Pre-mRNA Processing: Introduction:
Ribose Nucleic Acids (RNAs), in eukaryotes, are synthesized by three different RNA polymerases and use different promoters and different transcriptional factors. Most of the RNAs are synthesized in the nucleus and all of them are longer than the functional RNAs, so they are called as precursor RNAs i.e. precursor ribosomal RNAs, pre tRNAs, pre 5s RNAs and pre mRNAs and many small RNA are also produced as pre NC RNAs.
In the chapter Ribose Nucleic Acid, with the exception of mRNAs, processing of other rRNA and tRNAs has been discussed. In this section we consider various aspects of pre-mRNAs and their mode of processing. Pre-mRNAs are encoded with vital information in the form of codons for specific polypeptide chains. Its integrity in terms size and organization of sequence, its secondary structure and stability determines the kind of protein it produces and ultimately determines the character and function of the cell, thus the fate of the cell depends upon various processing events of pre-mRNA.
Most of the eukaryotic mRNAs that are synthesized are many times longer than their functional mRNAs. They are of various sizes and characters, so they are called Heterogeneous RNA (hnRNA). Here the term functional RNA, of any kind, is used to denote they are in the final form in which state they are employed for certain functions. All mRNAs are meant for translation activity. Ribosomal RNAs are involved in organizing ribosomes; tRNAs are involved in decoding mRNAs. However pre-mRNAs are subjected to a variety of modifications, such as Capping, adding a poly (A) tail, splicing, and editing, before they are transported out of the nucleus into cytoplasm. More than 70% of the pre-mRNAs synthesized in the nucleus are degraded because they are not processed. The required factors and enzymes for processing are found within the nucleus. Once they are transported as mRNPs they are translated and some are deadenylated to 25-40nts and remain untranslated as informosomes. They will be activated when required.
Size of Hn RNAs and mRNAs- [sample of few of them]:
|
Gene |
Hn RNA size (Bases) |
mRNA size in ntds |
Intron/ Exons
|
|
B-Globin |
1382 |
800 |
2/3 |
|
|
Insulin: Human Rat-insulin1 Insulin2 IGF |
1700 1500 1500 2100 |
400 400 400 |
2/3 2/3 1/2 |
|
|
Protein kinase |
11000 |
1400 |
7/8 |
|
|
Ovalbumin |
6000 |
1100 |
7/8 |
|
|
Albumin |
25 000 |
2100 |
14/15 |
|
|
Catalase |
34 000 |
1600 |
12/13 |
|
|
LDL receptor |
45 000 |
5500 |
17/18 |
|
|
Factor VIII |
186 000 |
9000 |
25/26 |
|
|
Apolipo protein |
13900 |
ApoA-2400 ApoB 8000 |
28/29 |
|
|
Thyroglobulin |
300 000 |
8700 |
36/37 |
|
|
Dystrophin |
>2.4 MB |
14-17000 |
>60 |
|
|
Ovomucoid |
|
5-6000 |
7/8 |
|
|
DHFR |
31 000 |
1600 |
5/6 |
|
|
Actin |
|
1500 |
12/13(?) |
|
|
Pyruvate kinase |
9500 |
L 2.1 and 3.6 |
9/10 |
|
|
Rat fatty acid synthase (FAS) |
++ 1.5kb |
|
42/43 |
|
|
Tropomyosin alpha |
|
2149 |
12/13 |
|
|
Chick collagen alpha |
40 000 |
|
50/51 |
|
|
Fibronectin |
75 000 |
|
31/32 |
|
|
Tyrosine kinase |
|
|
12+A+A |
|
|
Slo-pre mRNA,in inner ear of vertebrates |
|
~6120-11000 |
21/22 |
576 forms in inner ear |
|
Dscam in Drosophila,gene~60,000bp |
|
~7800 ? |
95+20constitutive (4 clusters- 12, 48,33,2 each) |
38,016 forms in retinal neuron connection with brain neuronal cells |
|
Titin-> 300 domains, 3-4MD |
|
115,635 ntds (21k-102kntds |
363/367(177/178)
|
27000-34500,>(81k to 102k ntds |
Smallest, largest and oldest proteins:
Proline- acts as a catalyst, some consider this as an enzyme. Glutathione-is a 3 a.a protein; Chignolin: is 10 a.a Protein. Titin: The largest protein known is found in muscle fibers; located in muscle fibers connects Z line and M line, 34500a.a; 244 domains 3.7-3.79MD;’ The PG5 protein is the longest man made protein; 10 nanometres and a mass equal to 200 million hydrogen atoms. Fossil egg shell proteins 3.8 million yrs old Incredible-David Nield.
