Protein_Synthesis-Regulation_of_Protein_Synthesis- Eukaryotic.
Translation in eukaryotes is regulated at various levels and it is very complex. In EKs transcription and translation events are separated in time and space. Availability of mRNA for translation is controlled by transcription, RNA processing and transport of the nucleus. Once it is transported to destined sites it can be translated or some of it can be stored as inactive mRNA and it can remain so; only to be translated at later point of time or when needed.
Initiation of translation takes place at one of the AUGs, if there are more than one AUGs in 5’ UTR space, which one of the AUGs is used and how it is controlled is determined by the location of Kozak sequence (G/ACC-AUGG) in right context of AUG. What determines, which mRNA to be translated and in what frequencies? How, certain mRNAs remain untranslated and how they are activated at other times? Protein synthesis also depends upon the stability of the mRNAs and their secondary structures, which may be found at 5’ UTR sequence or they may be present at 3’ UTR end of the mRNAs. Protein synthesis can also be regulated at chain initiation, elongation or at termination level. The final and functional protein production is also regulated at posttranslational level.

http://www.inkymousestudios.com/
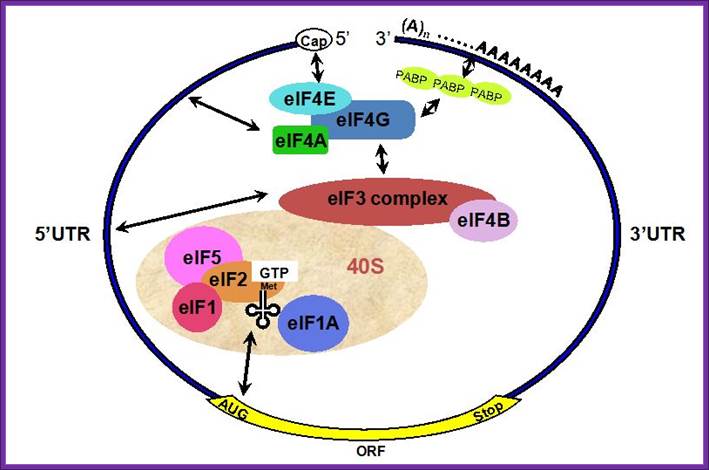
General view; A schematic drawing of a eukaryotic mRNA, illustrating some post-transcriptional regulatory elements that affect initiation of protein synthesis. www.intechopen.com
Autogenous regulation:
- Translation perse, however, is regulated by a variety of mechanisms which are unique to eukaryotes, but some regulatory mechanisms are common with prokaryotes, for ex. Synthesis of several proteins is controlled by what is called autogenous control.
- Tubulin genes are a family of genes in eukaryotic systems. Among the several autogenous regulatory systems, the best-studied system is Tubulin synthesis. Within cells, there is a dynamic equilibrium between free Tubulin monomers and its polymerized polymers. When the concentration of monomeric Tubulins exceeds than the concentration of polymerized Tubulins, the Tubulin protein bind to its own Tubulin mRNA and blocks its own translation, which leads to the degradation of the mRNA? It is also regulated at transcriptional level.
Regulation at chain initiation factor eIF-2a:
Initiation of Protein synthesis requires a host of factors, thus initiation is regulated. Initiation can be regulated by the biding of EBP1 to 4E, thus block initiation process.

A grand view of translational initiation complex formation; Pathway of translation initiation in eukaryotes. Eric Klann & Thomas E. Dever; http://www.nature.com/
A binary complex of eukaryotic translation initiation factor 2 (eIF2) and GTP binds to methionyl–transfer RNA (Met–tRNAiMet), and the ternary complex associates with the 40S ribosomal subunit. The association of additional factors, such as eIF3 and eIF1A (1A), with the 40S subunit promotes ternary complex binding and generates a 43S pre-initiation complex. The cap-binding complex, which consists of eIF4E (4E), eIF4G and eIF4A (4A), binds to the 7-methyl-GTP (m7GTP) cap structure at the 5' end of a messenger RNA (mRNA). eIF4G also binds to the poly(A)-binding protein (PABP), thereby bridging the 