Self-Splicing RNAs:
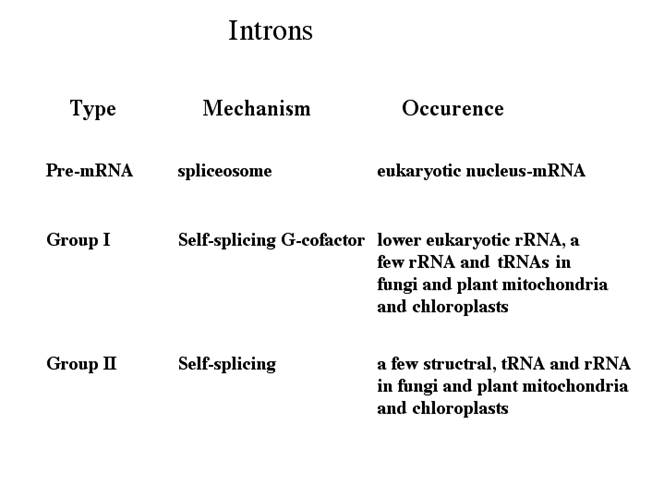
Group I, II, III RNAs and Twintron
· Group-I: Ex. trn-L, is similar to self-splicing rRNA of Tetrahymena or Maturase of yeast Cyt.b.ox. Group I are also found in Physarum fungal mitochondria and T4 phage. They require RNA binding proteins to act as chaperones to correctly fold the RNA into an active intermediate structure in vivo. Pre-tRNA introns in Bacteria and in higher eukaryote plastids are typical examples of self-splicing group I introns. In archaea splicing requires an endonuclease similar an endonuclease for tRNA splicing.
· Group-II: A majority of genes including trn-A, trn-L folds into a very complex sec. structures as in Cytochrome oxidase of maize and yeasts. They are generally found in bacteria (common), and in archaeal genes of Methano corcina. They are very common in lower plant chloroplasts.
· Group-III: The trn G, trn-k, trn-V, rpl 2, rpsl 2, rpsl-6 etc have conserved splice center sequences, at 5’ GTGCGNY and at 3’ ATCNRYY (N) YYAY, similar to those of EK nuclear genes. They are found in all eukaryotes, more common in Crown groups. They are very common in ‘crown group’of eukaryotes- smallest monophyletic group, or "clade".
· Group IV Introns: They are seen in tRNAs. Splicing requires ATP and an endonuclease.
· Group (?) Twintrons and Mirtrons; These are developed from labile introns integrating into existing introns. There are different types such as Group II introns in Group II introns and group I introns in group I introns as Twintrons. Many miRNA segments are located within an intron, which are released after intron processing, they are called Mirtrons.
Splicing of RNAs found in the Intron regions of eukaryotic mRNAs. It requires certain specific structural features at Exon and Intron splicing junctions. Splicing, Cis and Trans, in general requires a host of specific RNAs (for hybridizing) and associated proteins to bind to specific sequences in Introns and the splice junctions. in addition, few more specific factors are required for splicing in tissue specific manner for alternative form of splicing. Yet one finds that some RNAs by themselves, with assistance of some small molecules or without them, by assuming certain structural conformation, are capable of catalyzing cutting and joining by trans-etherification reactions. The mechanisms by which some of these RNAs perform self-splicing activities differ from one another. One kind belongs to Group I introns, and the other belongs to Group-II introns; and new kind of introns are added, they are group III and group IV. Data base search showed the presence of introns within an intron called ‘Twintrons’ have been discerned. Some Viroidal RNAs also show self splicing features, but use slightly a different mechanism in their splicing process, like hammer headed Ribozyme. It won’t be out of context if one calls these RNAs as Catalytic or enzymatic RNAs for they don’t have any other known functions except for locating in intervening sequences of mRNAs. The dogma that only proteins can perform catalytic activities is no more tenable. It is true that proteins can fold, undergo conformation into 3-D structure, they can provide an active site, a binding site for a specific substrate with a specific geometrical shape, they can be activated and they can be inactivated, and they obey certain kinetic laws and bring about catalytic activities. Yet the RNAs which have catalytic activities may not have those sophisticated structural and functional features; still perform some simple but very effective chemical reactions.
The samallest catalytic “enzyme is Proline or a Catalyst” Drs. Mohammed Movassaghi and Eric N. Jacobsen of Harvard University; even one can include ‘Lithium’. Dissecting the whole reaction cycle, authors note three features basic to enzymatic catalysis, namely specificity, speed and turnover. First is the binding or capture of the reactants on the protein surface. There is a built-in specificity to this. A large number of metals act as ‘catalysts’. Proline catalyses the aldol condensation of acetone to various aldehydes with high stereo-specificity (proline as an aldolase), that has set the pace. Is catalyst and enzyme are same or different?
Most of the pre mRNAs contain internal introns. The said introns are spliced with the help of spliceosome structures. Structurally some introns differ from the normal and prevalent introns which undergo cis-splicing. Most of the mRNA introns contain 5’ GU –AG3’ as splicing joints, but some plants have AU-AC introns.
