Protein_Synthesis-Regulation_of_Protein_Synthesis-Prokaryotic.
Autogenous regulation:
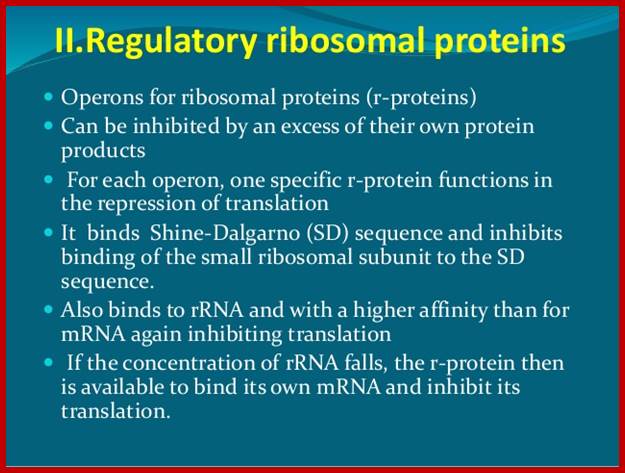
In prokaryotes, transcription and translation events are coupled. In E.coli, genes for ribo-proteins are organized into six operons and specific promoters control each of them. Each operon can code for specific number of proteins in polycistronic mode.
Larry Moran; http://sandwalk.blogspot.in/
The operons are Str, Spc, S10, Alpha, L11 and Rif. Some of the operons, in addition to riboproteins, contain other genes, whose products are the constituents of RNAP and translational factors. The operons are named after the first protein product of the polycistronic mRNA. Elongation factors EF-G and EF-Tu are found in the str-Operon. The RNAP subunit alpha is located in the operon alpha; and Beta and Beta-prime are located in rif operon.
Each of the polycistronic mRNA is translated by ribosomes; termination at the end of each cistron and reinitiation takes place at each of the intercistronic regions. They don’t have internal ribosome entry sites (IRES). Intercistronic spacer provide sequences for reinitiation for they have S/D sequences upstream of the start AUG of the next cistron.
Each of the polycistronic mRNAs, at one of the cistrons, contain sequences which generate structure which simulate the structural motif of 16s rRNA.
At least one of the protein product from each polycistronic mRNA has structural features, which show affinity to its own mRNA and bind to its own mRNA at a particular translation site, because of the mRNA in that region has structural motif similar to one of the protein binding sites in rRNA.
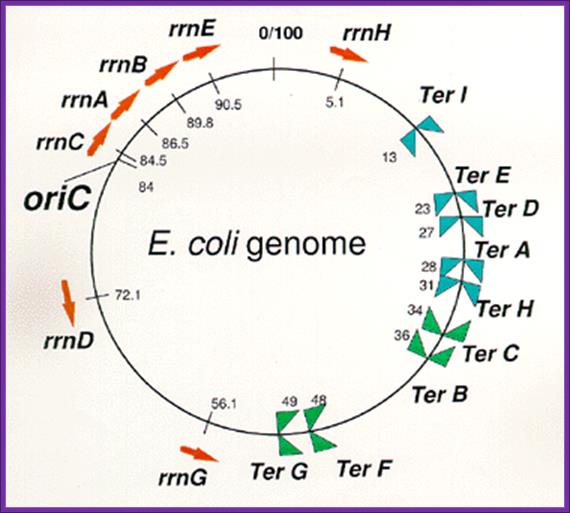
Arrangement of rrn operons and nine Ter sites on the E. coli genome.
Reds arrows indicate locations and transcriptional direction of rrn operons. ![]() means
DNA replication terminus (Ter) site which can block replication fork
approaching from the right, Professor: Takashi Horiuchi; www.nibb.ac.jp
means
DNA replication terminus (Ter) site which can block replication fork
approaching from the right, Professor: Takashi Horiuchi; www.nibb.ac.jp
|
Operon |
Poly-cistronic mRNAs |
Protein that binds |
|
Str: |
I--------I-s12---s7—EF-G—EF—Tu---I |
S7 protein binds to its own Initiating sites.
|
|
Spc operon |
I-------I—L14—L34—L5—s14--s8-L6---L55--S5-L30---L15---Y---X---I
|
The protein S-8 binds to the initiating site of L5 and Inhibits its own translation.
|
|
S10 operon |
I------I—s10---L3---L2---L4---L23—sL2---L22—s19--s3--L16—L29—I |
The L--4 protein binds to s10 region And inhibits Translation. |
|
Alpha operon |
I------I—s13—s11—s4—s9----alpha—L17—I |
The s4 binds to s13 and Prevents its translation.
|
|
L11 operon |
I------I-L11---L1—I |
The L1 binds to L11 and Inhibits its translation. |
|
Rif operon |
I-----I---L10----L7—beta—beta’—I |
The L10 binds to its own Initiator region and blocks translation.
|
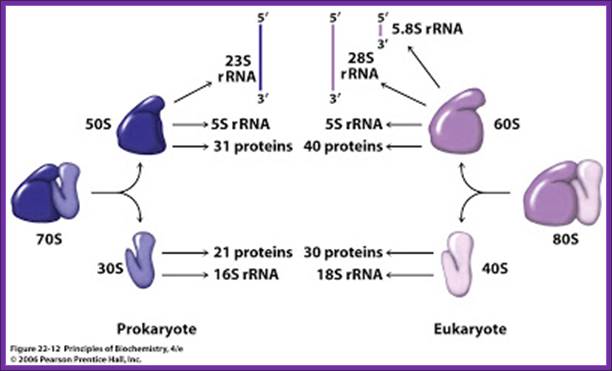
Biogenesis of ribosomes requires, at least, equimolar concentrations of both riboproteins and rRNAs. For example small ribosomal subunit formation requires one 16s-rRNA and one each of the 21 s-riboproteins for the assembly.
The autogenous regulation works, when there is insufficient number of rRNA to the excess production of riboproteins. In such situations translation of riboprotein mRNA is blocked by its one of the riboproteins, till the availability of rRNA is restored to the level that is required.
Blocking of riboprotein synthesis is achieved when specific riboproteins are in excess. One of the proteins from a specific mRNA binds to its own mRNA at a specific site.
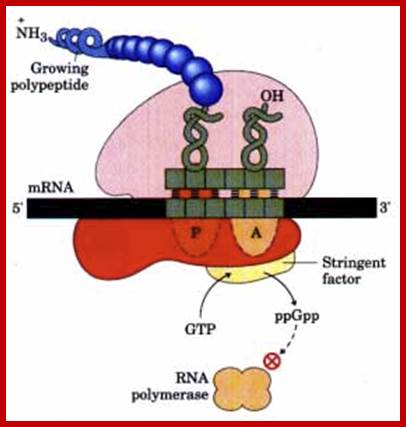
The stringent response to amino acid starvation in E. colfi is triggered by binding of an uncharged tRNA in the ribosomal A site. A protein called stringent factor binds to the ribosome and catalyzes the synthesis of ppGpp. The signal ppGpp inhibits RNA polymerase by an unknown mechanism, reducing rRNA synthesis. http://www.bioinfo.org.cn
https://www.slideshare.net/

Structure of some ribosomal protein operons in the mRNA transcripts:
The r-protein acting as translational repressor is shaded red in each case, and its site of action is indicated. Each translational repressor blocks the translation its own mRNA by binding to one of the site on the mRNA. Genes that encode subunits of RNA polymerase are shaded yellow; genes that encode elongation factors are shaded blue. (Recall that the r-proteins of the large (50S) ribosomal subunit are designated Ll to L34; those of the small (30S) subunit, Sl to 521.) https://flp.kdis.edu.cn211-xkx36/doc/ Structure of some ribosomal protein operons in the mRNA transcripts. The r-protein acting as translational repressor is shaded red in each case, and its site of action is indicated. Each translational repressor blocks the translation of all genes by binding to this one site on the mRNA. Genes that encode subunits of RNA polymerase are shaded yellow; genes that encode elongation factors are shaded blue. (Recall that the r-proteins of the large (50S) ribosomal subunit are designated Ll to L34; those of the small (30S) subunit, Sl to 521.) http://www.bioinfo.org.cn/
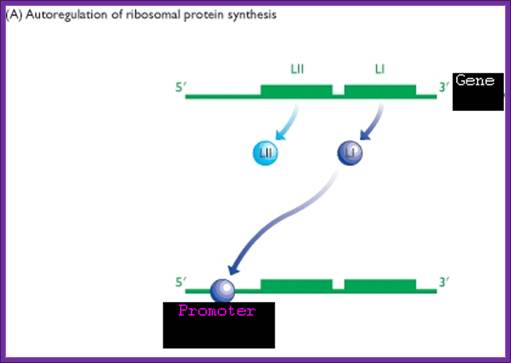
One of the riboproteins of each operon has structural feature that has great affinity to one of the structural domains of mRNA, which is similar to that of rRNA domain or motif. Binding of riboproteins to specific regions block translation, thus prevent over production of riboproteins.
When the concentration of RNA increases, because of high affinity towards rRNA structural motifs, they dissociate from their respective mRNAs and bind to rRNA to assemble ribosomes. Such a regulation is deemed as autogenous regulation.
Autogenous regulation is not just restricted to bacterial systems, but it is also prevalent in other systems.
E.coli +ŕ-L11—L1—3’
Translational regulation: one of its riboproteins L1 (general) binds to its own mRNA and blocks its translation- Sue Jinks-Robertson and Masayasu Nomura
![]()
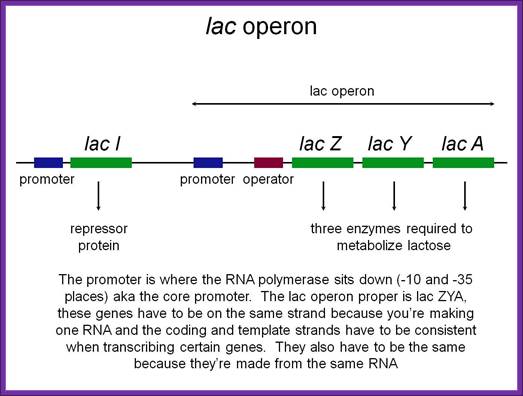
LacI gene produces a tetramer repressor that binds to LacZ operator and regulates lac operon expression. But when Laci protein is in excess, it binds to its own Laci gene promoter and blocks its expression; Flash Cards-www.study blue.com

Biology 572: Temperate phages; maintenance of lysogeny. www.science.ws
Lysogeny is induced and maintained by a repressor gene cI, The expression of it blocks many genes, but at the same time it blocks its own expression to maintain lysogeney.