5' and 3' ends of the mRNA. This mRNA circularization and the ATP-dependent helicase activity of eIF4A are thought to promote the binding of the 43S pre-initiation complex to the mRNA, which produces a 48S pre-initiation complex. Following scanning of the ribosome to the AUG start codon, GTP is hydrolysed by eIF2, which triggers the dissociation of factors from the 48S complex and allows the eIF5B- and GTP-dependent binding of the large, 60S ribosomal subunit. Although the precise timing and requirements for the release of factors from the pre-initiation complexes are not clear, the 80S product of the pathway is competent for translation elongation and protein synthesis; Eric Klann & Thomas E. Dever
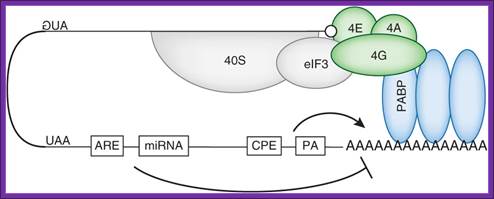
Although almost every step of this process is a target for regulation, translation initiation is rate-limiting112. Accordingly, it is the main target for translational control mechanisms115. miRNA binding sites and AREs target this loop by promoting deadenylation, thereby disrupting the binding of PABP on the poly(A) tail and mRNA circularization. These events greatly decrease the translation efficiency. The CPE and polyadenylation site promote cytoplasmic polyadenylation (see Box 2), although CPE can also contribute to mRNA deadenylation.; Laure Weill et al; http://www.nature.com/
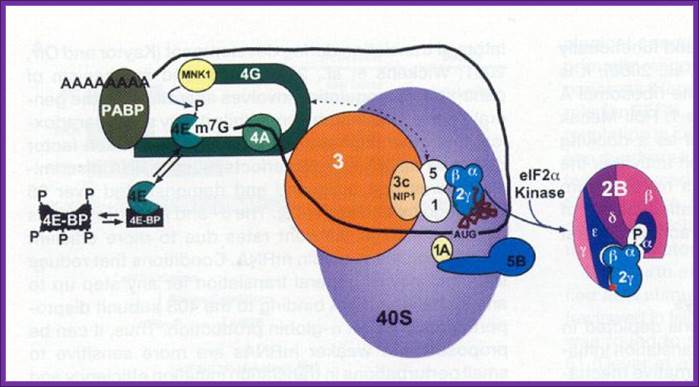
https://www.slideshare.net
Blocked eIF-E by 4EBP, on 4EBP phosphorylation, it dissociates from the 4G; thus it releases it from inhibition. The 5’end cap is bound by 4E, if phosphorylated the mRNA is active in initiating translation with 4FG and PAB1. But the 4E is bound by 4EBP, translational initiation is blocked, but if the 4EBP is phosphorylated it dissociates from 4E and translation is initiated.
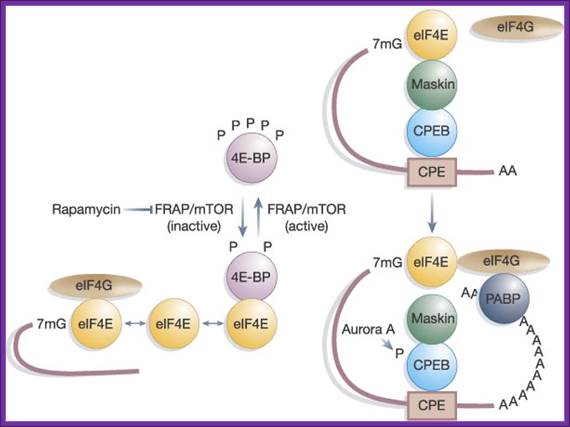
The kinase FRAP/mTOR hyperphosphorylates 4E-BP on several sites; this causes the liberation of eIF4E from 4E-BP, and the association of eIF4E with both capped mRNA and eIF4G. The inhibition of FRAP/mTOR by rapamycin leads to the hypophosphorylation of 4E-BP and enhanced binding to eIF4E. Maskin binding to eIF4E excludes the eIF4G–eIF4E interaction on CPE-containing mRNAs. The inhibition of translation by Maskin is abrogated by cytoplasmic polyadenylation, which is induced by Aurora A-catalyzed CPEB phosphorylation. The newly elongated poly(A) tail is bound by poly(A) binding protein, whose association with eIF4G helps disrupt the Maskin–eIF4E complex and facilitate initiation. The compartmentalization of Maskin- (or Cup-) bound eIF4E probably makes it resistant to further regulation by 4E-BP. http://www.readcube.com/
Another most prevalent event is phosphorylation of eIF2-alpha subunit; once it is phosphorylated, the eIF2alpha-GTP binds to initiator tRNA and loads onto small (40s) ribosomal subunit. This complex binds to 5’end of the mRNA and moves on the leader sequence with the assistance of helicase. When the ribosome with eIF2a-itRNA finds its initiator codon it settles. With binding of 60s subunit, the eIF2alpha-GTP hydrolyses and eIFaGDP dissociates. The eIFa-GDP gets recycled with eIFBGTP (GEF). If eIFa gets phosphorylated by certain kinases, eIFa-GTP cannot be regenerated for the eIFB-GTP binds it remains bound and the eIFa-GTP cannot be regenerated. So translation initiation gets blocked.