Half-life of mRNAs:
In general, mRNA species with short half-life were enriched among those genes with regulatory functions (transcription factors), whereas mRNA species with long half-life were enriched among genes related to metabolism and structure (extracellular matrix, cytoskeleton). The turnover of mRNA lacking a poly-A tail is much higher than mRNA containing a poly-A tail. Poly-A plays a role in translation of mRNA by increasing the stability of mRNA and allowing mRNA to function normally. Exponential equations and graphs make it easier for the half-lives of mRNA to be calculated and compared.
(1) genes possessing at least one intron produce mRNA transcripts significantly more stable than those of intron less genes, and this was not related to overall length, sequence composition, or number of introns; (2) various sequence elements in the 3′ untranslated region are enriched among short- and long-lived transcripts and their multiple occurrence suggests combinatorial control of transcript decay; and (3) transcripts that are microRNA targets generally have short half-lives.
It was also reported that the median half-life correlated with the length of the cell cycle of Escherichia coli, yeast, and human HepG2/Bud8 cells, ranging from 5 to 21 to 600 min, respectively. Transcript stability in Arabidopsis by transcription inhibition revealed that transcript half-lives varied from 12 min to >24 h. Recent study in mammalian cells revealed that the presence of a single intron stabilized a transcript irrespective of length or position, even though it contained two instability elements.
There are intron less, ex. IFNalpha1 and IFNbeta1, c Jun mRNAs, nearly 5% of the human mRNAs are intron less. In yeast most of the genes are intron less.
Degrading factors like miRNA and siRNA (micro, short interfering RNAs) are called to mRNA trigger cleaving of mRNA by endonucleases. Decapping at 5' end exposes mRNA message gradually, and loses protein expression in translation doesn't change mRNA directly modifies translation process. Decay is initiated when the poly (A) tail shrinks below a critical length and the 5' cap is removed by decapping machinery. Deadenylases shorten the poly (A) tail and can directly promote decapping. Certain proteins’ binding to mRNAs increase their stability. For example IRP’s binding to 3' IREs of mRNA increases stability. Presence ARE elements free from IRE binding proteins in 3’UTR makes them more unstable.
Analysis of this region identified three cytosine-rich (C-rich) segments that contributed to globin mRNA stability when studied in transfected erythroid cells. This sequence is bound by certain RNPs. During its approximately four-month lifespan, the human red blood cell (RBC) travels approximately 300 miles, making about 170,000 circuits through the heart, enduring cycles of osmotic swelling and shrinkage while traveling through the kidneys and lungs, and an equal number of deformations while passing through capillary beds. It has been speculated that accumulated damage to the RBC, especially to its membrane, renders the aging RBC unfit to circulate, leading to its destruction, via mechanisms which are poorly understood. RBC life time is ~110-120days. Globin mRNA half-life is 24hrs. Yeast Nrd1/Nab3 surveillance complex, which may recognize cryptic noncoding RNAs and cryptic unstable transcripts via a short poly-(A) tail of four adenosines and target them for rapid decay mediated by the TRAMP complex.
Long antisense noncoding RNAs and nascent rRNA transcripts of stalled RNA polymerase I complexes are also targets of the Nrd1/Nab3-TRAMP surveillance complex. Depletion of the major 5′–3′ exoribonucleases, XRNA, resulted in the stabilization of most mRNAs with half-lives less than 30 min. Thus, on a transcriptome-wide scale, degradation of most mRNAs is initiated by deadenylation.
Long lived protein- Crystallin (e), 27000 to 33000a.a; 3630,000kDa’, lives at least 60-70- yrs and even nuclear pore complex proteins are long lived. Extremely long-lived proteins (ELLPs), found on the surface of the nucleus of neurons, have a remarkably long lifespan; nuclear pore complex (NPC), transport channel proteins lived more than an year. The largest protein known to day is Titin. It consists of 34,350 a.a. The mol.wt, is 3,816,188.13 Da (3816188kDa). Crystallin is the longest living protein found in the lens of eyes of all animals; the multimeric protein mol.wt of alpha A 173a.a (mainly in lens) 19.9 kDa and alphaB 175a.a (19.25kDa).