Exon 1-I----------------------Intron-----------------------------I exon2
Mammals: ---I GURAGU----------------UNYURA*Y—py—--------YAG I---
Yeast: ---I GUAUGU—---UAYUAA*Y----py-------CAG I—
AT-AC: ---I AUAUCUU—UCYUUAA*Y—py—YCCAC I—
Trans: ---I GUAA-----------UAGUAA*Y—py—UUYAC I—
Plants : --IAUAUCCU---UCCUUA*A------------YCCACI
Group I: ----IUA ------------------XXXXXXX-----------------------UI----
Group II: ---IGUYG--------nnnnn—A*Y----py-------AG(Y) I—
Group III: ----IGu--// GU—nnA*Y---AG//---XXXA*X—AG I---
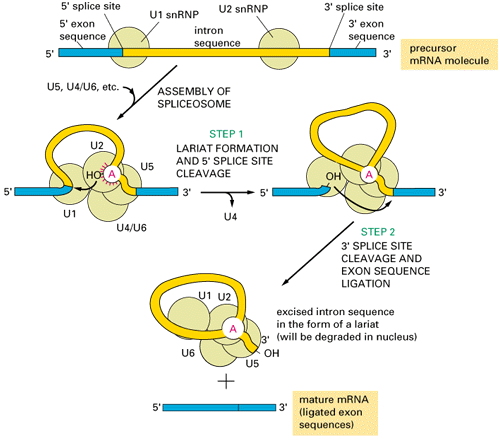
Pre mRNA cis-splicing with Gu—AG splice juctions:
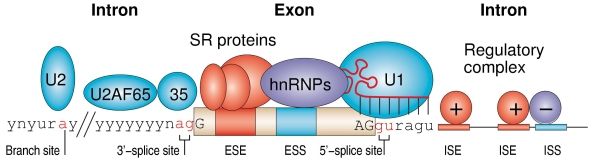
Splicing RNPs bound to specific sites along with sn RNAs/snRNPs ( snRNAs not shown except U1). Exon is very well defined by their 5’ and 3’ splicing sites, and ESE (SR), ESS (hnRNPs). On either side of exons one finds Introns with specific binding elements.
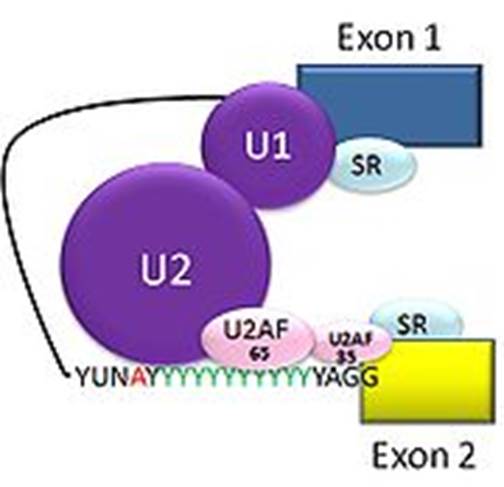
Cis splicing: Positioning of U1 and U2 RNAs and their associated RNps bring about exon-branch site close to 5’ splicing site firs phosphorolysis reaction and phosphor esterification reactions simultaneously for splicing ; which leads to the second reaction of splicing 3’ splicing site.
Cis splicing use snU1, U2, U4, U5 and U6 RNAs and their associated proteins. They assemble on to introns in sequence and perform splicing in sequential fashion.

Cis splicing of the ‘fas’ apoptosis receptor mRNA:
The figure just outlines Alternative Splicing components bound to mRNA
a) The 5' splice site downstream from exon 6 in the fas pre-mRNA has a weak agreement with the consensus sequence, and is not bound by the U1 snRNP. b) Binding of TIA-1 protein (T-cell intracellular antigen-1) to an intronic splicing enhancer site stabilizes binding of the U1 snRNP. The 5' donor site complex assists in binding of the splicing factor U2AF to the 3' splice site upstream of the exon. c) Binding of polypyrimidine tract binding protein (PTB) to the ure6 exonic splicing silencer in exon 6 prevents the 5' complex from assisting in U2AF binding.
In situations as in the above diagram a and c, exon 6 is skipped, giving an mRNA encoding a soluble protein product. In situation b, exon 6 is included, and the resulting mRNA encodes the membrane-bound isoform of fas protein, which stimulates programmed cell death (apoptosis).
Self splicing introns:
Group I introns (selfish Group introns):
(Lambowitz et al., 1999, (Haugen et al., 2005).
Discovery of self-splicing in T. Cech’s lab, 1981
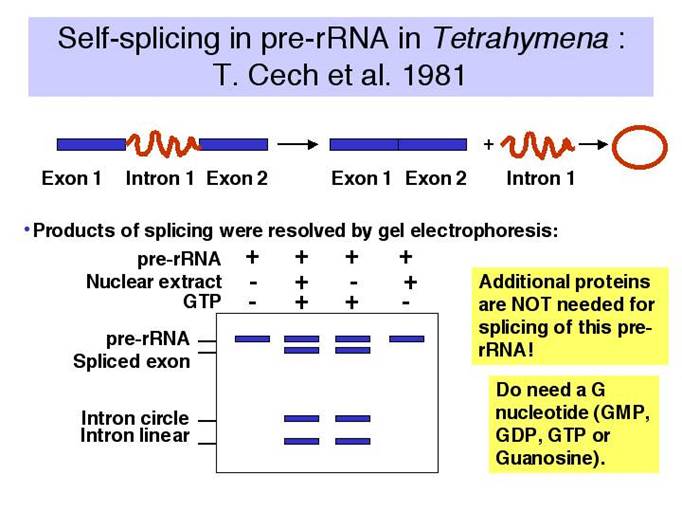
Tetrahymena thermophila, a ciliate and Physarum polycephalum (slime mold), transcribe their nuclear ribosomal RNA, whose size is 35S and consists of 16s and 5.8s and 26 s segments in the same order.
Group I intron splicing events; splicing requires just Guanosine nucleotide, which binds to specific sequence elements in the intron of the rRNA that act on the 5’ site of the intron and cleaves and forms a covalent bond at the 5’end to generate another splicing event.
Group I introns are found in bacteria, lower eukaryotes and higher plants. However, their occurrence in bacteria seems to be more sporadic than in lower eukaryotes, and they become prevalent in higher plants. The genes that group I introns interrupt differ significantly: They are found in rRNA, mRNA and tRNA genes in bacterial genomes, as well as in mitochondrial and chloroplast genomes of lower eukaryotes, but only invade rRNA genes in the nuclear genome of lower eukaryotes. In higher plants, these introns seem to be restricted to a few tRNA and mRNA genes of the chloroplasts and mitochondria. Both intron-early and intron-late theories have found evidences in explaining the origin of group I introns.