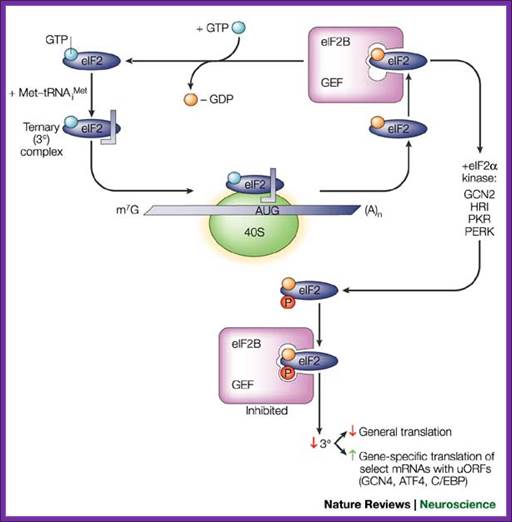

Top Fig. Recycling of eIF2 by eIF2B and regulation by the
eIF2 kinases; Eric Klann & Thomas E. Dever; http://www.nature.com/;
Bottom Fig. http://www.nichd.nih.gov/ Recycling of eIF2 by eIF2B and
Regulation of eIF2a kinase
kinases; Eric Klann & Thomas E. Dever; http://www.nature.com/;
Bottom Fig. http://www.nichd.nih.gov/ Recycling of eIF2 by eIF2B and
Regulation of eIF2a kinase
The
eukaryotic translation initiation factor 2 (eIF2)–GTP binary complex binds to
methionyl–transfer RNA (Met–tRNAiMet) and forms a ternary
complex that then associates with the 40S ribosomal subunit. After start-codon
recognition, GTP is hydrolysed by eIF2 and the binary eIF2–GDP complex is then
released. The guanine nucleotide-exchange factor (GEF) eIF2B converts inactive
eIF2–GDP to active eIF2–GTP, a process that is inhibited by phosphorylation (P)
of the ![]() -subunit of eIF2 on
serine 51 by one of the four known eIF2
-subunit of eIF2 on
serine 51 by one of the four known eIF2![]() kinases. Phosphorylation of eIF2
kinases. Phosphorylation of eIF2![]() converts eIF2 to a competitive inhibitor of eIF2B, and inhibition
of eIF2B results in lowered levels of ternary complexes, which reduces general
translation but increases translation of a specific class of messenger RNAs
(mRNAs) with upstream open reading frames (uORFs)22. ATF4, activating transcription
factor 4; C/EBP, CCAAT/enhancer-binding protein; GCN, general control
non-derepressible; HRI, haem-regulated initiation factor 2
converts eIF2 to a competitive inhibitor of eIF2B, and inhibition
of eIF2B results in lowered levels of ternary complexes, which reduces general
translation but increases translation of a specific class of messenger RNAs
(mRNAs) with upstream open reading frames (uORFs)22. ATF4, activating transcription
factor 4; C/EBP, CCAAT/enhancer-binding protein; GCN, general control
non-derepressible; HRI, haem-regulated initiation factor 2![]() kinase; m7G, 7-methyl-GTP; PERK, eIF2
kinase; m7G, 7-methyl-GTP; PERK, eIF2![]() kinase 3; PKR, protein kinase-RNA regulated, interferon-inducible
double-stranded RNA dependent; Eric Klann
& Thomas E. Dever;
kinase 3; PKR, protein kinase-RNA regulated, interferon-inducible
double-stranded RNA dependent; Eric Klann
& Thomas E. Dever;
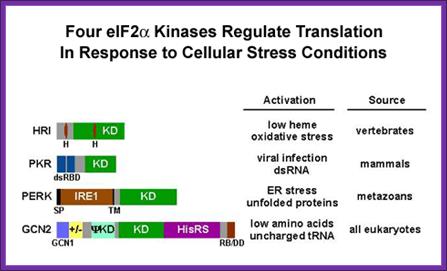
This recycling reaction is a key control point to regulate translation initiation and it is the target of the eIF2a protein kinases. Four protein kinases have been identified that specifically phosphorylate Ser-51 of the alpha subunit of eIF2. Factors responsible for kinasing eIF2a are GCN2, PKR, HRI and PERK.
