Capping and Poly-Adenylation:
The nascent pre mRNA in eukaryotes is long contains various elements which are to be cut and processed within the nucleus; and it also undergoes various modifications fit to be translated into a functional protein.
Pre mRNA goes through various processing steps to generate functional/translatable RNA; http://www.sequencecomputogy.org
Capping and its importance:
- In eukaryotes as the pre-mRNA emerges, during transcription, the 5’ end is free, if allowed it to be free; it will be soon degraded by 5’ exonucleases. It aids in mRNA transportation. It binds to Poly-A binding proteins through a factor called Cap binding factor G and enhance the rate of translation.
Capping complex associated with phosphorylated RNAPII-CTD tail, performs capping at the 5’ end of the pre-mRNA. The CTD tail has the repeats of seven amino acids- Y1S2P3T4S5P6S7 ; the S. Y and T are the sites for phosphorylation. This sequence is repeated 22 to 52 times in RNAPII enzymes.

a | TFIIH-mediated phosphorylation of Ser5 of the carboxy-terminal domain (CTD) of RNA polymerase II (Pol II) occurs on pre-initiation complex formation or before promoter-proximal pausing1. DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) probably facilitate Pol II pausing in the promoter-proximal region, and TFIIS also associates with the paused polymerase. TFIIS stimulates the intrinsic RNA-cleavage activity of Pol II to create a new RNA 3'-OH in the Pol II active site after backtracking of the polymerase. Capping enzyme associates with the Ser5-phosphorylated CTD and with Spt5), and the nascent RNA becomes capped during this first stage of elongation. The RNA cap is formed by addition of a methylated guanosine to the 5' end of the RNA through the action of capping enzyme and an RNA (guanine-7) methyltransferase. b | Positive transcription-elongation factor-b (P-TEFb)-mediated phosphorylation of DSIF, NELF and Ser2 of the Pol II CTD stimulates productive elongation, and the capping enzyme might contribute to this process by counteracting the negative effects of DSIF and NELF. TFIIS facilitates efficient release of Pol II from the pause site by aiding the escape of backtracked transcription complexes. NELF dissociates from the transcription complex and DSIF, TFIIS and P-TEFb track with Pol II along the gene. TFIIF, eleven-nineteen lysine-rich in leukemia (ELL), and elongin, which stimulate Pol II elongation activity, might also associate with the elongation complex; Abbie Saunders et al; http://www.nature.com/
As the 5’ end of the pre mRNA emerges out of RNAP II complex, the CTD bound capping complexes add Guanine (G-CH3-P`P) to the first pA or pG (N) of the precursor mRNA. The capping enzyme complex CEC binds to the RNP II before transcription gets initiated. As soon as the first nucleotide emerges out of the enzyme the CEC complex acts and transfer 7’methylated G-o-pop-oH. Eukaryotes have designed to have multisubunit complex associated with CTD complex tail. The complexes perform Capping, Splicing and poly-A addition as the mRNAs are transcribed. Capping of RNA with NAD+, NADH or 3’dephospho-coenzyme A is also observed in RNA population.
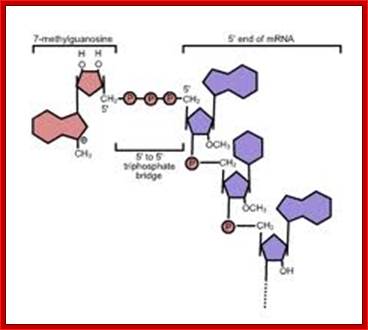
Process of 5’Capping- first 5’Phosphate is removed by 5’triphosphatase; then Guanylyl-transferase adds cleaved GTP i.e GDP resulting in 5’ to 5’ linkage; the 7’ nitrogen of the Guanine N7 methyl transferase using S-Adenosyl–L homo cysteine resulting in 5’cap called), this is called Cap-O. Adjacent nucleotides in the mRNA can be modified adding methyl adenosine to 2’O ribose cap cap1. Then similar methylation can be added to the second and third nucleotides of mRNA- thus called Cap1, cap2, and Cap3.
- Eukaryotes have devised processes to prevent mRNAs from exonuclease degradation from both 5’end and 3’end and stabilize the mRNA and to make it to be translation efficient.
- The latter is achieved by the addition of Guanine-phosphate to the 5’ end of phosphate of the first nucleotide (mostly Adenine), this is called the “cap”, then the Guanine is added with methyl group at 7’ position. Or 7’CH3-Guanine itself is added first.
- Capping regulates mRNA transport out of the nucleus, prevents 5’exonuclease degradation, promotes translation via binding to ribosomes and promotes 5’ proximal exon excision. Perhaps this process is facilitated by the cap looping around and interacting with spliceosome complex.
- The CBC cap binding complex binds to 7’CH3-cap of the 5’ mRNA; this recognizes the nuclear pore complex proteins and facilities mRNA export out of the nucleus.
- The complex is also recognized by the translation factors. The CBC eIF4E/eIF4G blocks the decapping enzymes. This increases the life time of mRNAs.
- Interestingly uncharacterized and undesirable mRNAs are transported into P-bodies for short time storage.

Pre-mRNA 5’ end maturation; Canonical 5’ RNA cap
synthesis pathway. RNA cap-first structures are conventionally synthesized by
the sequential γ-phosphate hydrolysis by an RTPase, GMP transfer by a GTPase,
N7-methylation by an N7MTase, and 2’O-methylation by a 2’OMTase. The
contribution of each substrate to the formation of the final 5’ RNA cap
structure is highlighted by a color code: pppRNA (black), GTP (blue) and SAM
(green and red); Frédéric
Picard-Jean, Maude Tremblay-Létourneau et al ; http://www.intechopen.com/
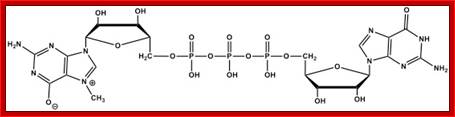
- 7’CH3 G 5’-P-P-P + P-P-P--5’A p N N N N—3’ + P~P and P.