P32 protein of T4 Phage:
The protein p32 of phage T4 plays a central role in its DNA repair, recombination and replication. The p32 protein has structural features for binding to ssDNA in sequence specific manner.
The p32 gene has a promoter whose sequence is compatible for the binding of p32 protein; by binding of p32 to this region of the DNA when replicating, it enhances the transcription of its own gene, thus more and more of the p32 proteins are produced.
When the concentration of these proteins exceeds to the optimum, it binds to its own mRNA at a particular site in sequence specific manner and prevents its own mRNA translation. This amounts to autogenous regulation.
- R17: This plasmid produces a protein called R17-replicase and it is responsible for the replication R17 DNA. The mRNA of R17 replicase produces a hairpin structure that attracts its own translated product and binds, thus the translation of R17 mRNA is blocked. This happens when there is excess of R17 protein.
- T4 Reg. A: Early T4 gene products bind to early mRNAs in sequence specific manner, including AUG sequences. Thus prevents the expression of T4 early genes.
T4 DNA polymerase: The DNA-pol of T4 virus is a monomer. When this protein production is in excess, it binds to Shine–Delgarno sequence of its own mRNA, thus block the synthesis of T4 DNA-pol.
RNA-phage: The Q-beta viral genomic RNA is a positive sense molecule. This on introduction into cells, it generates well-defined secondary structures. Translation of the genomic RNA starts at one of the initiating region called internal ribosomal entry site (IRES), that is the only region that a ribosome can access to and initiate translation. This is not the only genomic RNA exhibits IREs, there are others too. In this situation, because of extensive secondary structure, translation cannot be initiated in any other region. As the translation initiates at this site, leads to the opening of the secondary structure ahead in the other region; thus it facilitates the translation of the other regions of the mRNA.
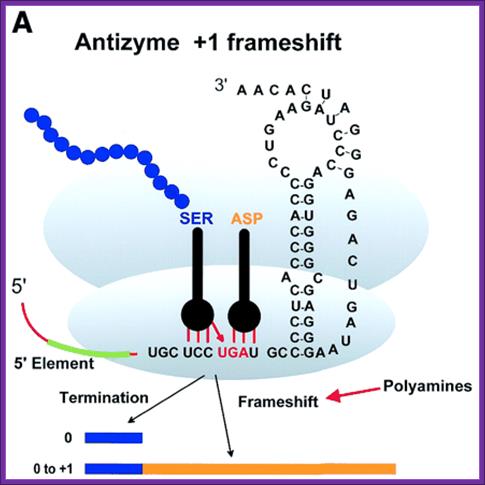
Regulation of RF-2 Synthesis: By Frame shifting:
The translation of chain releasing factor RF-2 mRNA, in E. coli, binds to its internal UGA, a TER codon and terminates protein synthesis.
The RF2 protein has terminal GGQ (mimics CCA of tRNA) as amino acid sequences which provide water for the chain release. The other end of the RF2 contains SSF which recognizes Ter codons.
The RF-2 is monomer of ~38kd (340 amino acids) protein coded for by the RF-2 gene. The RF-2 mRNA has a terminator codon at end of 340th codon. When translated it produces the functional RF-2 protein. The mRNA of RF-2 gene has another terminator codon UGA tucked within the reading frame at the end of 25th codon i.e at 26th codon.
When the concentration of the RF-2 protein is in excess, it binds to UGA at 26th codon position and terminates its translation. The 25a.a long protein is released and degraded. When the concentration of RF-2 is low, translation of the RF-2 mRNA halts at 26th UGA codon, for the reason that there are no RF-2 proteins. When the translation stalls at 26th UGA, with tRNA at its site, because of the lack of CUU UGA binding RF-2 factors, the ribosome shifts (wobbles) its reading position by one nucleotide forward from UGA to GAC, which is read as ‘leu’ by leu-tRNA, the next codon for Asp onwards the codons are read in three nucleotide each to produce functional RF-2 protein. In Chlorobium tepidium the Ter sequence is CUU UAA. This shift in the reading frame occurs with 100% efficiciency. In E. coli RF2 gene, an internal Shine–Delgarno (SD) sequence, just before the shift site, to be important for frame shifting. The frame shift actually changes the codons, yet the frame shift reading frame contains a terminator codon at the end of the ORF, RF2 terminates the translation. Inspite of the frame shift at 25th codon he codon sequences change yet it contains a terminator codon at end mRNA.
f.met- - - - - - - - - - - - -- - - --26th - - - - - - - - - - - - - - - -340th.
5’…AUG - - GGG UAU CUU UGA CUA CGA C-------UAG 3’
5’…AUG- - - - - - - - - - - - - - - -U GAC UAC GAC - --- - - - -
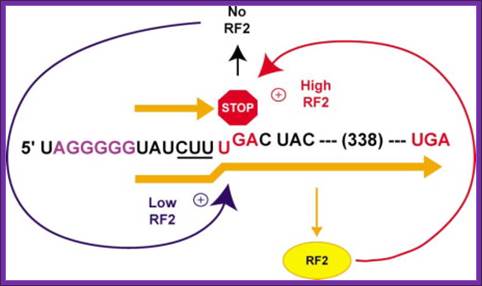
Autoregulation of RF2 Synthesis by +1 Frameshifting: The frameshift signal of the prfB gene is depicted, with the internal termination codon (UGA) highlighted (STOP) and the number of amino acids to the termination codon in the +1 frame shown. As RF2 becomes limiting (blue arrow) due to termination at this site, +1 slippage of tRNALeu occurs from the zero-frame codon CUU (underlined) to the overlapping +1 frame codon (UUU), enhanced by the upstream SD-like sequence (purple). However, as RF2 accumulates (red arrow), translation termination begins to predominate. Arial Unicode MS
Shifting reading frame by (+) 1: From: Recoding: Site- or mRNA-Specific Alteration of Genetic Readout Utilized for Gene Expression- (A) Antizyme frame shifting. The +1 shift at the last codon (UCC) before the termination codon of ORF1 of human antizyme 1 is stimulated by polyamines and by a 5′ mRNA element and a 3′ pseudo knot. Ivaylo P. Ivanov, et al; http://www.landesbioscience.com/
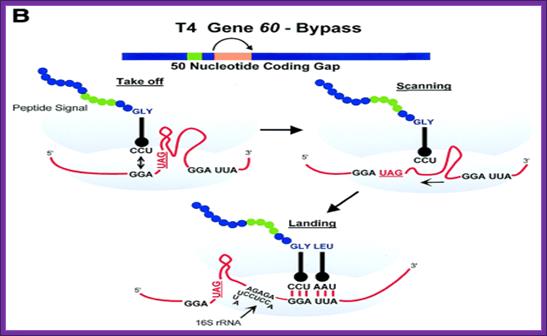
Gene 60 codons bypassing:
Fifty nucleotides between codons 47 and 48 of phage T4 gene there are 60 coding sequence; they are bypassed by ribosomes in response to matched take-off and landing site codons, a stop codon directly after the take-off site in a stem–loop structure and a nascent peptide signal that acts within the ribosome.
Skipping 60 coding sequences during translation; https://www.wjgnet.com

Internal ribosome entry site-based vectors for combined gene therapy: Efficient T4 gene 60 translational bypassing requires five bypassing signals. These signals include matching GGA codons bordering the gap, a stop codon, a short stem-loop structure, an optimal 50-nt spacing, and a region of the nascent peptide. https://www.ncbi.nlm.nih.gov

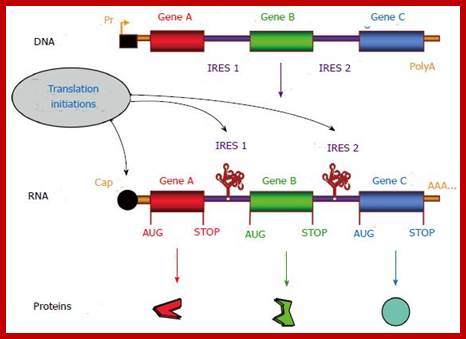
Cap-dependent and internal ribosome entry site-dependent initiation, two alternative mechanisms of translation. A: The so-called cap-dependent ribosome scanning mechanism predicts that ribosome 40S subunit binds to the mRNA 5’ end. Ribosome binding requires the initiation factor 4F (eIF-4F, composed of the three proteins eIF-4E, -4A and -4G). Then the mRNA is unwound under the control of the helicases eIF-4A and -4B, allowing the ribosome to scan the mRNA until recognition of an initiation codon (classically AUG)[11,12]; B: When an Internal ribosome entry site (IRES) is present in the mRNA 5’ untranslated region, IRES trans-acting factors (ITAFs) allow ribosome 40S internal recruitment, independently of the presence of cap and eIF-4F. The IRES-dependent mechanism occurs in the case of picornavirus uncapped mRNAs as well as for cellular capped mRNAs.
Gene therapy appears as a promising strategy to treat incurable diseases. In particular, combined gene therapy has shown improved therapeutic efficiency. Internal ribosome entry sites (IRESs), RNA elements naturally present in the 5’ untranslated regions of a few mRNAs, constitute a powerful tool to co-express several genes of interest. IRESs are translational enhancers allowing the translational machinery to start protein synthesis by internal initiation. This feature allowed the design of multi-cistronic vectors expressing several genes from a single mRNA. IRESs exhibit tissue specificity, and drive translation in stress conditions when the global cell translation is blocked, which renders them useful for gene transfer in hypoxic conditions occurring in ischemic diseases and cancer. IRES-based viral and non viral vectors have been used successfully in preclinical and clinical assays of combined gene therapy and resulted in therapeutic benefits for various pathologies including cancers, cardiovascular diseases and degenerative diseases. https://www.wjgnet.com/
https://www.wjgnet.com
Skipping 60 coding sequences during translation; Somogyl,P wet al;http://www.umich.edu/
Prokaryotic selenocysteine insertion and redefinition: Selenocysteine is decoded by UGA Ter codons in sel-cys encoding mRNAs found in prokaryotes and even in eukaryotes. The selenocysteine incorporating mRNA contain a stem loop sequence at the 3’ end of the UGA codon. The selenocysteine gets incorporated into the protein directly at UGA followed by a stem loop structure whose apical loop is bound by a selenocysteine tRNA and specific elongation factor SELBp, resulting in a tethered amino-acylated tRNA poised for the oncoming ribosome. These figures are adapted from Atkins et al.