Some group I introns encode homing endonuclease (HEG), which catalyzes intron mobility. It is proposed that HEGs move the intron from one location to another, from one organism to another and thus account for the wide spread of the selfish group I introns. It is true that no biological role has been identified for group I introns thus far except for splicing of themselves from the precursor to prevent the death of the host that they live by. A small number of group I introns are also found to encode a class of proteins called maturase that facilitate the intron splicing and transposition.
These sub rRNA segments are processed by RNase-III and RNase D and other enzymes, but the 26s RNA segment contains an additional 414 to 416 ntds long region which is not processed by the above said enzymes. In the presence of such 416 ntds long RNA within renders rRNA non functional nor has any meaning to the functional rRNA for forming ribosome, so this segment is considered as Intron and the segments found on either side of the Intron are called Exons.
· This part of the RNA i.e. Intron segment undergoes elaborate, but predefined secondary structural organization and uses some 5’ end of Intron conserved guide sequence to pair with certain conserved sequences of 3’ end of the first Exon.
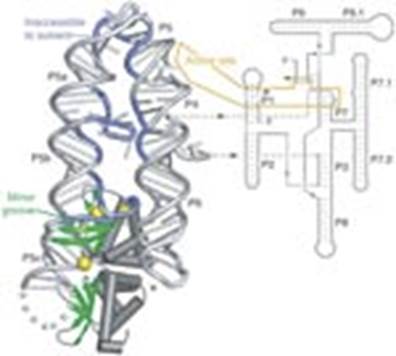
· Most of the group Intron-I generate at least nine secondary structure named as p1 to p9.
- Figure 3.3.11.

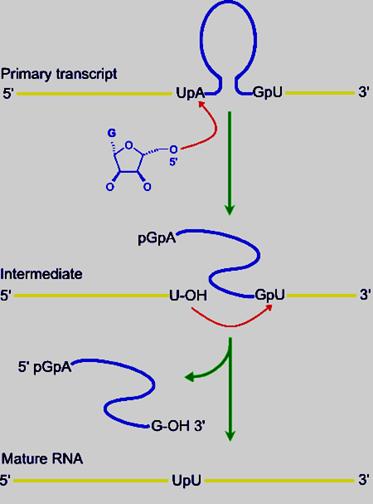
· RNA involvement in self‑splicing is stoichiometric, but the excised intron does have a catalytic activity in vitro. The 3‑D structure of the folded RNA is responsible for the specificity and efficiency of the reaction (analogous to the general ideas about proteins with enzymatic activity). The specificity of splicing is caused, at least in part, by base‑pairing between the 3' end of the upstream exon and a region in the intron called the internal guide sequence. The initiating G nt also binds to a specific site in the RNA close to the 5' splice site. Thus two sites in the pre-rRNA intron are used sequentially in splicing.
Exon 1 1 st---------------------//intron//---------416th (421)- Exon2
------CUCUCU [ UA-------UUUACCCU AUG----GAGG ] GU I U-------
Spliced rRNA: -------ex1.p U p U.exon2--------
1st 16 20 416th ntd
Released intron: 5’pppGAp-----------A----A----------G3’OH
The number of Ps at the 5’ end of the G can one, or two or three depending upon what type of G’ is used
· Furthermore this secondary structure also brings the 5’ end of the second Exon closer to the 3’ end of the first Exon. It also generate IGS Internal Guide Sequence that actually base pairs with 3’region of the first exon.
· In addition, the secondary structure also produces a pocket to hold a cofactor such as Guanine nucleotide (either in the form of GTP, GDP or GMP or a just a Guanosine) and the cofactor binds to the site (pocket).
· Because of the secondary structural conformation, the guanine nucleotide is virtually placed on the first nucleotide ‘U’ of the intron. The 3’OH^- group of the Guanosine with negative charge attacks nucleophilically and performs a cleavage of the bond between the last nucleotide of the first Exon and the first nucleotide ‘U’ of 5’ end of the intron; simultaneously trans-esterification reaction takes place between 3’OH group and the 5’P of A nucleotide of the 5’ end of the intron. In this reaction it generates first Exon with 3’OH group free.
· This reaction leads to further conformational readjustments in such a way the 3’OH of the first Exon is brought very close to the 3’ splice site. This brings about another trans-esterification reaction by the nucleophile in which the first Exon and the second Exon are spliced.
· Important features of Group I introns is that there are no conserved sequences in splicing sites. Some internal guide sequences help in pairing of a small segment of 5’ intron with 3’ the first Exon.
· Secondary structure produces an active site for G binding and the OH group of guanine nucleotide performs first trans-esterification and cleavage reactions.
· In this process, how the secondary structure develops is important and to generate such structure, it should be endowed with segments of complementary nucleotide sequences. The secondary structure further folds into 3-D to form a structure similar to ribozyme. This acts like an enzyme in splicing events of 413-416 ntds long intron.
· The first reaction cleaves at 5’ of the intron. Second cleaving at the 3’ end of the intron results in freeing the intron and also joining two Exons. Excised intron may become a lariat structure and again by autocatalysis it can become linear.
· Mutations in consensus sequences abolish splicing. If such intron sequences are introduced in the middle of E.coli beta Galactosidase gene, the intron in the transcript gets spliced out and functional βGalactosidase is produced as functional product. This happens in bacteria where there is no scope for eukaryotic type of splicing, which clearly proves the point that such intronic RNAs are capable of performing self splicing reactions anywhere.