yD crystallin; http://www.medsci.org/v11p0158.htm
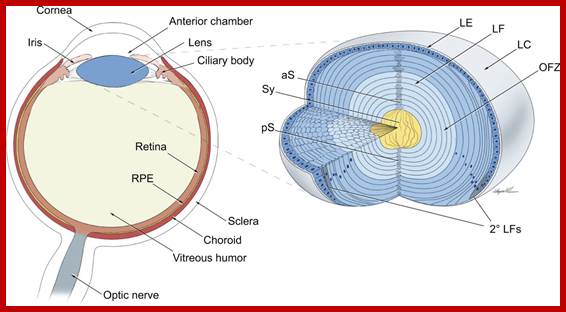
TS of vertebrate eye; Newly formed lens fibers are deposited on the older layers and produce concentric shells packed and radially aligned. http://dev.biologists.org/

Titin is the largest known protein; its human variant consists of 34,350 amino acids, (mouse 35,213a.a) with the molecular weight of the mature "canonical" isoform of the protein being approximately 3,816,188.13 Da with 244 individually folded protein.
Titin; 27000 to 33000a.a; 4200kDa’ with 363 exons coded; https://en.wikipedia.org; http://jcsciphile.com/molecule-of-the-month/
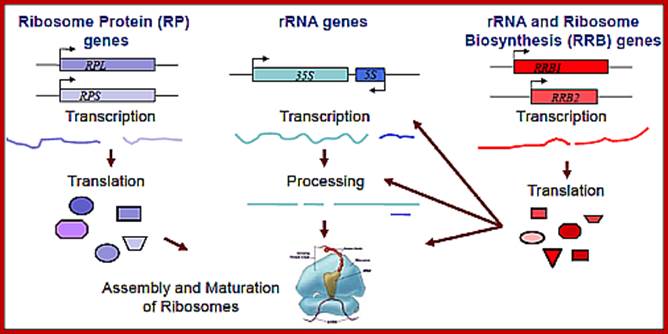
This figure holds good for both prokaryotes and eukaryotes with small differences i.e. tRNA synthesis is missing in the diagram; all the above RNAs and their products ultimately
produce ”Protein”. http://www.wesleyan.edu/; http://mmcalear.faculty.wesleyan.edu/
Pre-mRNAs:
Most of the RNA transcripts, whether it is rRNA, tRNAs or mRNAs, they are longer than their functional RNAs. All of them have many spacer or noncoding regions, so they are subjected to processing, which includes, molecular cutting, end modifications, joining of cut ends called molecular stitching, base modifications, like methylations, de-amination or de-methylations and others.
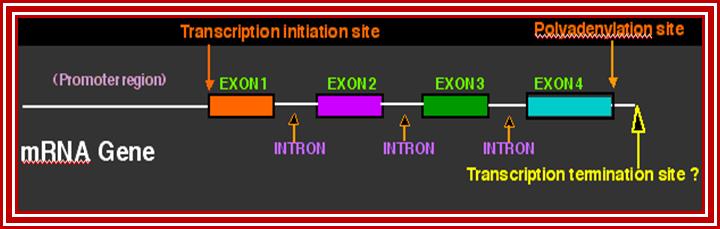
This lovely illustrative picture shows the anatomy of mRNA producing gene; http://dnaofbioscience.blogspot.in/
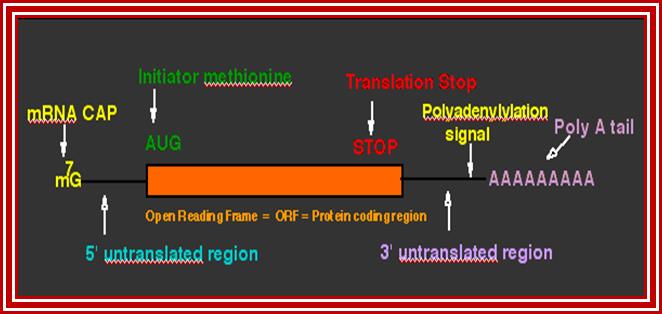
This diagram shows the fine details of the processed mRNA. http://dnaofbioscience.blogspot.in/

This diagram represents the anatomy of non-structural genes for rRNA, snRNA, tRNA and other NC RNA genes. Basically even mRNA genes are also considered as structural genes for they produce polypeptide chains. http://dnaofbioscience.blogspot.in/
Pre messenger RNAs synthesized in eukaryotes are, generally longer than their functional counter parts. They also contain specific segments of noncoding regions called introns and coding segments named as Exons. Different species of pre-mRNAs have different number of Exons and introns and the size of them also vary. But for a given species the number of Exons and Introns and their size is fixed and pre-determined. Having introns and Exons in fixed positions to produce one kind of defined protein in one tissue, the same can be differently cut and pasted to produce alternative form of proteins having different functions, that is what is called as alternative splicing. Even the spliced mRNA can be further modified at certain nucleotides for the protein to function differently in different tissues, which is named as editing. In some mRNAs some of the coding sequences have lost or missing and such mRNAs are added with specific nucleotides in specific positions to generate functionally correct mRNAs.