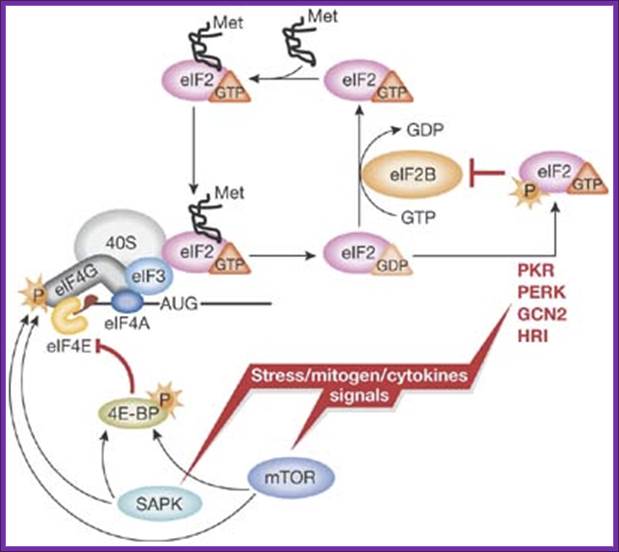
A simplified model of translation regulation by stress: Various stress, mitogen and cytokine signals evoke activation of distinct signalling pathways resulting in the reduction of protein synthesis. Activation of eIF2α kinases (PKR-like ER kinase (PERK); heme-regulated inhibitor kinase (HRI); double-stranded RNA-activated protein kinase (PKR); general control non-derepressible-2 (GCN2)) results in the reduction of available ternary complex, whereas activation of the mammalian target of rapamycin (mTOR) and/or stress-activated MAP kinase pathways (SAPK) impinges on the activation of messenger RNA and its recruitment into the 48S initiation complex. For simplicity, only selected factors are shown. Met, methionine, P, phosphate. http://embor.embopress.org
http://www.nichd.nih.gov/
These kinases are activated under different cellular stress conditions included heme-deprivation (HRI), virus infection (PKR), ER stress (PERK), and amino acid starvation (GCN2). Interestingly, phosphorylation of eIF2a on Ser-51 inhibits eIF2B’s (GEF activity) and thus impairs general translation, but it can also lead to translational activation of specific mRNAs including the yeast GCN4 mRNA and the mammalian ATF4 mRNA (see Dever, 2002). Current efforts are directed at identifying the residues in eIF2a that directs the kinases to specifically phosphorylate Ser-51. A combination of molecular genetic and biochemical tools is being employed to define the minimal substrate for phosphorylation and to identify the residues in yeast eIF2a that are critical for translational regulation and phosphorylation of Ser-51 by GCN2 and/or PKR.
Interferon Mediated Control:
- Reo, Polio, Influenza, Vaccinia, VSV and other viral infection, leads to the production of interferon in mammalian cells; it is the first line of defense.
- Interferons are glycoproteins, depending upon the type of tissues; they are classified as alpha-IFNs from WBCs, beta-IFNs from fibroblasts and gamma-IFNs from lymphocytes.
- Infection of cells takes it route through the receptor or through wounds or through oral pathway.
- Infection of cells, with RNA or DNA viruses, results in the synthesis of ds RNAs due to transcription, which happens during viral genome replication or transcription.
- The dsRNA in turn activates certain cytoplasmic IFN receptors and different Interferon genes are expressed.
- Interferons that are released to extra cellular medium, bind to uninfected cells via their cellular receptors and induce them to be in antiviral state. They are very effective even at very low concentrations such as 3x10^-14 M. They have wider specificity than antibodies.
- Interferon genes called alpha, beta and gamma, have been cloned and they are used in clinical treatments. Interferons prevent viral proliferation largely by inhibiting cellular translation events.
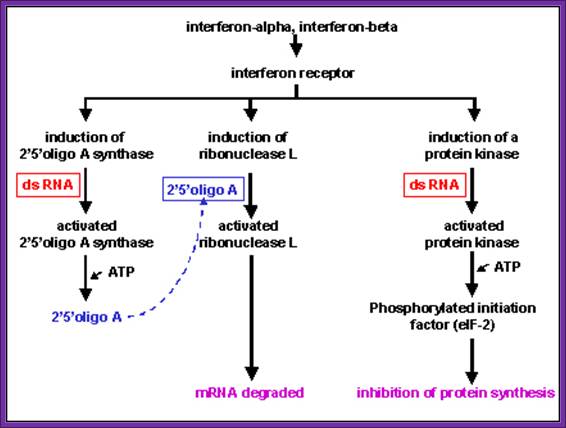
Effects of Interferon; Role of dsRNA and its effect on 2’5’oligo A, and ds RNA activated protein kinase inhibits eIF2 by phosphorylation; Microbiology and Immunolgy; Dr.Margaret Hunthttp://pathmicro.med.sc.edu/
- Interferons induce the synthesis of protein kinases, they get activated in the presence of dsRNA; this is dsRNA dependent kinases, and they specifically phosphorylate the serine-51 of alpha subunits of eIF-2a factor.
- Interferons also induce the synthesis of 2’- 5’Aoligo synthase, which in fact is activated by dsRNA. This catalyzes the synthesis of (ppp5’A-2’p5’A2’p5’A2’p)n oligos, (The oligos have repeat units of p5’-A-2’p ). The above said small molecules activate RNase-L, which acts upon mRNAs, and degrade them to its respective nucleotides. The 5’ppp2’A oligo-molecules by themselves undergo rapid degradation by 2’-5’-phophodiesterases.
www.slideplayer.com/
Interferon production and the spreading of the same to other cells keep them in antiviral state (innate Immunity); this is the first defense against viruses or any infectious agents before cell mediated or immunoglobulin mediated defense takes over.