- 7’CH3 G 5’-P-P-P-5’Ap-NNN- + P +P + P
- The enzyme that performs this reaction is Methyl-Guanylyl transferase. The capping involves 5’ to 5’ phosphate bonds.
- Enzymes required for this process are assembled on CTD tail of RNA Pol-II large subunit.
7’CH3 G-5’P-P-P-5’A- = Cap-0
2’O-CH3 A-- = Cap1
http://en.wikibooks.org/
7’CH3.G5’p.p.p+p.p.p5’A.n.n-à 7’CH3.G.5’ap.pb.pa.5’-A.n.n +PiPi + Pi
- In addition, more methyl groups are added to 2’OH group of the first nucleotide and then to the second and third.
- Added 7’CH3 G is termed as cap-O. Addition of methyl group to 2’OH of ribose of the first nucleotide is called cap-1, cap2 etc.
- Methylation of adenine at its 6’ position often takes place at a frequency of one in thousand molecules, which is also called cap-1.
- Any addition of methyl group to 2’OH of the ribose of the second and third is called cap-2, cap-3 and so on.
- The enzyme for adding methyl moiety to 2’OH group of the ribose is 2’-O-methylase.
7-CH3G Mono methylated Cap
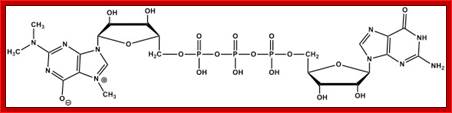
2’2’7CH3G Trimethylated Cap
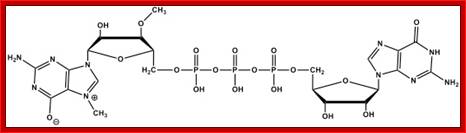
ARCA Anti reverse cap analogue.
In Orthomyxovirus viruses, addition of cap like structure to its RNA or (+) RNA is entirely different. Viral enzymes choose host’s capped mRNAs and cut the leader sequence with few more nucleotides at A or G, then adds this segment to its 5’ end of viral (+)-sense RNA; a process called “capping by molecular stealing”. In eukaryotes Cap structure is not just the part of pre-mRNAs; but cap structure is added many small Mol.wt. Sn RNAs though they are non-coding RNAs. Some viral RNAs use 5’VpG as the cap. Capping takes place very early during mRNA synthesis.
Cap Analogs –Enhance mRNA stability and translation efficiency (Jeno Bioscience):
Many eukaryotic and viral mRNAs are modified at their 5' ends by addition of 7‘-Methylguanosine (N7-methyl guanosine or m7G), known as "Cap". "Capping" of the mRNA structure plays a crucial role in a variety of cellular processes which include translation initiation, splicing, intracellular transport and turnover. Capped mRNAs are generally more efficiently translated in wheat germ and reticulocyte in vitro translation systems, and they are less susceptible to exonuclease degradation during microinjection experiments compared to uncapped mRNAs. However it is to be noted that the unmethylated cap analog shows no significant difference in its translation properties compared to the monomethylated cap analog.
In Ascaris lumbricoides transcription of the spliced leader RNA was catalyzed by RNA polymerase II, and the majority of the spliced leader RNAs synthesized in vitro possessed a trimethyl guanosine cap structure identical to that found on in vivo-synthesized spliced leader RNA.
It should be noted capping of mRNA can be RNA Cap Analog: 1. Unmethylated G, 2. G[5']ppp[5']G, 3. Monomethylated, m7G[5']ppp[5']G. Capping of mRNA can be done in an in vitro condition too.
Primitive eukaryotes like Caenorhabditis elegans produce mRNAs capped with either m7GTP or m32,2,7GTP. Caenorhabditis elegans also expresses five isoforms of the cap-binding protein eIF4E. Some isoforms (e.g. IFE-3) bind to m7GTP–Sepharose exclusively, whereas others (e.g. IFE-5) bind to both m7GTP- and m3 2, 2, 7 GTP–Sepharose; many viruses are have unconventional mechanism of Capping at the 5’ end of mRNA; Hiroshi Miyoshi2, Donard S. Dwyer3, Brett D. Keiper1, Marzena Jankowska-Anyszka4, Edward Darzynkiewicz, EMBO (2002).
Importance:
- Capping prevents 5’ degradation from 5’exonucleases.
- Capping provides stability to mRNAs.
- Capping facilitates the transport of mRNA into cytoplasm otherwise they remain in the nucleus.
- Capping enhances the efficiency of translation of mRNAs.
- Capping enhances the efficiency of splicing at 5’end introns.
- Capping with poly (A) provides synergism during translation.
- Luciferase mRNAs have been used to determine its half-life and translation efficiency with or with out cap and poly- (A) tail.
- Half-life of Luciferase mRNA without cap and without poly (A) is just 31 minutes, and translational activity is 2900 (as measured in terms of light emitted by ug of radioactive protein).
- But mRNAs without cap but with poly (A) tail shows half-life of 44 minutes. And its activity is 4480. The capped mRNA without poly (A) has half-life of 53 minutes and translation activity is 62000 a virtual 50% increase in its half-life and translational efficiency.
- The capped mRNA with poly- (A) tail, has a half-life of 100 minutes and its translational activity is 1,333 000; the relative effect of cap on its activity 200 fold.
- During translation mRNA cap and poly-A tail bind to each other through a protein eF4G and gets circularized.