Bioinformatics.upf.edu- glossory
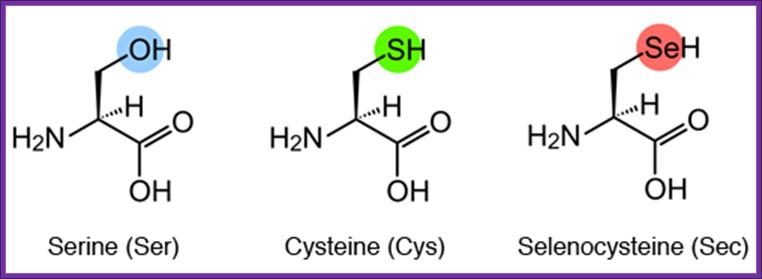
Structural formulae of serine (Ser), cysteine (Cys), and Sec
Sec is an amino acid in which the oxygen (O) of Ser or the sulfur (S) of Cys is replaced by Se. Cysteine is not converted to selenocysteins, but serine is converted to selenocysteine by incorporation od selenium, http://www.spring8.or.jp/
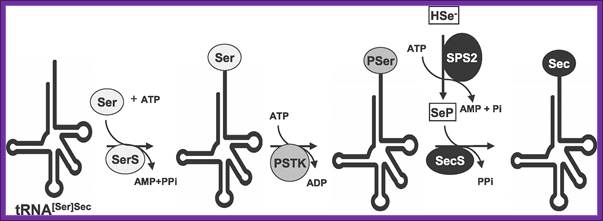
Sec Biosynthesis in Eukaryotes-The pathway of Sec biosynthesis is shown (see text for details and abbreviations are defined in the text with the exception of SerS [seryl-tRNA synthetase] and PSTK [O-phosphoseryl-tRNA[Ser]Sec kinase]), Xue-Ming Xu equal contributor, Bradley A Carlson et al, PLOS/Biology.
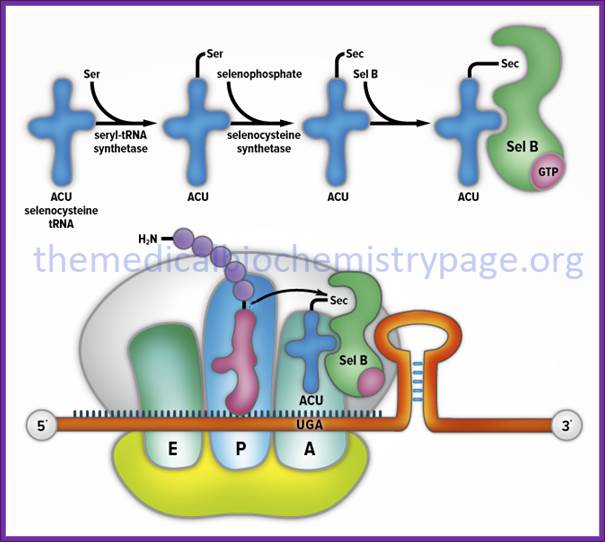
Selenocysteine biosynthesis and incorporation. The first steps
involve the activation of serine onto the (Sec)-tRNA followed by enzymatic
conversion to selenocysteine generating (Sec)-tRNA(Ser)Sec. Next the
(Sec)-tRNA(Ser)Sec is
bound by SelB and the complex is incorporated into the translational machinery
aided by SBP2 (not shown). The elongating protein is transfered to the
selenocysteinyl-tRNA via the action of peptidyltransferase as for any other
incoming amino acid and normal elongation continues. http://themedicalbiochemistrypage.org/
The tRNA is loaded with serine,
and then selenium is added to the OH group of serine thus seleno-cysteine is
produced. Cysteine tRNA is not involved in this process. The selenium carrying tRNA
contains 3’ACU5’ as anticodon sequence. Structurally this tRNA has different
base pair in the acceptor stem. This selenium cysteine tRNA is bound to stem
loop structure found at the 3’ end, and it is this that decodes UGA as selenium
cysteine. Several proteins are involved in the conversion of cysteine into
selenocysteine which decodes UGA as selenocysteine.
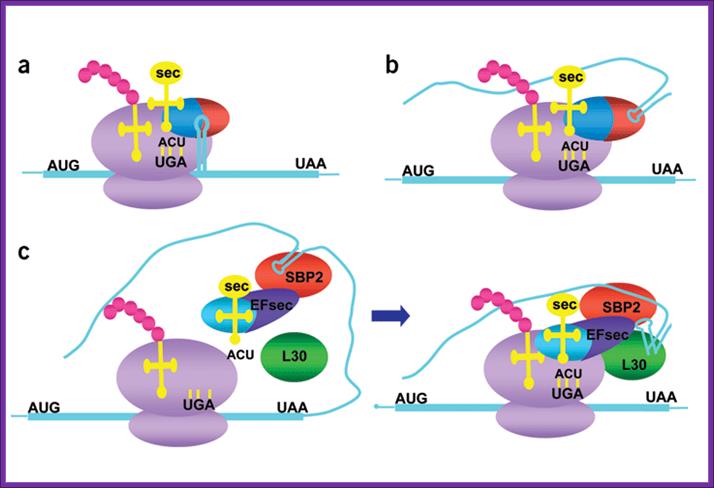
Knowing when not to stop; Models for selenocysteine incorporation:
(a,b) Models for prokaryotic (a) and (b) archaeal recoding machinery. mRNAs are
blue, ribosomes purple, tRNAs yellow, nascent peptide pink, and codons black.
SELB from prokaryotes and archae are shown as red and blue ovals (blue,
elongation factor domain; red, SECIS-binding domain). (c) Current model of eukaryotic recoding machinery. Eukaryotic
EFsec is blue and dark purple (blue, elongation factor domain; dark purple,
SBP2-interaction domain). SBP2 is red and L30 green. The kink-turn in the SECIS
element is depicted in the right panel.
Mechanism of co translational incorporation of selenocysteine:
Artemy Beniaminov, Akiko Takeuchi, Laurence Wurth and Christine Allmang
In prokaryotes incorporation selenocysteine into proteins require a number of proteins such as selB, SECIs element and stem loop binding proteins SBP1 and SBP2.
In eukaryotes, decoding of UGA selenocysteine codons also requires a complex molecular mechanism containing RNA and protein factors, some of which are already characterized while others are still unknown. Our goal is to identify all the components and to obtain a detailed understanding of their function.
The tRNASec and the Selenocysteine insertion elements SECIS (an RNA stem-loop in the 3’UTR of selenoprotein mRNAs) play crucial roles in UGA Sec decoding. Our laboratory proposed earlier structure models for the tRNASec and SECIS RNA, has isolated and functionally characterized the mammalian specialized translation elongation factor eFSec as well as the human SECIS-binding protein SBP2. To better understand the principles governing the SBP2-SECIS RNA interaction and the function of SBP2, which are at the heart of the selenoprotein synthesis mechanism, we set out to determine the crystal structure of this complex, in collaboration with the group of Philippe Dumas. Moreover, we are actively attempting to identify and characterize the molecular partners of SBP2.
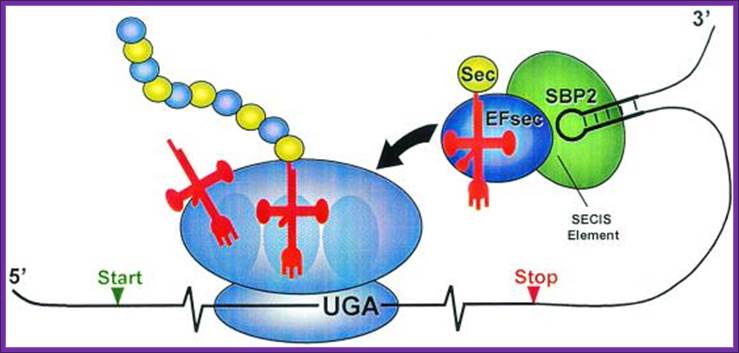
Mecanismo de inserción de Sec en eucariotas.; http://bioinformatica.upf.edu/
In
eukaryotes, seven gene products were found necessary for selenocysteine
biosynthesis and its co-translational incorporation. It is very likely that
other, as yet unidentified partners are involved. Key-steps are still to be
discovered and dissected, and this precisely constitutes our objective.
Selenocysteine is a cysteine analogue with a selenium-containing selenol group in place of the sulfur-containing thiol group. It is present in several enzymes for example glutathione,peroxidases, tetraiodothyronine5'deiodinases, thioredoxin reductases, formate dehydrogenases, glycine reductases, selenophosphate synthetase 1, methionine-R-sulfoxide reductase B1 (SEPX1), and some hydrogenases; there are 25 human protein containing selenocyateine amino acid.
Selenocysteine was discovered by biochemist Theresa Stadtman, wife of Earl R. Stadtman, at the National Institutes of Health –WIKIPEDIA;
Stringent Response in Bacteria and other systems: Production Guanosine tetra phosphate;
Under optimal requirement
situation, if there is a slowdown in the synthesis of rRNA, the riboproteins
accumulate and it is not desirable. What is intriguing is that the regulation
of rRNA synthesis to the requirement of ribosomal protein is not very well
answered. Under amino acid deficiency pppGpp is produced and the same is rapidly
converted to ppGpp by a pppGpp-5′-phosphohydrolase. This is often
referred to as ‘stringent response’. These and further observations suggested
that ppGpp is involved in the control of rRNA synthesis. Subsequently, it was
observed that ppGpp specifically inhibits rRNA synthesis in vitro , presumably
by reducing the affinity of the RNA polymerase to stable RNA promoters,
Patrick P. Dennis1,*, Mans Ehrenberg2
and Hans Bremer3-2004.
When bacterial cells cultured under deficiency of nutritional inputs or deficiency of certain amino acids, cells reduce rRNA and tRNA synthesis by 10-20 fold and mRNA synthesis by 3 fold. Even carbohydrate synthesis, lipid synthesis and nucleotide synthesis is reduced considerably. This kind of response is called stringent response. In such conditions molecules such as Guanosine tetra phosphates are produced (5’ppGpp3’). These molecules are also called magic spot molecules, for they can be detected by paper chromatography.
· When a particular amino acid or amino acids are deficient or if there is any mutation in aminoacyl-tRNA synthase, uncharged tRNA binds to the ‘A’ site and idles the translation process for the lack of charged tRNAs.