- Interferons released from the viral infected cells bind to the cell surface receptors and induce immunity against further infection of viruses. Interferon induces the synthesis of a protein kinase, which is ds RNA dependent for its activity (called double stranded RNA- activated inhibitor of protein synthesis or DAI).
- The dsRNA dependent kinase, when activated by dsRNA, phosphorylates the alpha subunit of eIF-2a
- The eIF-2 consists of three subunits such as alpha, beta and gamma.
- The eIF-2-GTP complex loads the initiator met-tRNA on to 40s ribosome at ‘P’ site. Once the met-tRNA is placed to its proper site the factor hydrolyses GTP to GDP and inorganic pi. Then the eIF-2-GDP dissociates from the ribosomal surface.
- In order to regenerate eIF2a, the eIF-2 B, which is also called as GEF, (GTP Exchange Factor) should interact with eIF2 abg complex and binds to it. In this reaction GDP is released and eIF-2-eIF-2B form a complex witheIF2abg. In this complex, eIF-2B facilitates the binding of GTP to eIF-2. The exchange of GDP to GTP is the function of eIF-2B.
- When the eIF-2, under certain conditions, if its alpha subunit gets phosphorylated at serine 51, when eIF-2-alpha-GDP binds to GEF, they bind so tightly they don’t dissociate from each other, for the eIF-2B has great affinity to the phosphorylated alpha subunit of eIF-2. With out the displacement of GDP, GTP cannot be loaded onto the eIF-2 factor. Till the eIF-2 is loaded with GTP, initiator met-tRNA doesn’t bind to the eIF-2 and the initiator eIF2GTP-met.tRNA complex is not regenerated.
- The above mechanism of phosphorylation and dephosphorylation of alpha subunit operates in viral infected cells where interferon mediated cell immunity operates.
- Such phosphorylation of eIF-2a subunits also takes place when cell suffers from heat shock or from other nutritional deficiencies.
- In interferon mediated cell immunity against fresh infection operates via interferon-mediated mechanism. In such cells dsRNA are produced due to interferon action. Cells do contain a host of kinases. One such kinase induced by interferon is a ds-RNA dependent kinase.
- In the presence of dsRNA, the kinase gets activated and this kinase phosphorylates the alpha subunit of eIF-2 factor. Phosphorylation of the alpha subunits prevents the regeneration of functional eIF-2, thus it blocks chain initiation of all mRNAs including those of virally produced mRNAs.
GCN 4p, a Master Regulator of Gene Expression, Is Controlled at Multiple Levels by Diverse Signals of Starvation and Stress:
- A similar situation is also observed under heat shock and amino acid starvation conditions.
· Upstream of open reading frames (uORFs) are frequently present in the 5′-leader regions of fungal mRNAs. They can affect translation by controlling the ability of ribosomes that scan from the mRNA 5′ end to reach the downstream genic reading frame.
- The translation of uORFs can also affect mRNA stability. For several genes, including Saccharomyces cerevisiae GCN4, S. cerevisiae CPA1, and Neurospora crassa arg-2, regulated by uORFs and it controls expression of mRNAs in response to specific physiological signals. The role of many uORFs that are identified by genome-level approaches, as have been initiated for Saccharomyces, Aspergillus, and Cryptococcus species, it remains to be determined. Heather M. Hood,1 Daniel E. Neafsey,2 James Galagan,2,3 and Matthew S. Sachs4
· Some uORFs may have regulatory roles, while others may exist to insulate the genic reading frame from the negative impacts of upstream translation start sites in the mRNA 5′ leader.
- In yeast cells, eIF-2 mediated regulation is observed with respect to the factor called GCN4, leucine zipper protein is a transcription factor for General Control of Nitrogen metabolism.
- The GCN4 that is synthesized under unusual conditions regulates the expression more than 40 genes related to amino acid metabolism.
- It activates the synthesis of its own protein by 100 fold when yeast cells are starved of essential amino acids.
- Its level of expression depends upon repressor specified by GCD 1, 2 and 3 loci.
- The GCN4 mRNA transcript is about ~1903 ntds long, the coding region starts from 578th ntd and terminates at 1325 th ntd.
- The mRNA has a cap and a poly-A tail. It has a 577ntd long 5‘ UTR sequence, which contains three uAUGs upstream from the actual iAUG; that is it has four uORFs upstream of the correct GCN4 ORF.