Pre mRNA Transcription Termination Site and Processing 1:
The 3’ end of the mRNA genes has many regulatory elements which perform functions like stability of mRNA, elements for degradation of mRNA, transport to certain loci in the cell, elements that make mRNA as informosomes remain untranslated, making mRNA inactive for the storage; also has site for the binding of mi/siRNA for inactivating or degradation of mRNA and poly-A addition elements. But transcription of mRNA is terminated beyond these elements. The termination site of transcription varies from one gene to the other. The distance from poly-A site and termination site varies. Most of the pre mRNAs are cut at the 3’ region at specific site and poly-As are added. In some such as histone, 3’ processing is different for they don’t contain poly A tail but one of the Histone mRNA is added with Poly(A) tail, very unusual.
Eukaryote transcription termination distance from Poly-A site
|
Gene |
Distance from poly-A signal site |
|
D-globin |
600bp |
|
A-globin |
1500bp |
|
Globin-mice |
100-300 |
|
Globin-human |
100-300 |
|
DHFR-mice |
1000-2000 |
|
a-Amylase |
2000-4000 |
|
Ovalbumin |
800-1000 |
|
Gastirin |
192 |
|
|
|
Addition of poly- (A) Tail and Its Importance:
- With the exception of histone mRNA all other mRNAs contain a poly- (A) tail.
- Poly-A addition takes place while the 3’ end of the pre-mRNA is produced on the template.
- Termination site of transcripts of structural genes is 1000 and odd nucleotides downstream of 3’ translation terminator TER site.
- Transcriptional termination and poly-adenylation event are not linked and they are independent events. While the transcription termination at 3’ end provides facility for cutting at a particular sequence and poly-adenylation; this happens while the transcription is at its end, perhaps even after termination.
- The enzyme components required are again are found assembled at CTD tail of the large subunit of RNAP II.
- The pre-mRNA at its 3’ noncoding region contains certain sequences such as AAUAAA, at about 20 to 30 ntds from the poly-A start of the functional mRNA. This sequence is considered as poly-A signal.
- From data base of vertebrate mRNAs, certain consensus sequence has been established; any change in the sequence affects poly-A addition. The sequences A98%, A86%, U98%, A98%, A95%, A96%.
- In addition to poly-A signal, at 24 to 30-34 ntds downstream from the poly-A signal, another sequence GU and Us are found in pre-mRNA. The GU and Us rich sequence in mRNA is required for processing.
- Yeast cells don’t have poly-A signal sites; instead they have UA rich sequence in the upstream of poly-A start site.
- Even, plants have poly-A signals such as AAUAAA, but prokaryotes don’t contain Poly-A sequences at their 3’ ends.
- To perform poly-adenylation, the following factors and enzymes are required.
- Polyadenylation can be inhibited by Cordycepin (3’-deoxyadenosine)
CPSF: Cleavage and poly adenylation specific factor is a complex of four subunits such as 160kd, 100kd, 70kd and 30 kDa each. The complex binds to AAUAAA sequence, by doing so it specifies a nearby position of poly-A (adenylation) start site. This complex also interacts with Poly-A polymerase and cleavage stimulation factor (CStF).
CStF: Cleavage stimulation factor identifies and binds to GU n U rich site. This identifies the site for cleavage. This complex consists of 77, 64, 50 kDa protein subunits. This interacts with cleavage factors such as CF-1 and CF 2.
CF1 and CF-2: Cleavage factors CF1 and CF2 associate with CPSF and CStF. The enzyme consists of 68, 59 and 29 kDa subunits. These are cleavage enzymes.
PAP: The molecular mass of Poly-A polymerase is 82kd, a monomer. It is required for facilitating cleavage and catalyzing poly-A addition. For its activity, it also requires poly-A binding protein-II (PAB-II). The enzyme is 689 aa long and contains, N-terminal, RBD domain (RNA binding domain), PM domain (polymerase domain), and NLS1 and NLS2 sites for nuclear localization (?). They also have serine and threonine rich sites for phosphorylation.
PAB-II: This33 kDa protein, is the counter-part of cytoplasmic poly-A binding factor (PAP-I). The binding of this factor stimulate PAPs activity and also it regulates the length of poly-A tail and provides safety from 3’exonucleases.