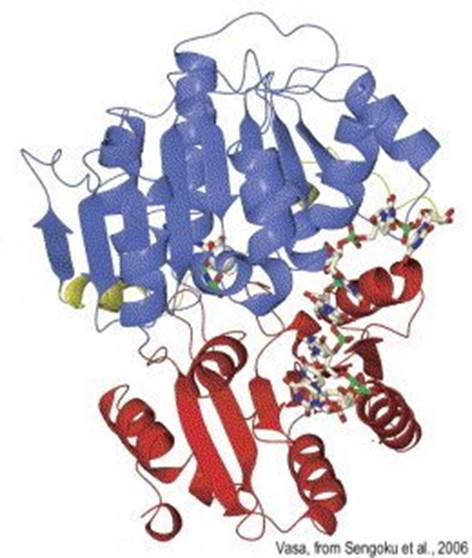
DEAD box proteins-RNA helicases are required for RNA conformational changes.
· Such group-I introns are also found in fungal mitochondrial genes.
· By introduction of introns in the middle of cloned eukaryotic gene, it has been found that the expression of the gene product enhances by 16 to 50 fold. This phenomenon is called Intron-enhancement.
· Intron-1, derived from maize alcohol dehydrogenase, when introduced in the middle of CAT (chloramphenicol acetyl transferase), a reporter gene, and into NPT-II gene with CAMV 35 promoter, the expression of their respective products has been found enhanced by 16 to 15 fold respectively.
· In Neurospora crassa, mitochondrial cytochrome-18 mRNAs have group-I introns. Mutations in this region have failed to produce functional enzyme. In this mRNA, it is assumed that the intron assumes tyrosine tRNA’s secondary or tertiary structure. To remove such introns either tyr-tRNA synthase binds or mere binding may induce self-splicing activity.
· Yeast mitochondrial mRNA for Cytochrome oxidase-b, called Cyt.B-ox exhibits successive splicing reactions to produce an Exon with 144 codons. During translation it reads through into an intron of 840 bases (280 codons) up to 70 % of it, as an open reading frame. However translation is terminated when it encounters a TER codon in the last part of the intron. The translated protein is often called Maturase.
· It is believed that this Maturase is responsible for splicing reaction of its own intron. The intron is assumed to produce secondary and tertiary structure similar to that of intron of nuclear rRNA of Tetrahymena or Physarum. Changes in the intron sequences fail to splice. However, the protein produced is directly involved in splicing. The actual splicing reaction is performed by the secondary structure of intron RNA itself and the protein Maturase only helps the secondary structure to be stable.
Group-II introns:
Group II catalytic introns are found in rRNA, tRNA and mRNA of organelles in fungi, plants and protists, and also in mRNA of bacteria. They are large self-splicing Ribozyme and have two structural RNA domains (usually designated dI to dVI). The second domain contains six extensions as domains. This model and alignment represents only domains V and VI. A subset of group II introns also encode essential splicing proteins in intronic ORFs. The length of these introns can therefore be up to 3kb. Splicing occurs in almost identical fashion to nuclear pre-mRNA splicing with two transesterification steps. The 2’ hydroxyl of a bulged adenosine in domain VI attacks the 5’ splice site, followed by nucleophilic attack on the 3’ splice site by the 3’ OH of the upstream exon. Protein machinery is required for splicing in vivo, and long range intron-intron and intron-exon interactions are important for splice site positioning. Group II introns are further sub-classified into groups IIA and IIB which differ in splice site consensus, distance of bulged.
Group II intron is a class of self-catalytic ribozymes and retroelements found in rRNA, tRNA, mRNA of organelles in fungi, plants, protists, and bacteria. Self-splicing occurs in vitro (for a few of the introns studied to date), but protein machinery is probably required in vivo. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with a branch point strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is thought that pre-mRNA splicing (see spliceosome) may have evolved from group II introns due to the similar catalytic mechanism as well the structural similarity of the Domain V substructure to the U6/U2 extended snRNA.
The secondary structure of group II introns is characterized by six typical stem-loop structures, also called domains I to VI or D1 to D6. The domains radiate from a central core that brings the 5’ and 3’ splice junctions into close proximity. The proximal helix structures of the six domains are connected by a few nucleotides in the central region (linker or joiner sequences). Due to its enormous size, the domain 1 is divided further into sub domains a, b, c, and d. Sequence differences of group II introns were identified which led to a further division into subgroups IIA and IIB. Group II introns also form very complicated RNA Tertiary Structure.
Group II introns possess only a very few conserved nucleotides, and the nucleotides important for the catalytic function are spread over the complete intron structure. The few strictly conserved primary sequences are the consensus at the 5’ and 3’ splicing site (...↓GUGYG&... and ...AY↓...), some of the nucleotides of the central core (joiner sequences), a relatively high number of nucleotides of D5 and some short sequence stretch of D1. The unpaired adenosine in D6 marked by an asterisk (7 or 8 nt away from the 3’ splicing site, respectively) is also conserved and plays a central role in the splicing process.
In 2005, A. De Lencastre et al. found that during splicing of Group II introns, all reactants are preorganized before the initiation of splicing. The branch site, both exons, the catalytically essential regions of D5 and J2/3, and epsilon-epsilon’ are in close proximity before the first step of splicing occurs. In addition to the bulge and AGC triad regions of D5, the J2/3 linker region, the epsilon-epsilon’ nucleotides and the coordination loop in D1 are crucial for the architecture and function of the active-site.
Some fungal mitochondrial genomes, plastid genomes and some bacterial genomes show resemblances to some eukaryotic nuclear RNA’s splice end sequences or splice junctions, such as 5’GT------Tpy3’, and the branching sequence is TACTA A*C. The sequences are exercised as lariats. Splicing is autonomous. Group I introns require G residues, but group-II intron don’t require even that, but Mg2+ and high temperature enhances splicing activity.