5’cap…………[… GCN4-ORF……………….TER…]. poly- (A) n
5’Cap------uAUG---—uAUG----—uAUG----ACCAUGG---------3’
<------------------------5’ UTR------------GCN4 ------- ORF ----------
Scanning and AUG selection:

After translating uORF1, ~50% of the 40S subunits resume scanning downstream in both starved and nonstarved cells. In nonstarved cells, all of the subunits quickly rebind TC, reinitiate at uORF4, and dissociate from the mRNA after translating uORF4. When TC levels are reduced by eIF2(P), a fraction of ribosomes fails to rebind TC until scanning past uORF4, allowing them to reinitiate at GCN4 instead. Consistent with this, the bypass of uORF4 and induction of GCN4 translation in starved cells is suppressed by overproducing all three subunits of eIF2, or all four essential subunits of eIF2B, both conditions expected to elevate TC levels. Morever, GCN4 translation is constitutively derepressed (Gcd- phenotype) in mutants with defects in eIF2 or eIF2B subunits, or Met-tRNAiMet biogenesis, in which TC assembly is impaired. More recentlyit is observed that truncating the eIF1A C-terminal tail (C) or overproducing the N-terminal tail (NTT) of eIF3c/NIP1 (c/NIP1) also produce Gcd- phenotypes that are diminished by overproducing all of the components of TC from a high-copy plasmid (hc-TC), suggesting that these mutations delay TC loading on 40S subunits scanning downstream from uORF1, allowing a fraction to bypass uORFs 2-4 and reinitiate at GCN4 in the absence of eIF2(P), Cherkasova, Vera etal.
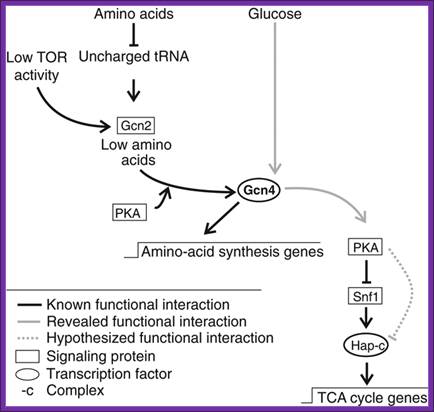
Signaling cascades involving Gcn4. Known signaling cascade involving Gcn4 and the here revealed Gcn4 signaling cascade of TCA cycle gene expression., (Schüller, 2003; Slattery et al, 2008; Zaman et al, 2008).
Fig: Gene4p, a master regulator; Summary of diverse regulatory mechanisms controlling Gcn4p expression or activity and the signals to which they respond. Activation is depicted with arrows; inhibition is depicted with bars. The environmental or physiological conditions regulating these responses are color coded according to the aspect of Gcn4p being regulated, including its expression at the level of translation, transcription, or degradation or its function as an activator. The role of Gcn2p in stimulating Gcn4p function in response to UV or glucose refeeding needs to be verified and thus is depicted with a broken line. Question marks indicate a lack of knowledge concerning the regulatory factors involved. X, hypothetical repressor of Gcn4p that is negatively regulated by the proteosome. See the text for details. http://ec.asm.org/
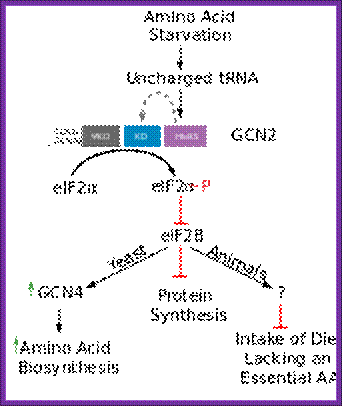
Overview over the functions of GCN2. (GCN1/GCN20=GCN1p/GCN20p binding site; PsiKD = unknown function; KD = Kinase Domain; HisRS = histidyl-tRNA synthetase) Adapted from; http://cell.com
Authors Tatyana Pestova’ et al, suggest that 98-101 also confers slower scanning between uORFs 1 and 4, which compensates for the delay in Transcription Complex TC loading and restores efficient reinitiation at uORF4, with impaired induction of GCN4 (Gcn-), obtained evidence supporting this model by determining the effect of 98-101 on scanning in a reconstituted mammalian system in which inhibition of primer extension (toe-printing) is used to map the locations of scanning PICs on specific mRNAs. We found that yeast eIF1A substitutes for the mammalian eIF1A in this assay and that 98-101 reduces the ability of PICs to both migrate from the cap and to bypass an upstream GUG.
- Under amino acid starvation conditions, uncharged tRNAs accumulate. Accumulation of uncharged tRNAs binds to another factor GCN2. The GCN2 has one domain for the binding of uncharged tRNAs and the other domain has kinase activity.
- When an uncharged tRNA binds to the GCN2’s tRNA binding domain, the kinase domain gets activated.
- The activated kinase domain phosphorylates eIF-2 alpha subunit at 51 th a.a, thus inactivates the initiating factor.
- Under the reduced concentrations of active eIF-2s, 40s ribosomes with their own initiator-met-tRNA, gain entry into the leader sequence of GCN4 mRNAs, but they bypass all other AUGs and initiate translation at GCN 4th ORF and synthesize GCN4 protein.
