to 555 ntds in some cases and mostly rich in U s. All U3sn RNA s are included in this group.
· Sno-RNAs are associated with several sno-RNPs. Several sno-RNPs contain 34 kDa proteins called Fibrillarins.
· One of the other sno-RNPs called MRP is also required for processing.
|
Species |
Base CH3 |
2’O-CH3 |
Pseudourine |
Total |
|
E.coli |
22 |
4 |
10 |
36 |
|
S.solfataricus |
~8 |
67 |
9 |
88 |
|
S.cervisiae |
10 |
55 |
44 |
112 |
|
X.laevis |
10 |
99 |
~98 |
207 |
|
H.sepiens |
10 |
107 |
~95 |
212 |
|
|
|
|
|
|
The vertebrate rRNAs contain more than 100 and so also Pseudouridine sites and methylation sites. As soon as the 5’end of the rRNA transcript is made available, methylation and pseudouidinylation starts at specific sites, which is determined by sequences and also the secondary structures develop on their own or aided by sno RNAs. Position of them is absolutely conserved.
· Methylations require specific sno RNAs called C/D RNAs and short motifs called C&D segments (actually they contain C and D and C’ and D’ regions), they are called so, for they have short conserved sequence motifs called C and D, whose sequences are complementary to certain regions of 18s and 28s rRNA, and even 5.8sRNAs.
The sno RNAs assume certain secondary structure which are superimposed by the association of sno-Rnps. There are two classes of sno-Rnps, CD sno-Rnps and H/ACA sno-Rnps.
The number of sno RNA detected and identified estimated to be more than 200. Some of the common sno-RNAs are U3, U26, U31, U48, u50, U73 U74, U80 and U81 and many more.
CD RNAs have stem and two open bulges; from the 5’ end they contain segment called C which contains a sequence- RUGAUGA. Continuation of this bulge leads to another bulge called D’ segment and contains sequence-CUGA. This continues with another segment called C’ and D. The rRNA sequences base pairs with D’ and D segments.
Similarly H/ACA sno-RNAs contain two stem loops separated by linear region with sequence called ANANNA and at the end of the second loop it has ACA sequence. Each of the stem loops contains middle bulge and terminal bulge. The rRNAs base pair with middle bulge region of the sno-RNA. Among the two unpaired ones, one of the U ntds gets converted to Pseudouridine. The enzyme involved performs two steps, cut N-sugar bond and join 5’C to the sugar to generate pseudouridine.
CD sno RNA associated proteins:
|
Yeast |
Vertebrates |
functions |
|
Nop1p |
Fibrillarin |
2’o methylation |
|
Nop56p |
Nop56 |
Assist base pairing |
|
Nop58p/Nop5p |
Nop58 |
Nap65 |
|
Snu 13p |
15.5p |
They are like riboproteins |
|
65kd, 68kd |
|
|
|
|
|
|
H/ACA sno-Rnps
|
Yeast |
Vertebrates |
Functions |
|
Gar1p |
GAR1 |
RNA binding |
|
Cbf5p |
Dyskerin/Nap57 |
Pseudo Uridine synthase |
|
Nhp2p |
Nhp2 |
RNA binding |
|
Nop10p |
Nop10 |
|
|
|
|
|
Processing factors associated with Sno-RNPs:
Rat1p—>5’-3’ exonuclease
Xrn1pŕ 5’-3’ exonuclease
Rnt1p—dsRNA endonuclease
Sen1p—RNA helicase,
Proteins assisting Sno-RNA biogenesis:
Rvb1p, Rvb29, from yeast- act as RNA helicase,
Srp40p from yeast, biogenesis and trafficking,
p55, p50, from vertebrates act as RNA helicase,
SMN protein in vertebrates -assist in assembly,
Nopp140 in vertebrates help in processing and trafficking
· Loss or mutations in some sno-RNAs prevents methylation at specific sites because the methylation site is complementary to sites found in sno RNAs.
· At the time methylation, rRNA base pairs to C and D regions of sno RNA. When they base pair properly from the 5’ end of the base paired rRNA, the fifth nucleotide get methylated by specific methylases and the donor of the methyl group is SAM (S-Adenosyl methionine).
· There are 40 or more such sno RNAs. Sno-RNA mediated methylations are not well characterized so far (?).
· Another group of Sno-RNAs is involved in converting Uridine to pseudouridine. There are 43 pseudo uridines in yeast rRNAs.
· There are groups of 20 or more H / ACA group of sno-RNAs, which are involved in conversion of Uridine to pseudo-uridines. The ACA group name has been derived from ACA sequence invariably found at 3’end of sno-RNAs. The ACA-sno RNA themselves fold into pre-defined hair pin like structures, where at 3’ end ACA, base pairs with its own 5’ end and stabilizes the structure. It also contains certain unpaired regions called by the names A and B boxes. The unpaired regions of H /ACA sno-RNA pair with precursor rRNAs, wherever the sequences are complementary.
· The Uridine present exactly at 15th position from ACA is converted into Pseudouridine. This conversion is not like any changes in the base, but in this, the ‘N that is bound to ribose, is cut and reconnected to 5th carbon of the Uracil. It is now discerned, few Uridine synthases, use sno-RNAs to convert Uridine to pseudo-uridines at specific positions.
Why pseudouridines are made at specific positions and what are their functions? Perhaps identify the sites for endonuclease to cut precursor rRNA into fragments for further processing.