Secondary structure of group II intron RNA.
The secondary structure of the RNA consists of six domains radiating from a central wheel. The ORF is located in the loop of domain 4. Exons are blue; intron is red; RT ORF is green. The exons pair with three sites in the intron to position them for splicing.
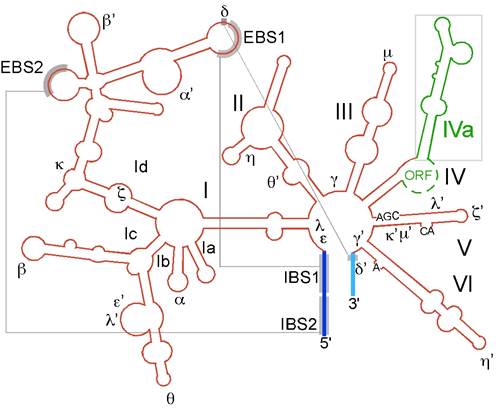
The largest domain of the ORF is the reverse transcriptase (RT) domain; if such group-II intronic RNAs are incubated under in vitro conditions, the RNA gets spliced out as a lariat automatically, that too with out any additional factors. They have certain conserved sequences located at just apposite to splicing sites and branching sites; this leads to base pairing, which facilitates splicing.
When conformational structures form, they fold in such a way the 2’OH group of the penultimate nucleotide of the branch site, which is invariably the A, is brought into close proximity with the 3’ end of the first Exon.
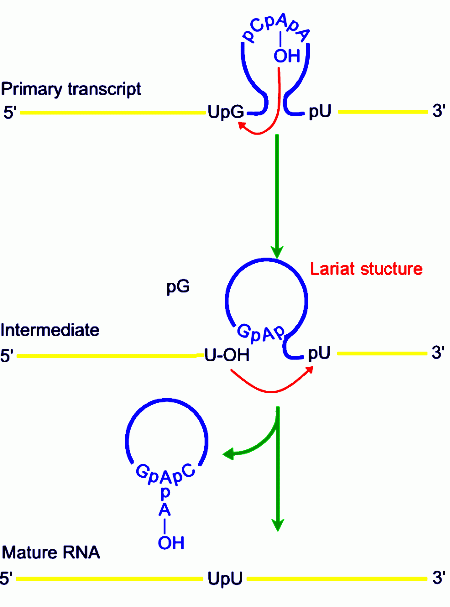
· This positioning of the 2’OH to the first Exon activates nucleophilic reactions like cleaving and trans-esterification, where 5’end of the intron forms covalent bond with 2’OH of branching site to produce a lariat.
· This cleaving furthers conformational adjustments, which bring the 3’OH group of the first Exon closer to the 5’ end of the second Exon, which results in another cleavage and trans-esterification reactions, resulting in the release of intron in the form of a lariat and Exons spliced in frame.
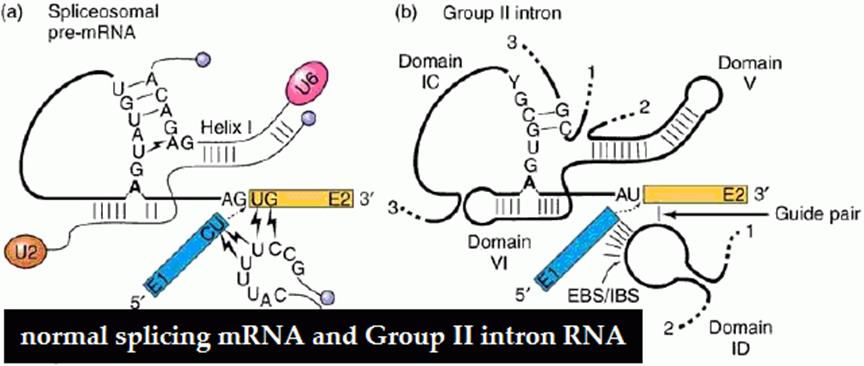
Compare the structural features of Group II and normal introns and splicing reaction; which shows they are more or less similar and shows an evolutionary trend.
Precursor:
---exon1----I GU------TACTA A* C---A py or AG I—exon2----
Spliced: -----Ex1------------I-P-I------ex2--------
· Some, group-II introns found in yeast ribosomal RNA genes have reading frames, if they are translated they generate a protein, which has a site specific endonuclease activity, as well as reverse transcriptase property and maturase functions, which help in splicing out the intron and reverse transcribe it and insert it into another gene; thus they exhibit intron mobility. Here the protein generated has N-terminal Reverse Transcriptase domain, central Maturase domain and Endonuclease domain at C-terminal. These are involved in homing Introns to other sites.
Protein generated:
H3N—RT—Maturase---Endonuclease—COO-
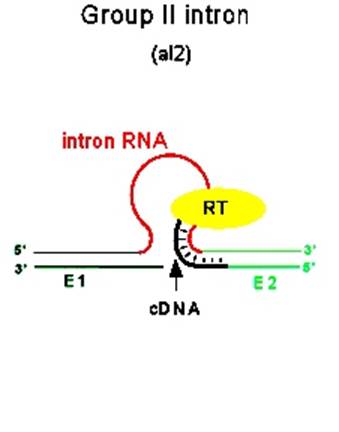
Introns are used for reverse transcription and the same (i.e.) copied one is used for transposition; both the diagrams one above and the other below depict the same event.
· In yeast w^+/W^- have such introns, that is why, when w^+ and W^- strains are mated, only w^+ strains are produced as offspring.