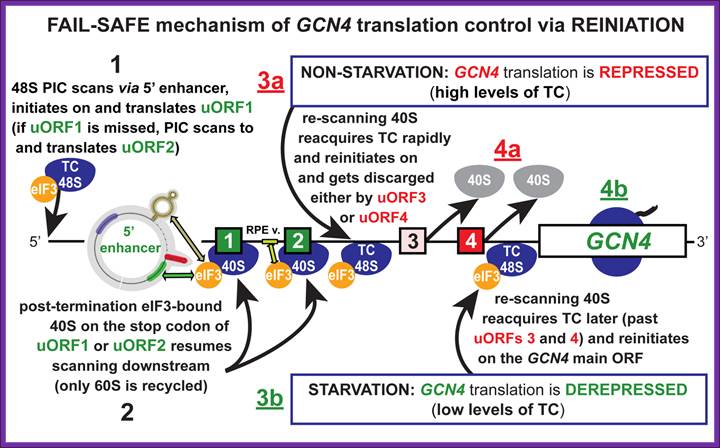
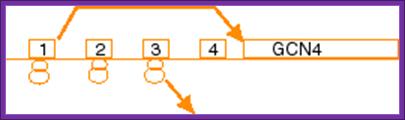
An unstarved cells, where TC levels are high, after the first uORF is translated the 40S subunit regains a TC and reinitiates on one of the two inhibitory, REI-non-permissive downstream uORFs (uORF3 or 4). This prevents complexes from reaching the GCN4 start codon and initiating there. When cells are starved for amino acids, the kinase GCN2 is activated to phosphorylate the alpha subunit of eIF2. Phosphorylation of eIF2-alpha leads to inhibition of TC formation. The low TC levels allow the 40S subunit to scan past the start codons of inhibitory uORFs 3-4 before reacquiring a TC, thus increasing the frequency of initiation at the GCN4 start codon. Mutations that constitutively activate GCN4 translation, allowing survival of amino acid starvation even in gcn2delta cells, produce the Gcd- phenotype(general control derepressed). Mutations that prevent activation of GCN4 translation by amino acid starvation in GCN2+ cells produce theGcn- phenotype (general control nonderepressible). We and others have shown that many of the Gcd- mutations reduce the efficiency of TC binding to the 40S subunit. Translation-born Gcn- phenotypes can arise from a variety of reasons such as for example: 1) reduced rate of mRNA recruitment; 2) reduced initiation at uORF1 (leaky scanning phenotype; PIC complexes miss the correct start codon with increased frequency and continue scanning downstream); 3) failure of the 40S ribosome to resume scanning after terminating at uORF1; 4) reduced rate of scanning (slow scanning phenotype); 5) instability of re-scanning ribosomes on the mRNA (defect in processivity of scanning). http://www.biomed.cas.cz/
Thus the translated product GCN4 activates several genes related to amino acid synthesis.
- Under normal situations chain initiations that occurs at upstream positions gets terminated.
- In such situation translation is initiated at ORF-1 and gets terminated and again it is reinitiation fails to translate and mRNAs falls off of the ribosome.
- When amino acids level is high or optimal, GCN2 dissociates from tRNAs and its kinase domain becomes inactivated. Hence no phosphorylation of the alpha subunit of eIF-2.
Iron Regulated mRNA stability and instability:
Heme Controlled Translation at eIF-2 Level:
Reticulocytes are exclusively engaged in hemoglobin synthesis. When Reticulocyte lysate is used for in vitro protein synthesis, synthesis goes on for some time and then suddenly stops, but the addition of heme, again reinitiates protein synthesis. Thus it can be concluded that heme has an important role to play in Goblin protein synthesis. However inhibition of goblin synthesis can be reversed by the addition of eIF-2 and GTP.
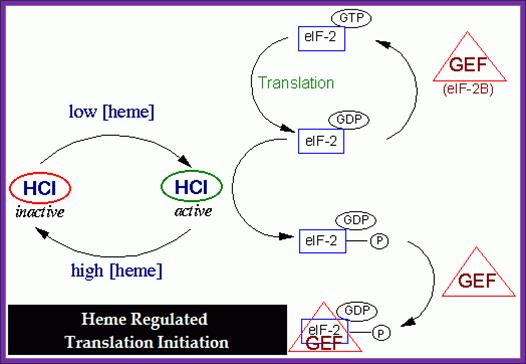
http://bricker.tcnj.edu/
The above diagram depicts how heme-hemin regulates the initiation of protein synthesis via regulating chain initiating factor.