· One of the riboproteins of 50s, called L-11 activates another associated protein called Rel-A. This protein is associated with ribosomes, one for every 200 ribosomes.
· Rel-A protein is an enzyme, when activated it catalyses phosphorylation of GTP that is bound to EF-Tu, to generate Guanosine pentaphosphate-5’pppGpp3’. It uses both ATP and GTP as substrates for the synthesis of G tetra phosphate
· Then one of the 5’ P is dephosphorylated by a specific phosphatase to generate 5’ppGpp3’ molecule.
· Before the synthesis of another molecule of 5’ppGpp3’ the first tRNA-EF-Tu factor should be displaced by new EF-Tu-tRNA to generate one more 5’ppGpp3’.
· Ribosomes during stringent response conditions, were able activate the synthesis of ppGpp molecules only when an uncharged tRNA is bound to a specific codon at ‘A’ site.
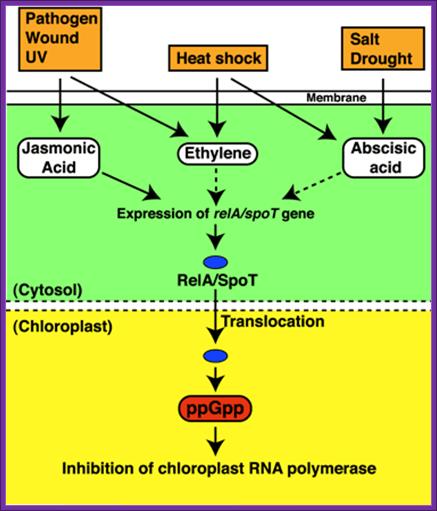
· A Variety of factors induce Rel A in plants. ex. Heat shock, salt draught, pathogen wounding, UV radiations; the Rel A is then transported into chloroplast and binds to ribosomes and induce the synthesis of 5’ppGpp3’.
ppGpp and pppGpp
5’pppG-OH3’ + 2ATP ---> 5’pppG-pp3’ + 2ADP
5’pppGpp3’ -----> 5’ppGpp3’ + pi
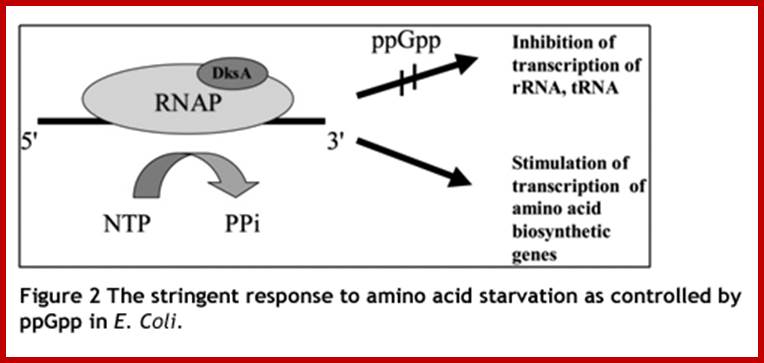
Following amino-acid starvation, the cells reduce themselves to a
minimal numbers, sufficient for surviving the starvation period. Thus, they are
able to recover quickly when nutrients become available again. https://www.trilinkbiotech.com
A proposed model for (p)ppGpp synthesis during amino-acid starvation:
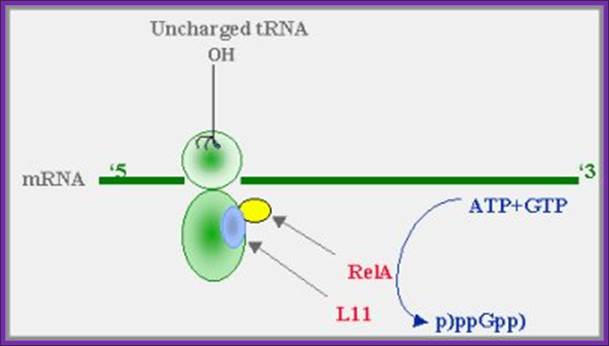
When aminoacylation of any tRNA cannot keep up with the demands
of protein synthesis, transient stalling of ribosomal elongation occurs. The
product of the relA gene (relA), which is bound to the ribosomes,
in association with the ribosomal protein L11, is activated and catalyzes the
synthesis of pppGpp from GTP. (guanosine 5’diphosphate, 3’diphosphate), The (p)ppGpp then "turns on" the stringent response.
Liitle is known about Rel P, Q and V; ftp.mcs.anl.gov/pub/compbio/
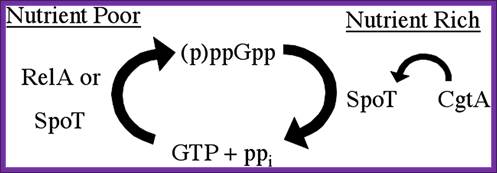
Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholera.Simplified model for the essential CgtA activity. Nutritional stress induces RelA or SpoT to generate ppGpp by phosphorylation of GTP. SpoT is required to hydrolyze ppGpp to prevent growth inhibition. CgtA is required to maintain normal SpoT ppGpp hydrolysis activity. http:// www.PNAS.org- David M. Raskin
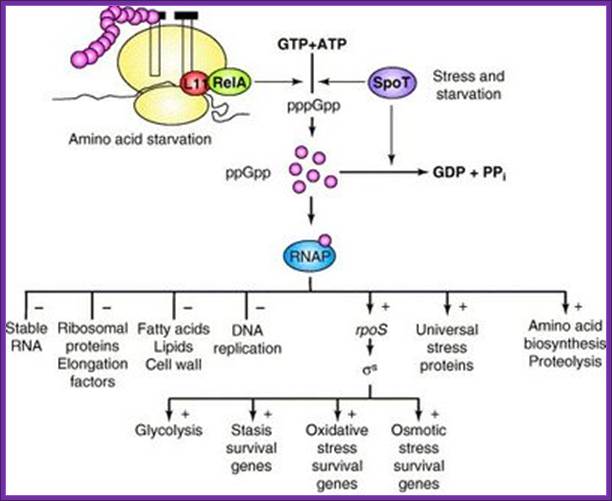
Diagram of the bacterial stringent response system. Two
parallel pathways synthesize pppGpp (which is subsequently
con-
verted into ppGpp) from ATP and GTP in response
to any of several
stress signals. Through interactions with RNA
polymerase (RNAP),
ppGpp alters much of metabolism (– indicates
inhibition; + indicates
stimulation). Source.
http://schaechter.asmblog.org/.a/6a00d8341c5e1453ef0133f386a181970b-400wi; Fred Neidhardt
· When conditions return to normalcy, a gene product called SPO-T in B.subtilis quickly degrades these magic molecules by certain specific phosphatase reactions where one of the 5’ P is removed. It is suspected that EF-G or EF-Tu is involved in degradation of the magic molecule.
Mechanism of RelA-mediated (p)ppGpp synthesis:
Under conditions of amino acid starvation, large pools of uncharged tRNAs are generated that can bind to the A-site of the ribosome with low affinity and as a result block the ribosome. (a). RelA detects a blocked ribosome thereby stimulated by 3’mRNA extensions protruding from the ribosome (b). RelA binds the ribosome and mediates (p)ppGpp synthesis (c). Concomitantly with (p)ppGpp synthesis, the uncharged A-site tRNA, is released. RelA hops to the next ribosome (d) and the process is repeated leading to levels of (p)ppGpp required to activate the stringent response. Replenishment of charged tRNAs binding with higher affinity to the A-site following post-starvation conditions enables replacement of uncharged tRNAs which rescues blocked ribosomes and reactivates translation (e). E, P and A refer to the corresponding sites on the ribosome. Different colors represent different tRNAs. (Braeken et al., 2006)
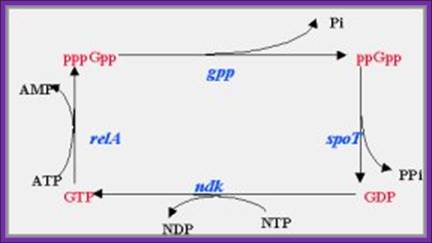
Action of SpoT
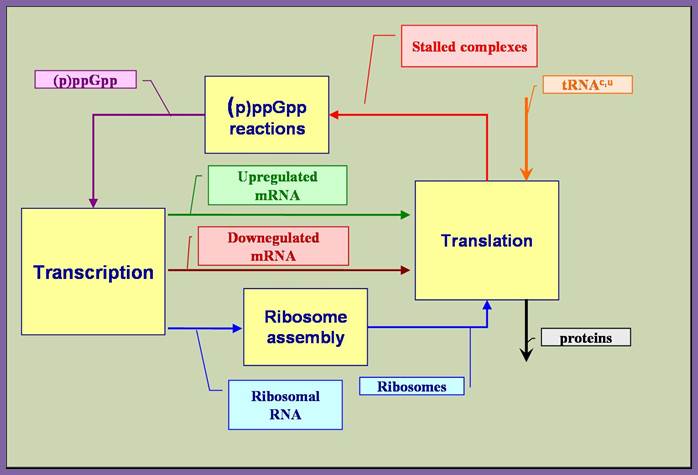
The above diagram illustrates the main feedback loop involved in the stringent response mechanism. While the main features of the stringent response mechanism outlined above are generally accepted, there is disagreement regarding the specific mechanism that achieves the differential regulation; http://biocomp.cis.upenn.edu/
The magic molecules are believed to act on the promoters of rRNA genes and reduce initiation and elongation of transcripts, it may also act on RNAPs and stall the reaction or it can interact with the template itself, thus it can have multiple effects on cellular processes including metabolism. These molecules are considered as global regulators. Costanzo and Ades (2006) have demonstrated that Sigma-E, independently misfolded protein, also is activated by the stringent response mediated by ppGpp. The ppGpp had previously only been known to activate Sigma-S and Sigma-54 following nutritional stress. When the cell is starved of an amino acid this nutritional stress is sensed and mediated by the stringent response. The stringent response has an impact on a vast amount of operons and including a strong down regulation of rRNA and tRNA to save energy. During exponential growth more than half of the RNA polymerases are involved in transcription of rRNA and tRNA so down regulation of these genes will save a lot of transcriptional power for alternative operons. If an uncharged tRNA binds to the A site in the ribosome, this will cause the activation of ribosome-bound enzyme RelA (RELaxed control gene A). Activated RelA enzymes will synthesize ppGpp which then binds to two active sites of RNA polymerase. ppGpp’s function is not fully understood but the RNAP preferences for different promoter sequences change upon binding to ppGpp. In the presence of a sufficient amount of amino acids the protein SpoT normally degrades ppGpp, hence stops the stringent response.”