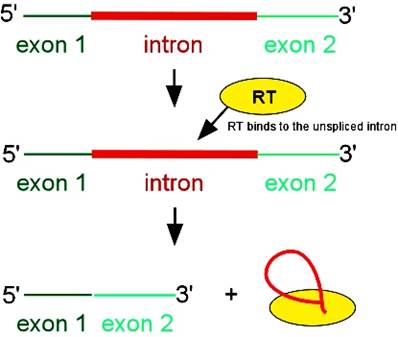
· They exhibit self-propagation property and also home introns in places or sites they chose. Such copying and transpositions can result in duplication and if such transpositions happened to be into one of the functional gene, then it can be made non functional.
· This type of intron activity is also observed in T4 phage ‘td’ intron, where its ‘td’ intron gets inserted into a 24 bp region upstream of its insert.
Maturase
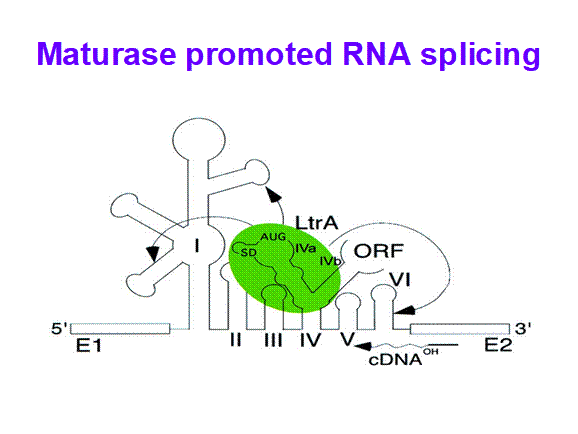
· In plants, some chloroplast genes for rRNA, tRNA and several protein coding genes are split.
It has been estimated that at any given time there are around 150,000 mRNAs in each mammalian cell. Since the average human pre-messenger RNA has about 7-8 introns, and considering the rapid rate of turnover of the cellular mRNA pool, it is hardly surprising that a major portion of cellular resources are devoted to ensure the accuracy and speed of splicing reactions. Indeed, each cell contains roughly a million copies of each of the snRNPs (small nuclear ribo-nucleoproteins), the basic functional subunit of the spliceosome, which perform the removal of introns from pre-messenger RNAs.
Spliceosomes are highly complicated machines: they contain close to 200 different components that assemble in a highly dynamic and elaborate fashion to perform the splicing reaction. Despite their critical role in cellular physiology and their involvement in many human diseases, the mechanism of function of this complex cellular assembly remains largely unknown. This is mostly due to the daunting complexity of the spliceosome, which prevents the traditional biology methods from being applied to its study.
To solve this problem, and at the same time, to gain insight into the mechanism of human pre-mRNA splicing, authors took a highly minimalistic approach. Previous studies had shown a critical role for the spliceosomal snRNAs in splicing catalysis, and similarities to ribozymes found in lower eukaryotes suggested that the spliceosome, also, might be an RNA enzyme. We chose just two of the snRNAs (out of over 200 RNA and protein factors in the spliceosome) which were shown to be at the heart of the active spliceosomes and tried to reconstitute the catalytic active site of the spliceosome in vitro. We could show that these two RNAs, U6 and U2, are inherently able to form the base paired structure they form in vivo without the help of any additional factors. Thus, the ability to form the functionally active structure is embedded in the sequence of these two RNAs. Similarity of self-splicing group II intron ribozymes (left) and the spliceosomal RNAs (right). Functionally similar domains are highlighted in the same color.
Next, authors of this informative article, tested them to see if they show biologically relevant activity, that is, if they can perform splicing. A number of activity assays indicated that the U6/U2 base paired complex could indeed show catalytic activity that was related to the splicing reaction, but differed from the authentic splicing reaction. Very recently, authors succeeded in obtaining splicing-like catalysis from these two RNAs: upon incubation with suitable pre-mRNA-like substrates, the U6/U2 base paired complex catalyzed two consecutive reactions on the substrates that resembled the first and the second steps of splicing
The catalytically active U6/U2 complex allows to pursue a number of questions: how does the spliceosome active site recognize its substrates? Can the same active site catalyze both splicing steps? What factors other than the snRNAs are involved in recognizing the splice sites and ensuring the accuracy of the reaction? Since the reaction we observe in our system is very inefficient, it provides us with an ideal system to study the effect of other spliceosomal factors on the efficiency of the reaction in a simple, minimal functional system that is amenable to detailed study. Authors of this passge believe that the in vitro-assembled U6/U2 complex opens the door to the study of many fundamental questions in splicing that otherwise could not be addressed.

Similarity of self-splicing group II intron Ribozyme (left) and the cis-spliceosomal RNAs (right) can be observed in the above diagram. Functionally similar domains are highlighted in the same color.
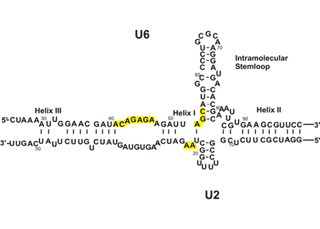
The catalytically active U6/U2 complex allows pursuing a number of questions: how does the spliceosome active site recognize its substrates? Can the same active site catalyze both splicing steps? What factors other than the snRNAs are involved in recognizing the splice sites and ensuring the accuracy of the reaction? Since the reaction we observe in our system is very inefficient, it provides us with an ideal system to study the effect of other spliceosomal factors on the efficiency of the reaction in a simple, minimal functional system that is amenable to detailed study. It is believed that the in vitro-assembled U6/U2 complex opens the door to the study of many fundamental questions in splicing that otherwise could not be addressed.
GroupI and GroupII act as ribozymes, yet they require RNPs for efficient folding to act as catalytical active structures in splicing; Alan M. Lambowitz and Mark G. Caprara, Steven Zimmerlyand and Philip S. Perlman 1999.