- Basically each species of pre-mRNA contain a 5’ noncoding leader sequence (5’UTR) from 5’ end to the first initiator codon, whose 5’ end is capped. The size and the sequence vary from one species to another.
- At the 3’ end, from the last terminator codon to the end, there is another noncoding region (3’UTR). This end is polyadenylated.
- Between the first initiator codon and the last terminator codon, there is a coding sequence, which is split into coding and noncoding sequences, which are respectively named as Exons and introns (introns are know as intervening sequences).
- The 5’ UTR has several regulatory sequences such as internal ribosomal entry sequences (IRE), shine Delgarno sequences in bacteria and Kozak’s sequence in eukaryotes required for proper translation.
- Similarly 3’UTR and its adjacent region contains several sequences for regulation-such as ARE loops, antisense RNA binding sequences, mRNA transport and localizing sequence etc.
- From comparative studies of more than 2500 proteins of eukaryotic systems (including their amino acid sequences) combined with computational analysis, protein biologists have discerned that there are only few Exons, say about 1000 to 5000 among living species. From this small number of Exons it is possible to construct all the known cellular proteins and more.
- If one consider that there are ten Exon coding for 40 amino acid residues, using permutation combination, one can generate 10x10 or more different types of polypeptide chains. If one uses combination of several different types of Exons one can easily generate more than 10^52 different kinds of protein with several Exons segments per a protein.
- A single Exon represents a motif in a protein. Example beta Globin exon-1 produces two helices A and B, exon-2 produces two helices and exon-3 generates two more helices. So each of the Exons can generate one or two similar or different structural motifs of a protein.
- If one carefully evaluates the correlation between an Exon and a protein structural motif, one finds same or similar types of motifs in several different proteins, which have similar functions. It is possible to visualize that most of the proteins are built out of a small number of motifs having certain common structural features and then different constructs are designed and added to generate different kinds of proteins with different functions.
- Molecular selection has operated and still operating in protein evolution.
- If a mRNA contains 10 different Exons with different sequences, one can generate not one kind of protein module but many hundreds and thousands of structural modules by random combination of intron exclusion and random inclusion of Exons. However nature has used one linear combination of Exons to generate proteins, but it can also be used for alternative combination of Exons and even introns to generate more than one protein from the same set of Exons found in a single mRNA. Nearly 75% of the transcripts in vertebrate systems do undergo alternative splicing. This overcomes the limited number protein coding gene sequences, which in humans is just 1-1.2% of the human genome; this unique devise can generate any number of different proteins. This molecular evolution is a paradigm for progressive evolution. It is now known that human genome contains genes with heterozygosity more than 1% of the genomes and nearly 45% of the genes are heterozygous (according Craig Ventor, a great adventurist among the present day molecular biologists).
The size and the number of the Exons and the introns vary from one species of mRNA to another, but invariably the size of introns is several times larger than the size of the Exons, which explains, to some extent, the C-Value paradox.