Stringent control mediated by the bacterial Alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) is a key regulatory process governing bacterial gene expression. By devising a system to measure ppGpp in plants, we have been able to identify ppGpp in the chloroplasts of plant cells. Levels of ppGpp increased markedly when plants were subjected to such biotic and abiotic stresses as wounding, heat shock, high salinity, acidity, heavy metal, drought, and UV irradiation. Abrupt changes from light to dark also caused a substantial elevation in ppGpp levels. In vitro, chloroplast RNA polymerase activity was inhibited in the presence of ppGpp, demonstrating the existence of a bacteria-type stringent response in plants. Elevation of ppGpp levels was elicited also by treatment with plant hormones jasmonic acid, abscisic acid, and ethylene, but these effects can be reversed completely by another plant hormone, indole-3-acetic acid. On the basis of these findings, we propose that ppGpp plays a critical role in systemic plant signaling in response to environmental stresses, contributing to the adaptation of plants to environmental changes
Causative factors for the synthesis of ppGpp; Proposed model for ppGpp signal transduction in plants. The dotted-line arrows represent pathways that are not yet demonstrated experimentally. http://www.pnas.org
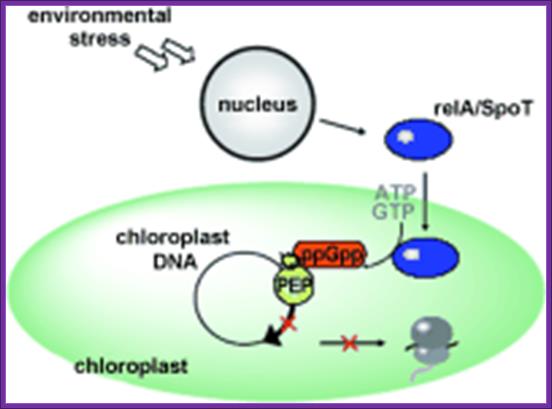
Rel-A induces the synthesis of ppGPP in chloroplast
A hyper phosphorylated guanosine nucleotide, (p)ppGpp, was initially identified as the effector molecule responsible for the stringent response in Escherichia coli under conditions of amino acid starvation (Figures). However, a rapidly growing number of reports shows that (p)ppGpp-mediated regulation is conserved in many bacteria and even in plants. It is now clear that (p)ppGpp acts as a global regulator during physiological adaptation of the organism to a plethora of environmental conditions. Adaptation is not only essential for surviving periods of stress and nutrient exhaustion but also for the interaction of bacteria with their eukaryotic host, as observed during pathogenesis and symbiosis, and for bacterial multicellular behavior.
Responses to chemical, physical and biological stresses: involvement of extracellular alarmones, pheromones and varisensors.
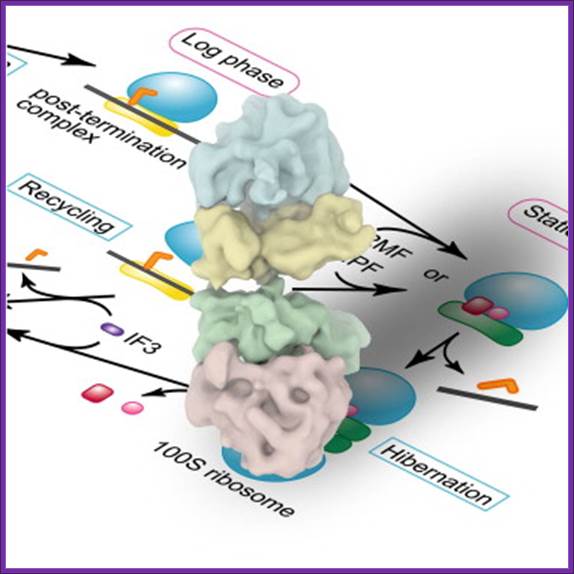
The above said agents do more than this, however, in that they both protect organisms from potentially lethal chemical stress agents in the stationary-phase, but also from the same and other agents in exponential phase. Ribosome Modulation Factor RMF motif located in 5’UTR of the RMF gene, which encode RMF factor. The RMF has two major properties namely: (1) leading to ribosome dimerization; the active 70S particles dimerising to form inactive 100 S ones, whilst (2) this ribosome binding component also influences rRNA degradation. It appears that the latter property is more important as a means of protecting organisms from stress. Of interest is the finding that ppGpp controls RMF synthesis, probably explaining why this component, which is not influenced by the sigma factor, Rpo S, nonetheless increases in level in stationary-phase. Interaction between 70s ribosomes involve proteins like small subunit proteins, S2, S3 and S5, appear to be critical for the dimerization. The RMF is a basic, small protein (pI = 11.3; MW = 6507 Da), and its expression level remarkably increases during the transition from the log to the stationary phase. Another protein factor expressed during the stationary phase and binding to ribosomes is the Hibernation Promoting Factor (HPF), which promotes the formation of the 100S ribosome by the RMF. It has been reported that the RMF inactivates ribosomes by covering the peptidyl transferase center and the exit of the peptide exiting tunnel (Yoshida et al., 2002 and Yoshida et al., 2004).
It has also been suggested that the HPF binds to the 30S subunit at the subunit interface (Maki et al., 2000 and Ueta et al., 2005). These results suggest that the RMF and HPF bind to the interface between the 50S and 30S subunits. Our (Takayuki Kato1, 2, 5, Hideji Yoshida3, 5,Tomoko Miyata) present study suggests that the scheme of ribosome cycle should be modified as shown in Figure 5. The binding of the RMF and HPF to the ribosome for the formation of the 100S ribosome must occur before the binding of IF3 to the ribosome. It is speculated that the transition from the 70S ribosome to the 100S ribosome starts before or after the recycling stage by RRF and EF-G. In both cases, tRNA and mRNA must be released by some factors other than IF3, which may be RMF and/or HPF, or other protein factors.
First, although studies of stationary-phase stress have not been made, for all the others the switching-on of the response involves extracellular components (ECs). For each, a pair of ECs, controls response triggering. Thus, unstressed cells produce, in their media, the first EC pairs, namely the extracellular sensing component (ESC). Different ESCs are produced for each type of stress tolerance response, and activation of the ESC by interaction with the stressor forms the second EC, the extracellular induction component, EIC. It is this latter that attaches to a receptor on the sensitive organism and induces tolerance, probably after the entry into cells.
The 100S ribosome was purified from Escherichia coli in the stationary growth phase ► The structure was analyzed by electron cryomicroscopy and single-particle image analysis ► The 100S ribosome is composed of two tRNA-free 70S ribosomes ► The 100S ribosome is formed through contacts between 30S subunits and has two-fold symmetry.
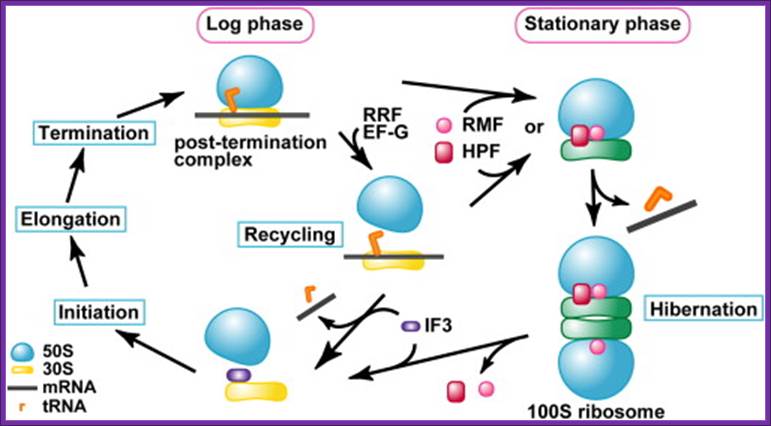 Schematic Diagram Showing the Formation Process of the 100S
Ribosome upon Transition from the Log Phase to the Stationary Growth Phase Protein
factors involved in the process are: RRF, ribosome recycling factor; EF-G,
elongation factor G; RMF, ribosome modulation factor; HPF, hibernation
promoting factor. Takayuki Kato1,
2,
5,Hideji Yoshida
Schematic Diagram Showing the Formation Process of the 100S
Ribosome upon Transition from the Log Phase to the Stationary Growth Phase Protein
factors involved in the process are: RRF, ribosome recycling factor; EF-G,
elongation factor G; RMF, ribosome modulation factor; HPF, hibernation
promoting factor. Takayuki Kato1,
2,
5,Hideji Yoshida
The RMF factor binds to PTC region and dimerizes ribosomes, and makes it functionally inactive.
Alarmones: Diadenine Tetra Phosphate, 5’AppppA3’: Kosaku Takahashi, Koji Kasai, and Kozo Ochi *
Under alarming conditions such as specific severe deficiencies, another kind of regulatory molecules are generated which act as hormones, hence the name Alarmones (Alarm + hormones = Alarmones); ppGpp are also called Alarmones.
One such molecule is Diadenine Tetra phosphate, 5’AppppA3’. Synthesis of the said molecules is catalyzed by bacterial aminoacyl tRNA synthetases. This enzyme gets activated and acts in the absence of specific tRNA. Diadenine Tetra phosphate acts like a signal molecule. Another Alarmone molecule is 5’ amino 4-imadazole carboxiamide ribose 5’phosphate (ZTP). It acts as second messenger stimulates phospholipase D and changes intracellular Ca2+ ion concentration and induces Nitric oxidase and inhibits adenosine kinase activity.

Diadenine tetra phosphate:
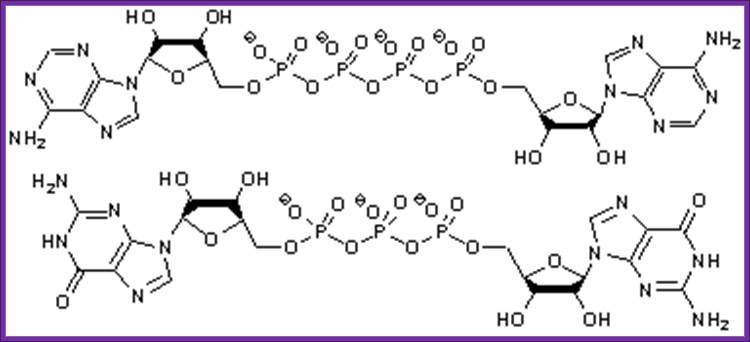
Diadenine Tetra phosphate and tri phosphate
Some of the alarmones produced in eukaryotes can diffuses from one cell to the other, like any other hormones and bring about reaction. In EK one such Alarmones acts on DNA pol alpha subunit and replication of DNA molecule is disrupted.