Notwithstanding of what described above, there are group II introns in bacterial systems. Database for bacterial group II introns by Manuel etal (august 30, 2011). The Database for Bacterial Group II Introns (http://webapps2.ucalgary.ca/~groupii/index.html#) provides a catalogue of full-length, non-redundant group II introns present in bacterial DNA sequences in GenBank. The website is divided into three sections. The first section provides general information on group II intron properties, structures and classification. The second and main section lists information for individual introns, including insertion sites, DNA sequences, intron-encoded protein sequences and RNA secondary structure models. The final section provides tools for identification and analysis of intron sequences. These include a step-by-step guide to identify introns in genomic sequences, a local BLAST tool to identify closest intron relatives to a query sequence, and a boundary-finding tool that predicts 5′ and 3′ intron–exon junctions in an input DNA sequence. Finally, selected intron data can be downloaded in FASTA format. It is hoped that this database will be a useful resource not only to group II intron and RNA researchers, but also to microbiologists who encounter these unexpected introns in genomic sequences.
· Group Twintrons: Intron within an Intron, ex. Group III Twintron in Euglenoid species..
· Such introns are found in chloroplast genome of Euglena gracilis, which actually contains74 group II introns (277-671ntds) and 47 group III introns (91-119ntds).
· Most of the group III introns are small with 95-110 ntds in size.
· They are degenerate version of group II introns.
· They lack conserved secondary structural features of Group II such as domains from I to VI, but contain V branches where one finds A’ branching site.
· They are located in euglena chloroplast DNA.
· These are rich in A/T sequences, they have conserved 5’ boundry sequences
· They are spliced through lariat formation.
· Interestingly some of the group III introns are located in the intercistronic polycistronic gene of euglena gracilis chloroplast genome.
· The factors such as U1, U2, U4, U5 and U6 snRNA-RNPs are required for splicing.
· Splicing is done by two transestirification reactions similar to cis splicing entrons.
Group III intron splicing:
Compare the figures for the basic mechanism and the components involved in self splicing

Group III Introns
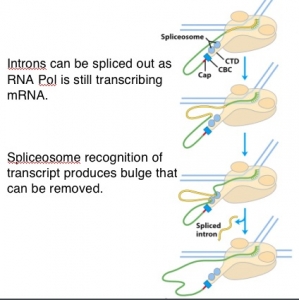
Group III intron slicing (not clear from the figure), they are processed while they mRNA is transcribed by splicing complexes found bound to CTD tail of RNA pol II
Group II splicing requires spliceosomes.
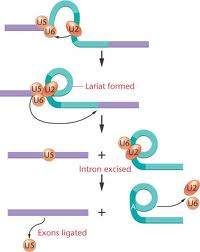
Splicing uses snRNAs required for cis splicing and generates lariat form introns
Splicing of group III require splicing components such as U1, U2, U4, U5 and U6 snRNA and their associated protein complexes. Number of branches is mostly one similar to group II VI branch and the branching site is in the sixth branch.
Twintrons:
Twintrons: Intron within an intron: Twintrons are presumed be developed by the insertion of mobile introns into any of the existing intron-so the name intron within an intron. Twintron was discovered by Donald W. Copertino and Richard B. Hallick as a group II intron within another group II intron in Euglena chloroplast genome , In 1993 a new type of complex twintron composed of four individual group III introns has been characterized (Wikipedia). Twintrons are introns-within-introns excised by sequential splicing reactions. A new type of complex twintron comprised of four individual group III introns has been characterized. The external intron is interrupted by an internal intron containing two additional introns. This 434 nt complex twintron within a Euglena gracilis chloroplast ribosomal protein gene is excised by four sequential splicing reactions. Two of the splicing reactions utilize multiple 5'- and/or 3'-splice sites. These findings are evidence that introns with multiple active splice sites can be formed by the repeated insertion of introns into existing introns (R.G Drager and RBHallick). Some labs have found that an intron found tucked with in an intron; their splicing is similar to normal splicing, but the internal intron is removed first and the external segments of introns are joined. Then the outer intron sequences as a segment is removed. A very good example is rpo C1 subunit of RNAP. Such introns are found in Euglena gracilis PSII gene. A twintron of 1605 ntds long consists of two group III introns coding for a 458aa long protein mat1. It is not surprising that some myxomycetes contain GroupI twintrons. A group I twintron consists of two distinct ribozymes (catalytic RNAs) with different functions in RNA processing, and an open reading frame encoding a functional homing endonuclease--all with prospects of application as molecular tools in biotechnology. Updated RNA secondary structure models of group I twintrons, as well as an example of in vitro ribozyme activity, are presented ( Einvik C, Elde M, Johansen S (1998).
I---exon1--1---------I-----Twintrons----I-----I—exon2----
Intron with in an intron is removed, and then the remained intron segment is removed as done in normal splicing
I---exon1—I---------IpI------I—exon2----
I---exon1—-p—exon2---I—
In plants, the size of introns generally of 100 to 150 ntds long, it is same with Drosophila and Caenorhabditis. But introns contain AT rich (74%) sequences and Exons also contain some AT rich (54%) sequences. In monocots introns are 59%AT rich and Exons 44% AT rich.
Richard J. Roberts and Philip Allen Sharp are the discoverers of ‘introns’, for which they were awarded Nobel Prize in 1999.