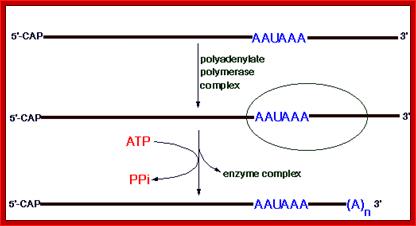
This diagram shows the process of Polyadenylation. http://www.web-books.com/MoBio; http://themedicalbiochemistrypage.org/
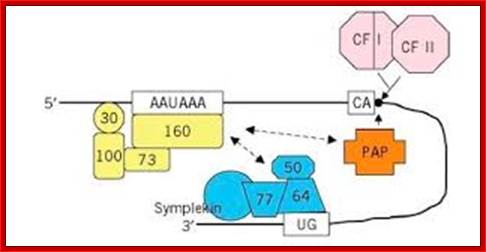
Poly-A adenylation components assembled at the site; http://what-when-how.com/
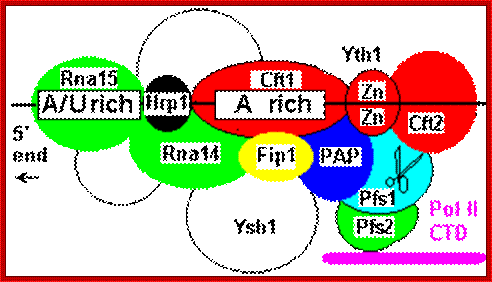
Poly adenylation in yeast cells: Yeast lacks a distinct AAUAAA sequence, and polyadenylation is associated with less well defined A/U rich and A rich sites. The CFPS equivalents are Cft1, Cft2 and Yth1. A factor comparable to CstF, Rna15/Rna14/Pfs2, binds upstream of CPFS a number of factors appear to be involved in binding downstream of the cleavage site as for mammalian CstF. Pap 1p is the polyA polymerase. Two-hybrid screens have shown it to interact with protein Fip 1p which in turn bridges to the CstF-like factor Rna14/Rna15, which then binds to a protein complex associated with an additional upstream A/U rich sequence marker. An hnRNP-like protein Hrp1 binds in between the CstF and CPSF equivalent factors. http://www.hixonparvo.info/
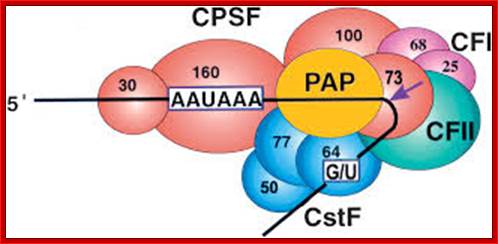
CPSF-73 as the 3′ processing nuclease: The core polyadenylation machinery (e.g., RNAP II is not included) is illustrated as it would assemble in a precleavage complex on an mRNA precursor, with CPSF-73 at the site of cleavage. The AAAUAA and GU-rich signal sequences are indicated, and the arrow denotes the cleavage site. The protein–protein and protein–RNA interactions depicted are consistent with available data; Kevin Ryan et al; http://rnajournal.cshlp.org/
Cleavage factors PfsIA (CstF), IB are involved in cleavage, but functions do not correspond exactly to the mammalian counterparts.
Mechanism:
- As the 3’end of the pre-mRNA is generated which is generally 200 or more nucleotide from the poly-A addition site, CPSF and CStF bind to their respective sequence sites i.e. poly-A signal and GU/U rich sequences respectively.
- These two interact in such a way the RNA in between is looped out and both CPSF and CStF are brought together. This interaction also brings cleaving sites together.
- At the same time CF1 and CF2 bind to CPSF and CStF in such a way they can easily locate the cleaving site and have access to RNA for cleavage.
- At this point, binding of the PAP to the complex stimulates the cleavage factors to cut.
- Then cleavage factors cut the RNA and CStF with the rest of the 3’ end of the RNA are released.
- Cleavage and poly-A addition is simultaneous and it is so quick that no one has isolated mRNAs with out poly-A tail so far.
- For PAP to be active, it requires stimulation from PAB-II, and then it leads to the addition of adenine nucleotides one by one. There is no template for synthesis of Poly-A RNA strand as an extension. This enzyme behaves just like DNA terminal nucleotide transferase.
- Initially adenylation up to 10 to 12 ntds is slow, and then poly-A addition is fast.
- As more and more poly-A are added more number of PAB-II associate.
- When the length of the poly-A tail reaches about ~250 ntds size, it stops, but PAB-II remains bound to the poly-A tail. In cytoplasm the PAB II is replaced with PAB I.
- Nuclear poly-A tail is about 220 to 250 ntds long and cytoplasmic tails are less than this.
- Poly-A tails present in eukaryotic mRNAs was discovered B. Darnel. When the RNA was digested with either RNase-A or RNase-T1, what was left was only ‘A’s. RNase-A cuts RNA after C’s and U’s, and RNase-T1 digests nucleotides after G’s. Poly-A addition is not inhibited by Actinomycin-D, which inhibits DNA dependent RNA synthesis.
- There are no such stretches of poly-Ts to 200 to 250 ntds long in DNA of a gene.
- Splicing of hnRNA starts even before poly-A addition initiation.
- Poly-A segment is subjected to constant turn over.
- There are many mRNA in which intron is used for poly adenylation.
Pre mRNA Transcription Termination Site and Processing 2:
Processing of Histone precursor mRNAs:
Transcriptional processing of histone transcripts requires specific factors and specific small molecular weight RNAs. Histone pre mRNA contain a cap at the 5’end and no poly (A) tail. Histone transcription is replication dependent. Histone mRNA is stored in frog oocytes and its translation is activated with fertilization. As the transcription of histone mRNA proceeds towards the end region of the gene, complementary base pair sequences generate a small stem loop (SL) structures followed by a specific sequence of nucleotides. However in the oocytes of Xenopus short oligo-As are added (by PRAPP) to histone mRNA at the end of stem loops. The presence of oligo-As and SLBP (31kDa) binding proteins (SLBP1 and SLBP2) prevent initiation of translation. The stored mRNAs in oocytes is bound by FRGY2 translation repressor proteins. SLBP2 is cytoplasmic and binds to histone mRNA when histone mRNAs are stored and remain inactive. It has RNA binding and RNA processing domains. However at meiosis the poly-A oligo’s are removed. It is how Xenopus histone mRNA is regulated in oocytes. Histone mRNAs of protozoa, plants, and yeast are polyadenylated (Chaubet et al. 1988). Interestingly histone mRNAs does not contain introns. In drosophila histone mRNA are stored in oocytes required for development. Even in frogs histone mRNAs are stored with SLBP2 bound. During early embryogenesis histone mRNA transcribed heavily for the development requirement. SLBP1 is required for histone translation activity. SLBP2 inhibits translation.
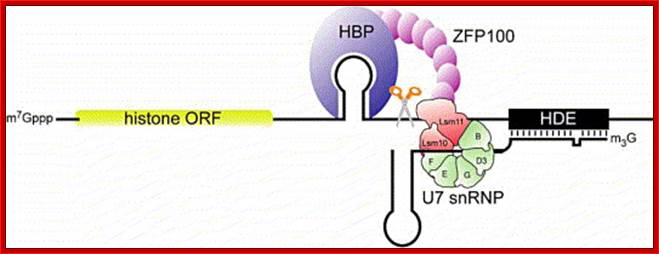
The U7 snRNA also contains a small stem loop and specific sequences at its 3’end, which are complementary to the terminal region of histone pre-mRNA transcript.