Dinucleoside polyphosphates are an unusual class of nucleotides discovered in the 1960s during studies into aminoacyl-tRNA synthases. These compounds are ubiquitous, but poorly understood with apparent involvement in a wide variety of intra- and extracellular mechanisms. Examples are Ap3-5A, Gp3-5G, Ap4G and Gp4U. Eukaryotic cell dinucleoside polyphosphate concentrations show a distinctive increase in response to stress and there are suggestions for a potential involvement in the onset of apoptosis and cell differentiation. More generally, such compounds appear to act as signaling molecules associated with blood platelet aggregation, neuro- and cardio protection, and vasoconstriction/dilation. Previous studies have been hampered by these compounds' tendency to be rapidly hydrolysed in vivo, as well as the unavailability of fluorescently-substituted, affinity activated or otherwise chemically modified analogues. Available chemical synthetic routes were difficult and expensive (low yields) with product purification being an additional obstacle.
The present study demonstrates that 99mTc-labeled adenine nucleotide analogs (Ap4A and AppCHClppA) that are stable in vitro and in vivo are easily prepared in high yield and purity. The competitive inhibition of Ap4A and AppCHClppA at ADP association sites on platelets makes them potentially highly selective pharmacological agents for vascular lesions where platelets show early localization and aggregation. These radiopharmaceuticals have potential both as radio diagnostic agents for the rapid detection of atherosclerotic plaques and for probing the fundamental patho physiology of atherogenesis.
One of the first established alternative functions for a class II AARS, the E. coli LysRS, was the biosynthesis of the dinucleotide polyphosphates, Ap3A and Ap4A.31 The general formula for these dinucleotide polyphosphates is abbreviated “ApnA,” where “n” represents the number of phosphates bridging the two adenosine moieties.

General structure of a diadenosine oligo-phosphate (ApnA). “n” represents the number of linked phosphate moieties.
Synthetic mRNA cap analogs with tetra phosphate 5’-5’ bridge in eukaryotes:Anna Maria Rydzik, Maciej Lukaszewicz, Joanna Zuberek, Joanna Kowalski, Zbigniew Marek Darzynkiewicz, Edward Darzynkiewicz and Jacek Jemielity, Org. Biomol.Chem.,2009,7,4763;DOI: 10.1039/b911347a
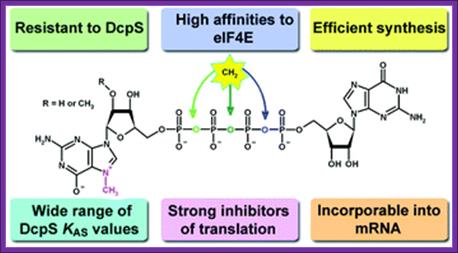
The cap analogs modified with a methylene group in the Tetra Phosphate Bridge is presented above. The compounds are potent inhibitors of translation in vitro with increased enzymatic stability and have high affinity for eIF4E proteins.
Four of the analogs were resistant towards enzymatic degradation by human Decapping Scavenger enzyme (DcpS). Binding studies of non-hydrolyzable analogs with DcpS revealed a broad range of KAS values for different analogs. All of the analogs were potent inhibitors of translation in a rabbit reticulocyte lysate system (RRL) and those resistant to DcpS turned out to be stable under an elongated time of pre-incubation while the inhibitory potency of standard was diminished in these conditions. For Anti Reverse Cap Analog (ARCA) dinucleotides (4–6), we have shown that they are effectively incorporated into mRNA and transcripts capped with these analogs undergo translation in vitro
Regulation of Bacterial Stress Responses: Carin K Vanderpool
In my laboratory we are interested in understanding how bacteria sense stressful environmental conditions and modulate gene expression in a way that allows them to adapt to or recover from the stress and continue to grow and compete with other cells in their environment. We are particularly interested in the roles of small regulatory RNAs in regulation of bacterial stress responses. Small RNAs (sRNAs) have diverse roles in regulation of cellular processes. The sRNAs are crucial for development in higher organisms. In bacteria, The sRNAs regulate responses to many environmental stresses. Global screens for sRNAs in Escherichia coli have uncovered more than 80 novel sRNAs of unknown function; a subclass of these bacterial sRNAs has a functional requirement for binding to the RNA chaperone Hfq. This type of sRNA has been found to regulate translation or stability of mRNA targets by a mechanism that requires sRNA: mRNA base pairing interactions (refer to the Fig).
![]()
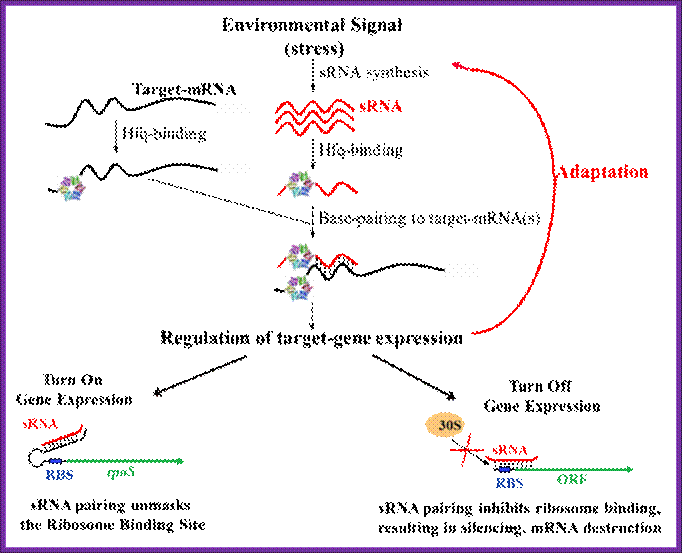
A large number of sRNAs are generated in bacteria. Hfq helps to stabilize the sRNA and binds both sRNA and mRNA, accelerating annealing in vitro. We are interested in how these small RNAs fit into regulatory circuits, and how Hfq and other proteins promote small RNA function; Susan Gottesman;
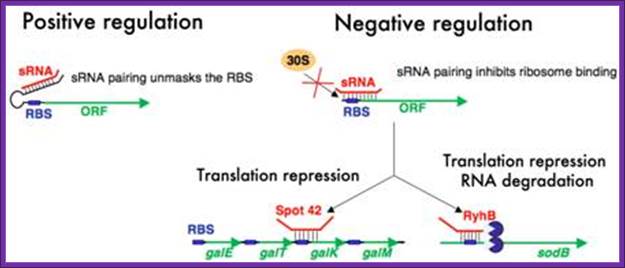
Mechanisms of regulation by bacterial sRNAs. Small RNAs can positively regulate translation of target mRNAs by binding to an upstream part of mRNA 5’ untranslated regions and preventing formation of a translation-inhibitory hairpin structure (left diagram). Negative regulation by sRNAs typically involves base pairing interactions that occlude the ribosome binding sites of mRNAs. This prevents ribosome association and thus represses translation (right diagram). In some cases translation inhibition is coupled to RNA degradation and in other cases it is not; Cari Vanderpool
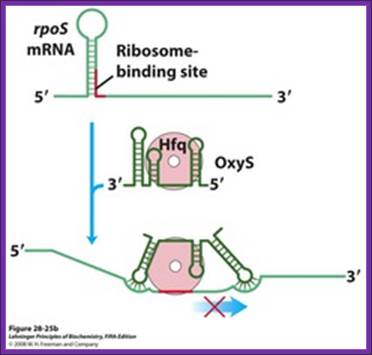
Regulation of bacterial mRNA function in trans by sRNAs. Several sRNAs (small RNAs)ムDsrA, RprA, and OxySムare involved in regulation of the rpoS gene. All require the protein Hfq, an RNA chaperone that facilitates RNA-RNA pairing. Hfq has a toroid structure, with a pore in the center. (b) OxyS blocks translation by pairing with the ribosome-binding site. http://quizlet.com

Houstan, Texas; Nicholas De Lay
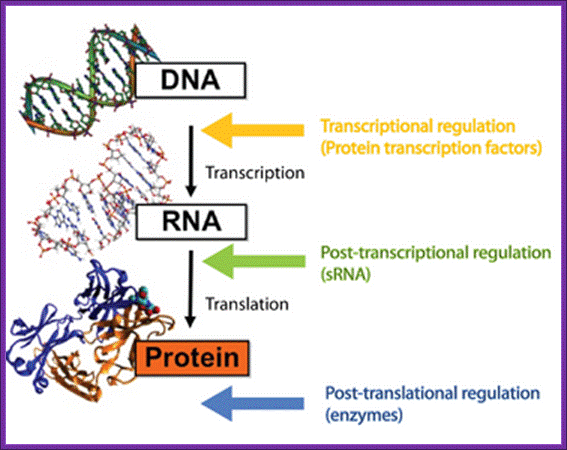
The central dogma in biology, where the different levels of regulation and the players involved are highlighted. Roger Wartell Seminar.
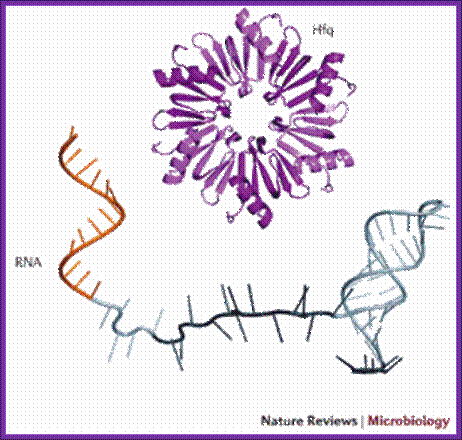
Hfq is RNA binding protein, Salmonella expresses~100 sRNAs; The Hfq-dependent sRNAs typically modulate protein synthesis by using short imperfect base-pairing with target mRNAs, thus altering translation and stability of the mRNA. We now understand that a single sRNA can regulate many target mRNAs using a highly-conserved short (≥7 nucleotide) seed sequence, yet how sRNAs act select with high specificity their targets in the background of thousands of other cellular transcripts is not understood. Equally, do proteins other than Hfq help mediate sRNA activity? Other fundamental questions which we are addressing are what are the benefits of using an RNA regulator versus a transcriptional factor in complex regulatory networks; how are the sRNAs themselves regulated; and how does this relate to virulence; Vogel
Mechanism of RNA silencing in Escherichia coli:
Escherichia coli have several trans-acting sRNA molecules repressing the expressions of their target genes, such as micC and sgrS 9sugar transport-related sRNA) and so on. Their mechanism has been becoming clear in recent years. (Fig.below) These sRNA molecules consist of complementary sequences to their target mRNAs and a scaffold sequence that recruits the Hfq (Host factor q protein or HF1) protein. Hfq protein, one of the RNA chaperones, facilitates the hybridization of sRNAs to mRNA by their complementary sequences and with RNase E. RNase E causes the degradation of the sRNAs by the target mRNAs. So the mechanism of gene repression by Hfq-dependent sRNAs is composed of the translational repression by their complementary sequences and Hfq proteins and the degradation of the target mRNAs by RNase E.