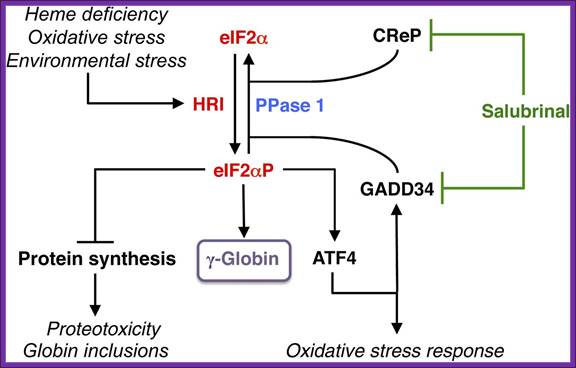
Among the family of eIF2α kinases, heme-regulated eIF2α kinase (HRI) is most predominant in the erythroid precursors. In heme deficiency, HRI is activated and phosphorylates eIF2α to balance globin synthesis with the intracellular heme concentrations ensuring that no globin is translated in excess of heme during erythroid maturation. HRI is also activated by oxidative stress and environmental stress, such as heat shock and osmotic shock. Under these stress conditions, the first action of eIF2αP is to inhibit general protein synthesis, especially globin to prevent proteotoxicity. Second, eIF2αP also increases translation of selective mRNAs, such as ATF4 mRNA to reprogram gene expression for adaptation to stress. GADD34 is a downstream target of ATF4 signaling, and it is responsible for bringing eIF2αP to type 1 phosphatase (PPase 1) for dephosphorylation to regenerate active eIF2, which is necessary for the recovery of protein synthesis of stress-induced gene expression that occurs late in the stress response. CReP has the same function as GADD34, except that it is constitutively expressed. Salubrinal, a selective inhibitor of eIF2αP dephosphorylation, interferes with the recruitment of eIF2αP to PPase1 through GADD34 and CReP, thus preventing eIF2αP dephosphorylation. Inhibiting eIF2αP dephosphorylation by salubrinal or by the knockdown of GAAD34 and CReP in differentiating human CD34+ cells induces γ-globin expression, leading to an increase in HbF production;http://bloodjournal.hematologylibrary.org/
- Heme is a porphyrin ring with a central Fe^2+ ion. When cells are starved of iron, heme is not produced.
- Under heme deficiency, a repressor called HCR, Heme Controlled Repressor) accumulates and also becomes active.
- This repressor contains a domain for the binding of hemin and another domain has a kinase function.
Hemin and Heme; Lopez Cervilla Zephyr; http://campusvirtual.ub.edu/
- When the hemin is absent the kinase domain becomes active. The activated repressor or kinase phosphorylates serine 51 of alpha subunit of eIF-2. Thus protein synthesis is blocked.
- If heme is made in excess, it is oxidized to hemin (note- hemin is an oxidized product of Heme). When hemin concentration exceeds certain level, it binds to HCR and makes it inactive, thus it cannot phosphorylate alpha subunit of eIF-2.
- At the same time heme reactivate eIF-2 by eIF-2 Phosphatase.
- It is fascinating to understand that there is an inherent relation between the levels of heme, hemin, and the synthesis of alpha and beta globins in RBC cells. Human or any mammalian Reticulocyte cells called red blood cells (RBCs) are highly specialized cells for they exclusively produce hemoglobin. Globin protein synthesis is actually regulated by the levels of heme and hemin.
· There is a correlation between the synthesis of globins and the levels of hemins. In the absence of heme, functional or what is called active HCR accumulates.
· When the concentration heme or hemin is high the HCR becomes inactive.
· When the concentration of Globin is higher than the levels of heme, all the heme molecules present are utilized and any more production of globins is futile. So the cellular regulation has devised a mechanism to stop the synthesis of globins.
· When there is deficiency of hemins, as heme is not produced to the desired levels, the HCR’s kinase domain gets activated, for there is no hemin to bind.
· The activated HCR kinase domain phosphorylates the alpha sub unit of the eIF-2, which in turn prevents the recycling and regeneration of functional eIF-2.
· This event leads to blocking of translational initiation of hemoglobin mRNA.
· When the heme is available in plenty and if it exceeds the concentration of Globin proteins, the heme is oxidized to hemin and it binds to the hemin-binding domain.
· The binding of Hemin to HCR inactivates the HCR and its kinase activity is totally suppressed, so no more phosphorylation of eIF-2 alpha subunit and all the eIF-2 are active and initiate translation of Globin mRNAs that is actually required for the optimal production of functional hemoglobins.
· The levels of HCR determine the synthesis of both alpha and beta Globin. Active HCR is produced when the level of heme is low. When the level of heme is low, there is no need for the synthesis of both types of Globin protein, if they are synthesized, it is a waste. So the HCR, in the absence or at low levels of heme (hemin), is active and catalyses phosphorylation of eIF-2 alpha subunit.
· This makes the recycling of the eIF-2GDP to eIF-2GTP. Thus translation of all mRNA is blocked.
In reticulocyte cells the major population of mRNAs is Globin mRNAs. On the contrary, if the concentration of heme increases, more globins have to be synthesized. In such situations, heme dependent specific phosphotases dephosphorylated the eIF-2 alpha subunit, which makes the eIF-2 to be recycled by eIF-2B very easily and Globin mRNAs are translated.