The U7 Sn RNA with its secondary structure base pairs with the 3’ region ACCCA of the transcript.
· As transcription proceeds beyond this region, a nuclear factor binds to 3’ end of the transcript and stops transcription and terminates the process.
The nuclear factor that contains a heat labile protein component binds to the stem loop structure of the transcript just upstream of the cleavage site. Transcriptional termination of histone transcripts requires specific factors and specific small molecular weight RNAs. As the transcription moves to the end region of the gene, complementary base sequences generate stem loop structures followed by a specific sequence of nucleotides.
Another factor U7 SnRNA/snRNPs which is 56 ntd long also contains a stem loop structure at its 3’ end and a 7’methylated Cap at the 5’end. It also contains sequences at its 5’end, before its stem loop, which are complementary to the terminal region called HDE (Histone Downstream Elements) of histone pre-mRNA transcript.

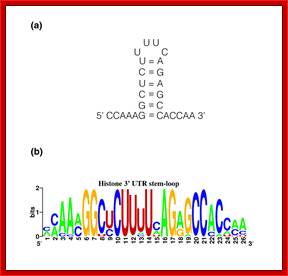

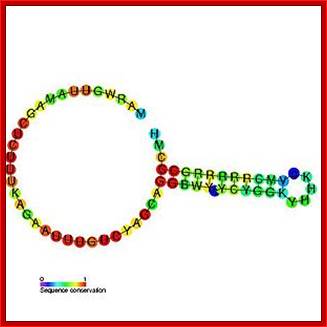
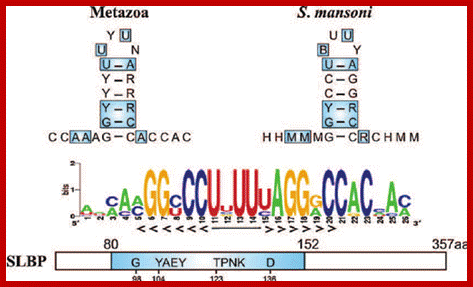
Top fig. http://emboj.embopress.org; Structure and conservation of the histone 3'-UTR stem loop; (a) Schema of the 3'-UTR stem loop structure present in metazoan mRNAs. (b) Sequence logo representation of the histone 3'-UTR stem loop. The sequences were obtained from the Rfam database. Mariño-Ramírez et al.; http://genomebiology.com/ and http://www.unc.edu/
The U7snRNA contains secondary structure and it is bound by SMN heptapeptide proteins. The snRNA base pairs with the ‘UGGGU’ sequence of the mRNA transcript at 3’ end next to stem loop region. This leads to the association of stem loop binding proteins SLBPs and CPSF and CsTFs to this stem loop structure. The bound factors cut the 3’ end of histone mRNA
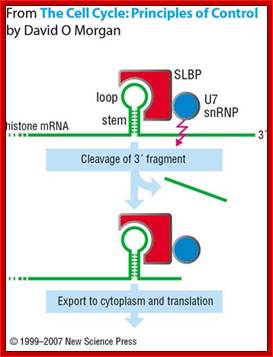
This pairing leads to the cleaving of the transcript 4-5 nucleotides downstream of the stem loop at CACCCA found in the mRNA. https://www.researchgate.net
Histone mRNA stem-loop. A schematic representation of the consensus sequence of histone 3 ′ mRNA hairpin in metazoans and S. mansoni . The interaction between the stem-loop and the stem-loop binding protein (SLBP) is indicated by blue boxes. The RBD motif is conserved in the parasite, forming a stable complex responsible for the histone mRNA processing and translation. The sequence logo shows the conservation among all the predicted S. mansoni histone hairpins. R = A or G (purine); Y = C or U (pyrimidine); M = A or C; H = A, C or U; B = C, G or T. https://www.researchgate.net/
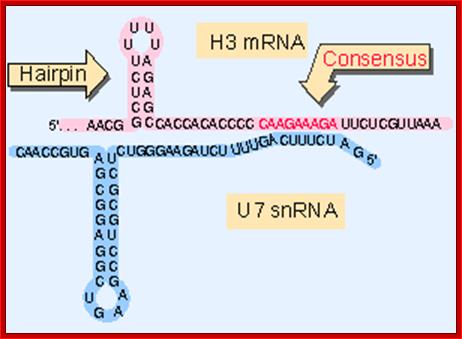
Generation of the 3 Fend of histone H3 mRNA depends on a conserved hairpin and a sequence that base pairs with U7 ;http://flylib.com/
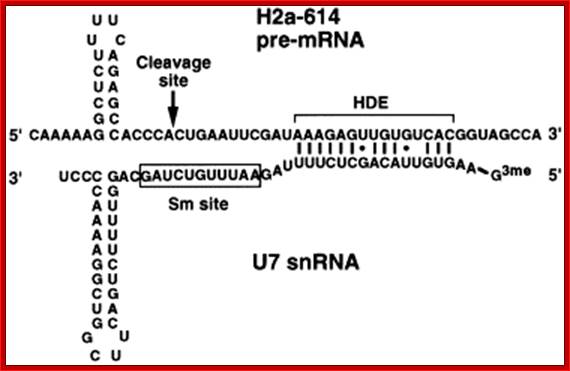
Representation of the processing reaction occurring at the 3′ end of replication-dependent histone precursor mRNA;. The endonucleolytic cleavage (indicated by the scissors) occurs between the highly conserved hairpin (see also Fig. 3A) and the purine-rich sequence HDE. Trans-acting factors are hairpin binding protein (HBP), the U7 snRNP and zinc finger protein 100 (ZFP of 100 kDa). Possible alignment of structural elements of the U7 snRNA and the histone pre-mRNA resulting from formation of a duplex between the U7 snRNA 5′ end and the histone downstream element (HDE). Sequences of the mouse H2a-614 pre-mRNA near the processing site and mouse U7 snRNA are shown. The possible Watson–Crick base pairs (vertical lines) and GU base pairs (dots) between the 5′ end of U7 snRNA and the HDE are indicated. Zbigniew Dominski, William F Marzluff.