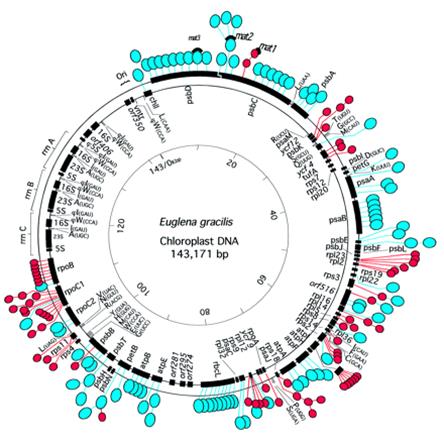
Introns in chloroplasts- red color regions contain introns
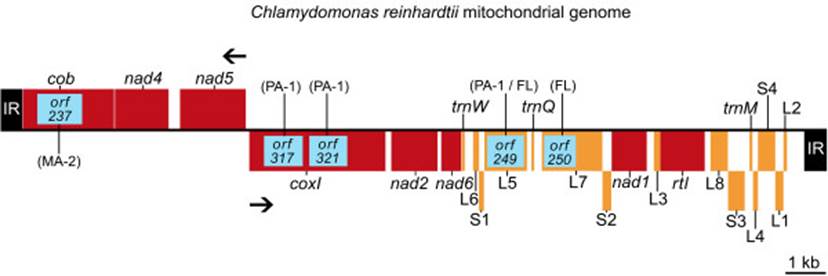
Introns in mitochondrial genomes- Odogonium cardiacum
Fig. The Prospero pre-mRNA is alternatively spliced. (A) Diagram (not to scale) of the twintron, which constitutes the second Prospero intron. U2- and U12-type splice sites and the PRE are indicated. (B) Sequences of the PRE RNA (Upper) and a mutant PRE RNA (Lower) used for the competition experiments shown in Fig. 2A.
Twintrons are introns-within-introns excised by sequential splicing reactions. Twintrons are presumably formed by the insertion of a mobile intron into an existing intron.
Twintron was discovered by Donald W. Copertino and Richard B. Hallick as a group II intron within another group II intron in Euglena chloroplast genome.[1] They found that splicing of both the internal and external introns occurs via lariat intermediates. Additionally, twintron splicing was found to proceed by a sequential pathway, the internal intron being removed prior to the excision of the external intron.
Since the original discovery, there have been other reports of Group III twintrons and GroupII/III twintrons in Euglena gracilis chloroplast. In 1993 a new type of complex twintron composed of four individual group III introns has been characterized. The external intron was interrupted by an internal intron containing two additional introns. In 1995 scientists discovered the first non-Euglena twintron in cryptomonad alga Pyrenomonas salina. In 2004, several twintrons were discovered in Drosophila.
Group IV- Introns found in tRNAs:
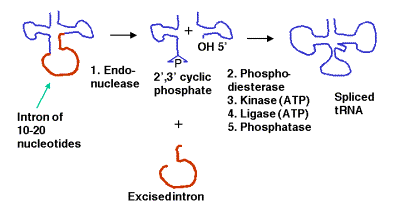
These short introns are removed in a series of steps catalyzed by enzymes. The enzymes include an endonuclease, a kinase and a ligase. Because the endonuclease generates a 2’, 3’ cyclic phosphodiester product, an additional phosphodiesterase is needed to open the cyclic phosphodiester to provide the 3’ hydroxyl for the ligase reaction. In addition, the 2’-phosphate (product of the phosphodiesterase) must be removed by a phosphatase.
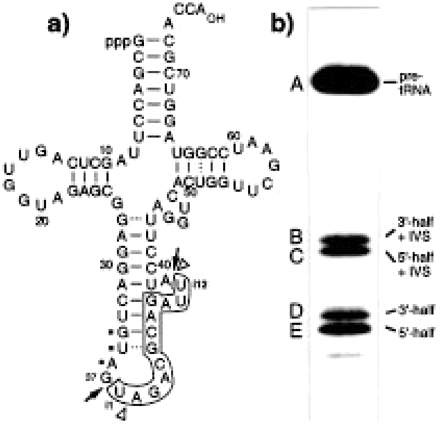
Fig:Structure and self-cleavage of pre-tRNATyr AtY3II*-T7 synthesized by in vitro transcription with T7 RNA polymerase. (a) The intervening sequence from i1 to i12 is boxed. Dots identify the anticodon. Arrows indicate the enzymatic 5′ (between nt 37 and intron position i1) and 3′ (between nt i12 and 38) splice sites. The sites of self-cleavage are indicated by arrowheads. (b) Self-cleavage of pre-tRNA AtY3II*-T7 in the presence of 100 mM NH4OAc, pH 7, 10 mM MgCl2, 0.5 mM spermine and 0.4% Triton X-100. The 32P-labelled pre-tRNA was incubated at 37°C for 4 h. A, pre-tRNA (88 nt); B, 3′-half of the tRNA with the intron without iU1 (50 nt); C, 5′-half of the tRNA with the intron (49 nt); D, 3′-half without the intron (39 nt); E, 5′-half without the intron plus iU1 (38 nt). Cleavage products were analysed on a 10% polyacrylamide-8 M urea-TBE gel. Another Heritage from the RNA World: Self-Excision of Intron Sequences from Nuclear Pre-tRNAs; Ute Weber, Hildburg Beier and Hans J. Gross.
Mirtron:
An addition to the above self splicing introns another called Mirtrons is added to the list Mirtrons contain contains miRNA insert in the intron found in Drosophila melanogaster and Caenorhabditis elegans. Mirtrons arise from the spliced-out introns and are known to function in gene expression (WIKI).
Canonical miRNA and mirtron pathways in Drosophila. Key protein families include RNase III endonucleases (Drosha and Dicer-1), double-stranded RNA-binding domain proteins (Pasha and Loqs) and Argonaute effectors (AGO1 and AGO2). Canonical miRNA precursors are cleaved by the Drosha/Pasha complex in the nucleus, cleaved again by the Dicer-1/Loqs complex in the cytoplasm, and predominantly loaded into AGO1. Mirtrons are short hairpin introns that use the splicing and debranching machinery to bypass the requirement for Drosha/Pasha but are subsequently processed by Dicer-1 to generate miRNA-class regulatory RNAs, A Drosophila pasha Mutant Distinguishes the Canonical MicroRNA and Mirtron Pathways.