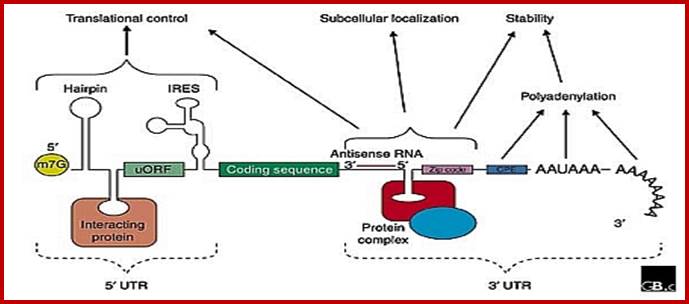
When the precursor mRNA is processed, it looks it has features as shown in the above figure. The processed mRNAs from 5’ end contains the “cap”, stem loops (regulatory elements), IRES-Internal ribosome entry site; ARE RNAse binding sequences, coding region with ORFs containing Kozak and AUG and terminate at Ter codons; at 3’ UTR the mRNA contains several sequences such as RTS (RNA transport) Zip code, cytoplasmic polyadenylation sequences called CPE, antisense region of si or mi RNA, ARE- stem loop structure for de-polyadenylation and degradation, poly-A signal sequence and poly-A tail. http://www.genomebiology.com
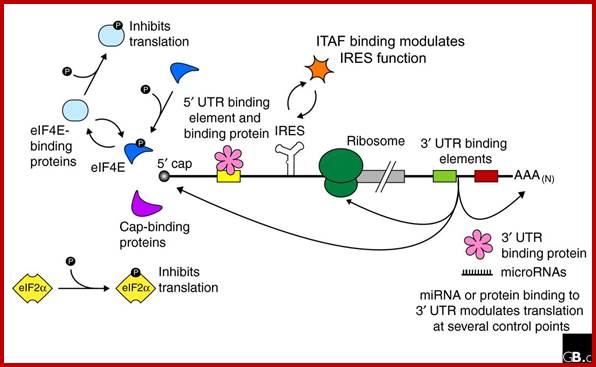
Structural organization of eukaryotic mRNAs and the different points of possible regulation of translation through various trans-acting factors;
5′-m7G, cap structure; eIF-4G eukaryotic initiation factor; IRE- Iron respone elements, Ire- Internal ribosome entry elements, CPE, cytoplasm polyadenylation element; EDEN, embryonic deadenylation signal; DICE, differential control element; Mmi RNA binding elements, mRNA localization signal elements, PABP, poly(A)-binding protein [?], possible sites of interaction of transacting factors (yet unknown) in the coding sequence. Regions of mRNA involved in subcellular localization and stability are also indicated.
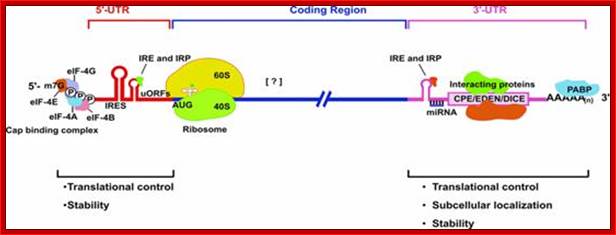
Structural organization of Eukaryotic mRNA; http://www.biocell.org
mRNA regulatory elements:
Regulation of eukaryotic mRNA translation occurs at numerous control points. Recognition of 3' UTR sequence or structural elements (green and red boxes) by RNA-binding proteins leads to either activation or repression of translation, often through alteration of the 3' poly(A) tail or through interactions with proteins that bind at the 5' terminal cap structure (that is, the initiation factor eIF4E or cap-binding proteins). Repression of translation by miRNAs can occur through inhibition of translation initiation or elongation, and may also lead to changes in the status of the mRNA 3' poly-(A) tail. Elements found within the mRNA 5' UTR (yellow box) can bind regulatory proteins that repress translation by inhibiting 48S ribosome scanning. Global regulation of mRNA translation is commonly achieved through modification of the translational apparatus (that is, by phosphorylation of the translation initiation factors eIF2α and eIF4E) and the ribosome itself, or modulation of protein partner binding affinities (such as the phosphorylation of the eIF4E-binding proteins). Translation can be initiated independent of the mRNA 5' cap through a structured internal ribosome entry site (IRES) in the 5' UTR whose efficiency in initiating translation is, in turn, modulated by trans-acting factors (ITAFs).