Recently, Utz.Fischer and his team has shown that the formation of spliceosomal U7-snRNPs requires assisting factors including the Survival Motor Neuron (SMN) proteins, mentioned earlier, the gene product that affects the neuromuscular and spinal muscular atrophy. SMN is part of a multiprotein (SMN-) complex that facilitates cytoplasmic assembly of U snRNPs in vivo in an ATP-dependent reaction. In this process, spliceosomal Sm proteins, which constitute the protein core of U snRNPs, must bind to the SMN-complex prior to their transfer onto the U snRNAs. The SMN-complex itself is functionally regulated by a methyltransferase, termed PRMT5-complex. This complex catalyzes the symmetric dimethlyation of arginines in some Sm proteins, thereby increasing their affinity for the SMN-complex. Among the seven Sm proteins 5 are common to all and two are specific to U7 snRNAs, they are sm-like called LSM10 and Lsm11 substitute for SMN proteins D1 and D2.
More recent studies indicated that not only the assembly of spliceosomal U snRNPs but also the histone-mRNA processing U7 snRNP particle requires assisting factors. Specifically, it is now known that a minor SMN-complex containing U7-specific factor mediates the formation of the U7-particle.
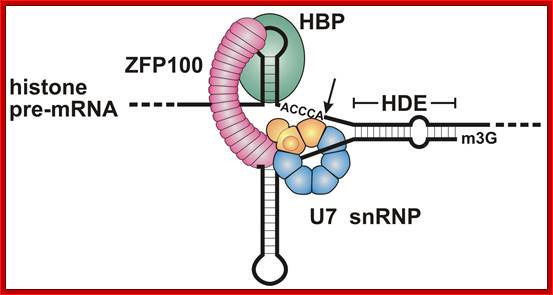
Molecular mechanism of histone pre-mRNA 3’ end processing by U7 snRNP.; http://www.ebi.ac.uk/ ; Kolev NG, Steitz JA. http://www.ebi.ac.uk/
Most metazoan messenger RNAs encoding histones are cleaved, but not polyadenylated at their 3' ends. Processing in mammalian cell extracts requires the U7 small nuclear ribonucleoproteins (U7 snRNP) and an unidentified heat-labile factor (HLF). The above authors describe the identification of a heat-sensitive protein complex whose integrity is required for histone pre-mRNA cleavage. It includes all five subunits of the cleavage and polyadenylation specificity factor (CPSF), two subunits of the cleavage stimulation factor (CstF), and Symplekin. Reconstitution experiments reveal that Symplekin, previously shown to be necessary for cytoplasmic poly(A) tail elongation and translational activation of mRNAs during Xenopus oocyte maturation, is the essential heat-labile component. Thus, a common molecular machinery contributes to the nuclear maturation of mRNAs both lacking and possessing poly(A), as well as to cytoplasmic poly(A) tail elongation.
A 3' exonuclease that specifically interacts with the 3' end of histone mRNA:
Dominski Z, Yang XC, Kaygun H, Dadlez M, Marzuluff WF.
Metazoan histone mRNAs end in a highly conserved stem-loop structure followed by ACCCA. Previous studies have suggested that the stem-loop binding protein (SLBP) is the only protein that binds to this region. Using RNA affinity purification, we identified a second protein, designated 3’hExo that contains a SAP and a 3' exonuclease domain and binds the same sequence. Strikingly, 3'hExo can bind the stem-loop region both separately and simultaneously with SLBP. Binding of 3'hExo requires the terminal ACCCA, whereas binding of SLBP requires the 5' side of the stem-loop region. Recombinant 3'hExo degrades RNA substrates in a 3'-5' direction and has the highest activity toward the wild-type histone mRNA. Binding of SLBP to the stem-loop at the 3' end of RNA prevents its degradation by 3'hExo. These features make 3'hExo a primary candidate for the exonuclease that initiates rapid decay of histone mRNA upon completion and/ or inhibition of DNA replication.
The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing: Dominski Z, Yang XC, Marzluff WF;Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599, USA.
During 3' end processing, histone pre-mRNAs are cleaved 5 nucleotides after a conserved stem loop by an endonuclease dependent on the U7 small nuclear ribonucleoproteins (snRNP). The upstream cleavage product corresponds to the mature histone mRNA, while the downstream product is degraded by a 5'-3' exonuclease, also dependent on the U7 snRNP. To identify the two nuclease activities, we carried out UV-cross linking studies using both the complete RNA substrate and the downstream cleavage product, each containing a single radioactive phosphate and a phosphorothiolate modification at the cleavage site. We detected a protein migrating at 85 kDa that cross linked to each substrate in a U7-dependent manner. Immunoprecipitation experiments identified this protein as CPSF-73, a known component of the cleavage/polyadenylation machinery. These studies suggest that CPSF-73 is both the endonuclease and 5'-3' exonuclease in histone-pre-mRNA processing and reveal an evolutionary link between 3' end formation of histone mRNAs and polyadenylated mRNAs.
Molecular mechanism of histone pre-mRNA 3’ end processing by U7 snRNP. Symplekin and multiple other polyadenylation factors participate in 3’end maturation of histone mRNAs: Kolev NG, Steitz JA.