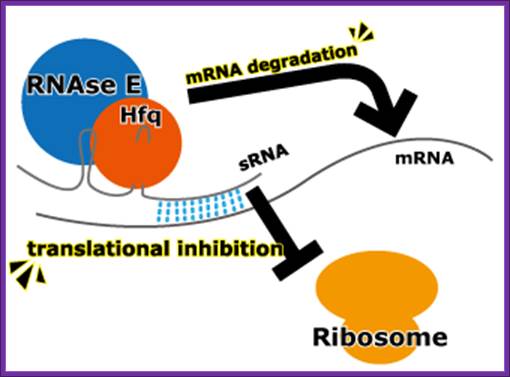
Mechanism of expression inhibition by Hfq protein-dependent sRNAs Design and use of synthetic regulatory small RNAs; http://2013.igem.org
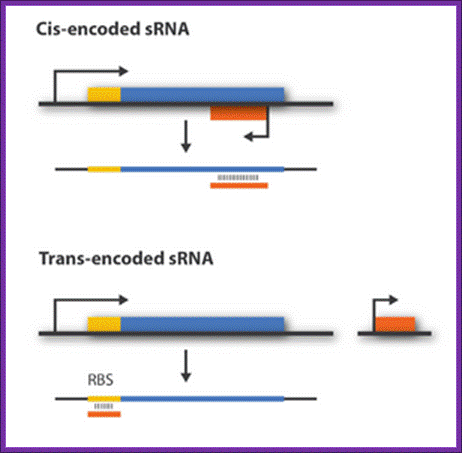
Regulatory sRNAs are divided into different sub-groups depending on their genomic locations with respect to its mRNA target. Cis-encoded or antisense sRNAs are encoded just opposite the one gene they regulate whereas trans-encoded sRNAs are located somewhere else in the genome and can regulate multiple target mRNA. Antisense RNA share extensive sequence complementarity with its target mRNA and hence do not require the RNA chaperone, Hfq, to stabilize the complex. This is in contrast to trans-encoded sRNAs which often only possess a complementary base-pairing region of 6-20 nucleotides and often require Hfq. We are focusing on trans-encoded sRNA in the project, so whenever we write sRNA we implicit mean trans-encoded sRNA; There a number of repressor sRNAs and equal number of activator sRNAs; http://2011.igem.org/Team
Hfq-dependent small RNAs can positively regulate translation of target mRNAs by binding to an upstream part of mRNA 5’ untranslated regions and preventing formation of a translation-inhibitory hairpin structure (left diagram). Negative regulation by Hfq-dependent sRNAs typically involves base pairing interactions that occlude the ribosome binding sites of mRNAs. This prevents ribosome association and thus represses translation (right diagram). In some cases translation inhibition is coupled to RNA degradation and in other cases it is not.
Understanding the role of small RNAs in glucose-phosphate stress
We study an sRNA called SgrS (sugar stress sRNA). SgrS is required to allow E. coli cells to recover from a condition that we refer to as glucose-phosphate (or sugar-phosphate) stress. Glucose-phosphate stress occurs when cells cannot efficiently metabolize glucose-6P due to a pgi mutation (mutant cells lack the first enzyme of glycolysis), or when wild-type cells are exposed to the non-metabolizable glucose analog α-methyl glucoside (αMG). When these phospho-sugars accumulate in the cytoplasm, the growth of wild-type cells is transiently inhibited. In contrast, mutant cells lacking SgrS show a severe and prolonged growth inhibition under glucose-phosphate stress conditions (Fig. 2). We want to understand how the activity of the SgrS small RNA allows cells to recover from glucose-phosphate stress.
Being Translated in Escherichia coli*:
Takafumi Sunohara, Kaoru Jojima, Hideaki Tagami, Toshifumi Inada and Hiroji Aiba.
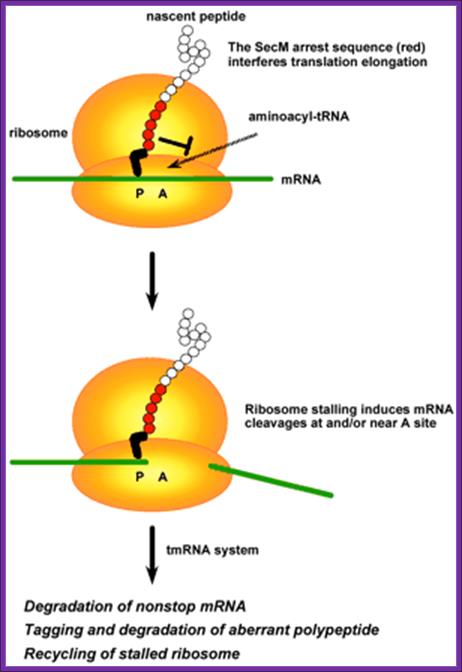
Recently, it has been found that ribosome pausing at stop codons caused by certain nascent peptides induces cleavage of mRNA in Escherichia coli cells. The question we addressed in the present study is whether mRNA cleavage occurs when translation elongation is prevented. We focused on a specific peptide sequence, derived from SecM that is known to cause elongation arrest. When the crp-crr fusion gene encoding CRP-AS17-IIAGlc was expressed, cAMP receptor protein (CRP) proteins truncated around the arrest sequence were efficiently produced, and they were tagged by the transfer-messenger RNA (tmRNA) system. Northern blot analysis revealed that both truncated upstream crp and downstream crr mRNAs were generated along with reduced amounts of the full-length crp-crr mRNA. The truncated crp mRNA dramatically decreased in the presence of tmRNA due to rapid degradation. The 3′ ends of truncated crp mRNA correspond well to the C termini of the truncated CRP proteins. We conclude that ribosome stalling by the arrest sequence induces mRNA cleavage near the arrest point, resulting in nonstop mRNAs that are recognized by tmRNA. We propose that the mRNA cleavage induced by ribosome stalling acts in concert with the tmRNA system as a way to ensure quality control of protein synthesis and possibly to regulate the expression of certain genes.
tmRNA:
A special bacterial RNA called tmRNA or SsrA RNA is a central player in a unique quality control system during protein synthesis. When a ribosome translates to the 3′ end of a broken or incomplete mRNA lacking a stop codon (nonstop mRNA), tmRNA charged with alanine enters the A-site of the ribosome to act first as an alanine-tRNA, then the 3’ end of the ala-tRNA extension itself acts short mRNA and it is translated and the product is released. This co-translation reaction (trans-translation) terminates at the stop codon that follows the tmRNA reading frame, releasing both the ribosome and the tagged polypeptide.
An additional important role of the tmRNA system is to facilitate the degradation of truncated mRNAs by removing stalled ribosomes and thus allowing 3′-to-5′ exonucleases to access the free mRNA 3′ end. Thus, the tmRNA quality control system not only degrades aberrant polypeptides once produced but also prevents production of aberrant polypeptides through a rapid elimination of damaged mRNAs.
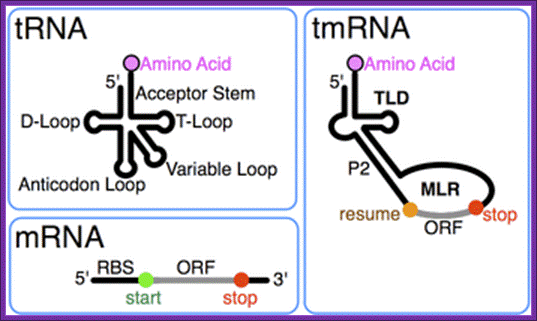
Takafumi Sunohara et al.
The tmRNA contains both tRNA and mRNA features, the tRNA structural feature carries Alanine a.a and mRNA carries coded information in the form of ANDENYALLAA in E.coli. Most tmRNAs are transcribed as larger precursors which are processed much like tRNA. Cleavage at the 5´ end is by ribonuclease P. Multiple exonucleases can participate in the processing of the 3´ end of tmRNA, although RNase T and RNase PH are most effective. Depending on the bacterial species, the 3'-CCA is either encoded or added by tRNA nucleotidyl transferase.
Similar processing at internal sites of permuted precursor tmRNA explains its physical splitting into two pieces. The two-piece tmRNAs have two additional ends whose processing must be considered. For alpha proteobacteria, one 5´ end is the unprocessed start site of transcription. The far 3´ end may in some cases be the result of rho-independent termination. WIKIPEDIA.
Model for mRNA cleavage induced in response to ribosome stalling caused by the SecM arrest sequence.
Riboswitches
Protein repressors and corepressors are not the only way in which bacteria control gene transcription. It turns out that the regulation of the level of certain metabolites can also be controlled by Riboswitches. A riboswitch is section of the 5'-untranslated region (5'-UTR) in some messenger RNA (mRNA) which has a specific binding site for the metabolite (or a close relative of a metabolite).
Some of the metabolites that bind to riboswitches:
- the purines adenine and guanine
- the amino acids glycine and lysine
- flavin mononucleotide (the prosthetic group of NADH dehydrogenase)
- S-adenosyl methionine (that donates methyl groups to many molecules, including
- DNA
- The cap at the 5' end of messenger RNA.
- In each case, the riboswitch regulates transcription of genes involved in the metabolism of that molecule. The metabolite binds to the growing mRNA and induces an allosteric change that –
- for other genes, enhances completion of synthesis of the mRNA.
- for some genes causes further synthesis of the mRNA to terminate before forming a functional product and,
- In both cases, one result is to control the level of that metabolite (a kind of feedback inhibition).
Some riboswitches control mRNA translation rather than its transcription.