U7 small nuclear RNA. https://en.wikipedia.org
Most metazoan messenger RNAs encoding histones are cleaved, but not polyadenylated at their 3' ends. Processing in mammalian cell extracts requires the U7 small nuclear ribonucleoproteins (U7 snRNP) and an unidentified heat-labile factor (HLF). We (Koli N.G et al) describe the identification of a heat-sensitive protein complex whose integrity is required for histone pre-mRNA cleavage. It includes all five subunits of the cleavage and polyadenylation specificity factor (CPSF), two subunits of the cleavage stimulation factor (CstF), and Symplekin. Reconstitution experiments reveal that Symplekin, previously shown to be necessary for cytoplasmic poly (A) tail elongation and translational activation of mRNAs during Xenopus oocyte maturation, is the essential heat-labile component. Thus, a common molecular machinery contributes to the nuclear maturation of mRNAs both lacking and possessing poly(A), as well as to cytoplasmic poly(A) tail elongation.
A 3' exonuclease that specifically interacts with the 3' end of histone mRNA: Dominski Z, Yang XC, Kaygun H, Dadlez M, Marzuluff WF.
Metazoan histone mRNAs end in a highly conserved stem-loop structure followed by ACCCA. Previous studies have suggested that the stem-loop binding protein (SLBP) is the only protein binding this region. Using RNA affinity purification, we identified a second protein, designated 3'hExo, that contains a SAP and a 3' exonuclease domain and binds the same sequence. Strikingly, 3'hExo can bind the stem-loop region both separately and simultaneously with SLBP. Binding of 3'hExo requires the terminal ACCCA, whereas binding of SLBP requires the 5' side of the stem-loop region. Recombinant 3'hExo degrades RNA substrates in a 3'-5' direction and has the highest activity toward the wild-type histone mRNA. Binding of SLBP to the stem-loop at the 3' end of RNA prevents its degradation by 3'hExo. These features make 3'hExo a primary candidate for the exonuclease that initiates rapid decay of histone mRNA upon completion and/ or inhibition of DNA replication.
At the end of translation SLBp proteins are phosphorylated and released. The released SLB proteins are ubiquitylated and degraded. The stem-loop of histone mRNA recognized by 3’exonucleases and the same are degraded.
Post-transcriptional control of animal histone gene expression—not so different after all:

Post-transcriptional control of animal histone gene expression;
Pamela Nicholson and Berndt Müller; http://pubs.rsc.org/
Histone
proteins are essential components of eukaryotic chromosomes. The expression of
histone genes is cell cycle controlled and coupled to DNA replication, to
ensure the packaging of replicated DNA into chromatin. The post-transcriptional
control of histone gene expression is a key element in this coupling to DNA
replication. It involves mRNA 3![]() end
formation by histone-specific nuclear RNA processing, which produces mRNAs
lacking a poly (A) tail, translation and mRNA stability control. This requires
several histone-specific trans-acting factors and was thought to be a special
case. Here we review recent observations that now reveal that many of the
factors involved are shared with processing, translation and degradation of
poly (A) mRNA
end
formation by histone-specific nuclear RNA processing, which produces mRNAs
lacking a poly (A) tail, translation and mRNA stability control. This requires
several histone-specific trans-acting factors and was thought to be a special
case. Here we review recent observations that now reveal that many of the
factors involved are shared with processing, translation and degradation of
poly (A) mRNA
Poly A-containing histone H4 mRNA variant (H4-v.1):
Isolation and sequence determination from bovine adrenal medulla:
Nathalie Gendron, Michel Dumont, Marie-France Gagné and Simon Lemaire*
A histone H4 cDNA variant (H4-v.1) was cloned from a bovine adrenal medullary phage library using PCR as a method of detection. The isolated clones contained a short 5′ untranslated region (UTR) followed by the histone H4 coding region and a long atypical 3′UTR. The 3′UTR comprised the palindromic and purine-rich sequences typical of cell-cycle dependent histone mRNAs, and a 1.1 kb extension downstream of the palindromic sequence ending with a poly (A) track typical of cell-cycle independent histone mRNAs. Northern blot and RT-PCR analyses indicate that the transcript is fully expressed in bovine adrenal medulla. Thus, bovine histone H4-v.1 mRNA represents the first example of a histone H4 transcript that contains both 3′UTR characteristics of cell-cycle dependent and cell-cycle independent histone mRNAs.
Negative coupling of processing to transcription. The model explains how processing might be inhibited by RNA polymerase II arrested downstream of the processing site. (A) RNA polymerase II paused at the arrest site leaves just enough RNA exposed to allow binding of SLBP and the U7 snRNP and processing of the transcript. (B) When the polymerase backslides and enters the arrested conformation, the HDE (black box) is no longer accessible to the U7 snRNP, and processing is inhibited. (C) Comparison of the 3′ regions of H4, H3, and H2b. Starting with the last two nucleotides of the stem-loop RNA, sequences from H4, H3, and H2b were aligned. Underlined text, purine-rich region; boldface text, T-rich region; boldface underlined text, pause-arrest site. http://mcb.asm.org/content
Functions of Poly (A) tail:
- It protects mRNA from 3’ exonuclease digestion, for the tails are bound by PAB-II in the nucleus or with PAB-I in cytoplasm.
- It provides stability to mRNA by interacting with cap bound factor and poly-A bound PABI; this leads to circularization of translating mRNA.
- Poly-A RNA produces more polysomes than poly-A less mRNA.
- Poly-A enhances translational efficiency for it is bound by CBP1 protein
- Poly-A neighboring region is used in transporting and localizing mRNAs in the cell interior. Microtubule fibers and Actin filaments and motor proteins like Dynein, Kinesin and Myosin proteins are used as motor proteins to transport mRNA to specific destination within the cell. Examples- Nano, Oskar and other mRNAs in developing drosophila egg. In cytoplasm, nuclear PAB-II is replaced by cytoplasmic PAB-I.
- It is believed that each time the mRNA is translated its poly-A size gets reduced, and it is called ticketing, but now it is known poly-A is degraded by a cytoplasmic enzyme called poly-A nuclease (PAN), which sequentially releases adenines. In the presence of PAB-I degradation is not pr3evented but slow till 20As, and then the degradation is blocked.
- Some poly-A RNAs are stored in cytoplasm as Informosomes, which contain a short chain of 20 or odd poly-As. At the upstream of poly-A site certain sequences called CPE (cytoplasmic polyadenylation elements) facilitate the binding of CPEB. Maskin binds to CPEB and Cap- bound eF-G at 5’end thus circularizes the mRNA. At the time of activation, the mRNAs are added with full-length poly-As.
- Addition of cytoplasmic Poly-As depends upon the sequence found at 3’ UTR sequences (upstream), at about 60 ntds from the first A of poly-A tail, which is called cytoplasmic polyadenylation element (CPE). This site is sensitive, in the sense; the presence of it facilitates the addition of poly-As. Substitution in this region is inhibitory to poly-adenylation and promotive to deadenylation.
- The upstream of AAUAAA, sequences like UUUUUAU and CPE are very important for stored mRNA. CPE is used for poly-A adenylation in cytoplasm. It is only after poly-adenylation, the hitherto inactive mRNAs become active and they are then translated. This kind of activation has been observed in xenopus Oocyte and other systems.
- The AUUUUUUA rich ARE sequences found in 3’ UTR are regulatory sequences used for degradation of mRNAs.
- Similarly there are antisense sequences for the binding of Si/mi RNA that leads to degradation of mRNA.
- When maturation specific synthetic mRNA and another mRNA with out such sequences are added to an Oocyte extract, the maturation specific element containing mRNA gets polyadenylated, but not the other. This UUUUUAU is called Cytoplasmic Polyadenylation Element (CPE). Poly-A addition to such mRNAs also requires the presence of AAUAAA signals.
Eukaryotic mRNAs get circularized/shows circular form:
The recruitment of eIF4F is stabilized by the binding of eIF4G to the 3′ PABP, and the resulting pseudo-circularization of mRNA molecules.
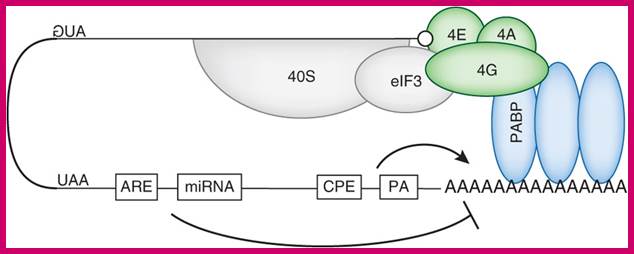
mRNAs contain miRNA binding site and AREs target this loop and promote deadenylation, thus disrupt PABP binding on poly(A) tail and mRNA circularization. http://www.nature.com/nsmb/

Translation factors hold mRNA in circular form; http://www.alpfmedical.info/ http://slideplayer.com/


Translational control of localized mRNAs: restricting protein synthesis in space and time; Protein synthesis can be divided into three steps: initiation, elongation and termination. Translation initiation (see figure) requires the association of the eukaryotic translation initiation factor-4F (eIF4F) complex (which consists of the cap-binding factor eIF4E (4E in figure), the RNA helicase eIF4A (4A) and the scaffolding protein eIF4G (4G)) with the methylated guanosine cap structure at the mRNA 5' end, and the subsequent recruitment of the 43S pre-initiation complex (which includes the 40S ribosomal subunit). Importantly, this recruitment is thought to be facilitated by the binding of eIF4G to the 3' polyadenine tail-binding protein (PABP) and the associated circularization of mRNA molecules. After scanning along the 5' UTR for an appropriate AUG start codon, the pre-initiation complex is then dissolved and the 60S ribosomal subunit joins the 40S subunit to form a translationally competent 80S ribosome. This process is facilitated by the factor eIF5B (5B) and initiates translation elongation.
The elongation phase is characterized by the addition of amino acids to the growing peptide and the translocation of ribosomes along the mRNA, a process that is partly controlled by the elongation factor eEF2. Finally, translation termination is associated with the release of the newly synthesized peptide and the dissociation of the ribosome from the mRNA. For a more detailed description of translation steps and regulators,F lorence Besse & Anne Ephrussi; http://www.nature.com/
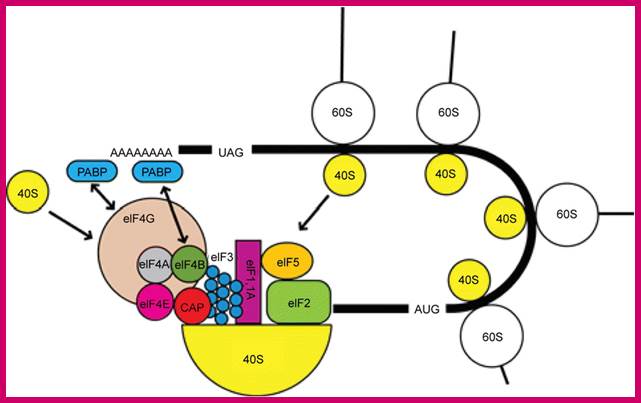
The interaction of PABP with the 48S initiation complex; The interaction between PABP, bound to the poly(A) tail, and eIF4G and eIF4B is indicated with arrows. The interaction results in the circularization of the mRNA. https://www.researchgate.net/