It has been suggested that these regulatory mechanisms, which do not involve any protein, are a relict from an "RNA world".
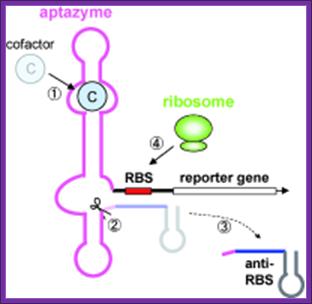
A cofactor bound to an RNA
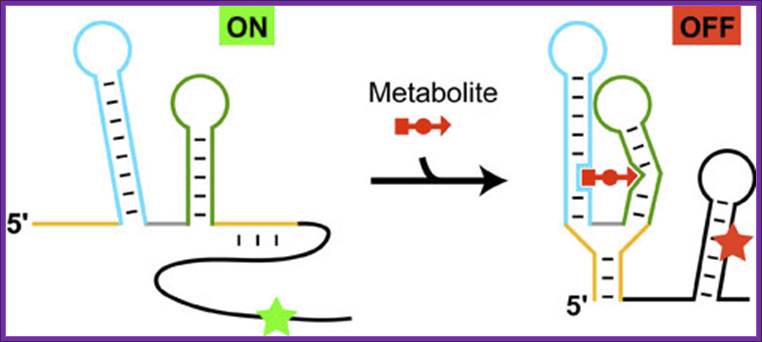
Schematic representation of the riboswitch's function exemplified by the thiamine pyrophosphate, specific riboswitch, which represses the initiation of protein biosynthesis in the presence of metabolite:
The structural elements of the metabolite-sensing domain are colored in yellow, blue, green and gray, and the expression platform is in black. In the absence of metabolite, the metabolite-sensing domain folds into the structure, that exposes the initiation signal of protein synthesis (green asterisk), thereby turning gene expression 'ON'. In the presence of metabolite (shown in red), the sensing domain folds into an alternative structure and causes the formation of the hairpin in the expression platform. As a result, the initiation signal becomes engaged in base pairing (red asterisk) and cannot function anymore, thereby shutting down gene expression, and acting as an 'OFF' switch.
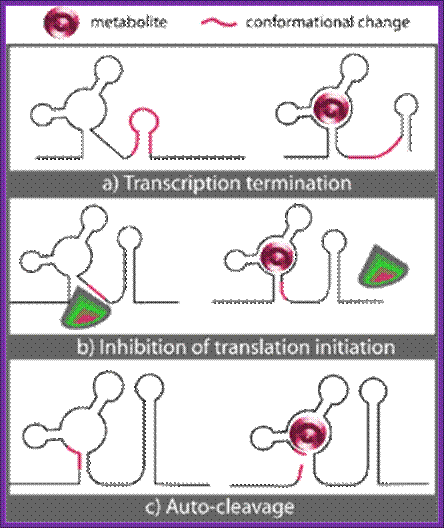
Three known mechanisms of riboswitch action upon binding of metabolite (M): a) Transcription termination. b) Inhibition of translation initiation. c) Auto-cleavage.
Toxin – antitoxin (TA) genes from prokaryotic cells.
Recently, it was shown that several bacterial toxins induce or exhibit endonucleolytic cleavage of mRNAs with the codon or sequence specificity. For example, RelE induces endonucleolytic cleavage of mRNAs bound to ribosomes at UAA and other codons in response to a stalled ribosome. However, the cleavage of ybeL mRNA at a stop codon was observed in various toxin-deficient strains including ΔrelE, ΔyoeB, ΔyafQ, ΔmazF, and ΔchpBK. We (Authors quoted below) also observed that the disruption of the relEB gene did not affect the mRNA cleavage in response to ribosome stalling at a stop codon and at a sense codon. In addition, we showed that the mRNA cleavage in response to ribosome stalling occurs in strains lacking RNase E, RNase G, or RNase III. Thus, neither bacterial toxins nor the major endoribonucleases seem to mediate the cleavage reaction. An attractive possibility would be that the RNA and/or protein components of the ribosome are directly involved in the cleavage reaction depending upon ribosome stalling. In any cases, the factors responsible for the mRNA cleavage in response to a stalled ribosome remain to be identified.
The Figure below shows the components of bacterial toxin–antitoxin gene loci, exemplified by the model system relBE of Escherichia coli. The relBE genes encode RelE toxin and RelB antitoxin. RelB and RelE proteins combine very efficiently and form a non-toxic RelBE complex. Thus, the cells can grow and survive even though they contain the RelBE complex. The RelBE complex binds to DNA in the relBE promoter region (prelB; symbolized by a broken arrow pointing left-ward) and negatively regulates transcription of the relBE genes. Lon is an intracellular protease that degrades RelB antitoxin in response to environmental cues (such as nutrient limitation). Degradation of RelB leads to activation of the RelE toxin. RelE is an enzyme (endonuclease) that cleaves mRNA positioned at the ribosomal A-site. Thus, during nutrient limitations, RelE inhibits translation. In turn, such metabolic regulation of translation increases cell survival during stressful conditions. Paradoxically, when RelE in excess, with RelB activates transcription of Rel-RelE-hokD operon. But under normal conditions both bind to operator region of rel-relb-hokD operon.
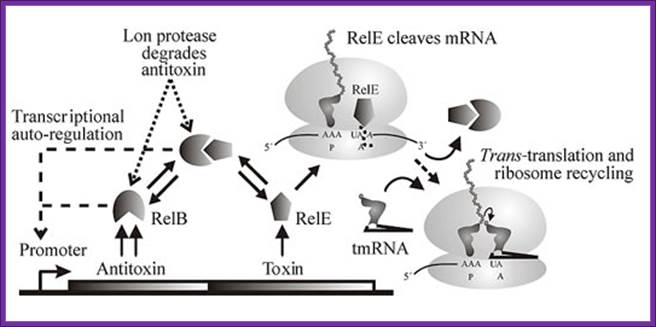
Genetic organization and components of the relBE locus from E. coli: During nutritional stress, Lon protease is activated by an unknown signal and cleaves RelB. In turn, this activates RelE that cleaves mRNAs positioned at the ribosomal A-site. Since RelE acts catalytically, translation can be very efficiently inhibited by RelE, Kim Pedersen1, Andrey V. Zavialov etal.
In a recent analysis of the completed genomes of 196 bacteria and 22 archaea, scientists have identified and annotated1240 toxin - antitoxin loci. The genomic patterns revealed that almost all free-living bacteria and all archaea have many toxin–antitoxin genes. The organisms investigated include major pathogens such as Mycobacterium tuberculosis, Staphylococcus aureus and Pseudomonas aeruginosa. On the genome of M. tubercolusis, we found 60 different TA loci as shown in the Figure below. The toxins of most TA complexes are intracellular enzymes that destroy mRNA, like RelE described above. Thus, activation of a toxin in a TA complex leads to efficient arrest of cell growth and eventually death of the bacterium. Thus, we believe that TA loci represent promising novel antibacterial drug targets.
Prokaryotic transcriptomes: A new view on regulation, physiology and pathogenicity: RNA enrichment: Rotem Sorek & Pascale Cossart; Nature reviews Genetics 11, 9-16 (January 2010)
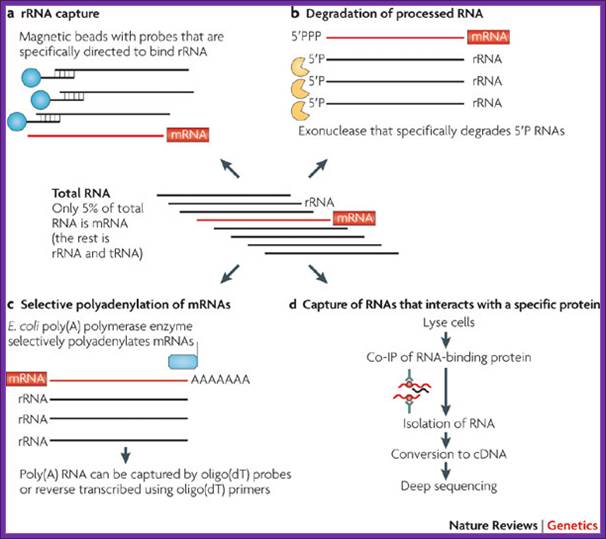
The figure outlines the ways and means of degradation of bacterial RNAs.
Among RNAs, mRNAs can constitute as little as 1–5% of total in the prokaryotic cell, so mRNA enrichment is recommended before transcriptomes’ sequencing. Although mRNA enrichment is not an absolute necessity, it can substantially increase transcript coverage and therefore increase the resolution of the resulting transcriptome maps. The downside of enrichment might be unanticipated biases in the sequenced transcriptome, but data is currently lacking with regard to such possible biases. The lower coverage in non-enriched samples could be easily compensated for by sequencing more cDNA, although this might increase the cost of the experiment. Several methods, which take advantage of the unique features of prokaryotic RNA, have been applied for mRNA enrichment.
Degradation of Processed RNA:
Most bacterial and archaeal mRNAs carry a 5′ triphosphate (5′PPP), which is analogous to the cap structure in eukaryotic RNA. Processed RNA molecules, such as rRNAs and tRNAs, carry a 5′ monophosphate (5′P). One of the exonucleases degrades 5′P RNA molecules, leaving the mRNAs intact. Preliminary analyses indicate that this approach can remove 10–20% of rRNA from Gram-positive and Gram-negative bacteria (R.S. and P. Hugenholtz, unpublished observations).
Antibody capture of RNAs that interact with a specific protein:
This approach targets a specific subset of RNAs. Co-immunoprecipitation (Co-IP) was used to isolate RNAs that are associated with Hfq, a protein that mediates between small RNA and their mRNA targets in bacteria. Sitka et al. reported that Hfq targeting reduced the fraction of rRNAs and tRNAs to about half of all sequenced cDNA reads.
mRNA degradation:
In bacteria, the lifetimes of mRNAs can differ by more than an order of magnitude, with profound consequences for gene expression. For many years it had been assumed that mRNA degradation in E. coli begins with endonucleolytic cleavage at internal sites. However, recent findings have challenged that view by showing that mRNA decay can instead be triggered by a prior non-nucleolytic event that marks transcripts for rapid turnover: the rate-determining conversion of the 5' terminus from a triphosphate to a monophosphate. This modification creates better substrates for the endonuclease RNase E, whose cleavage activity is greatly enhanced when the RNA 5' end is monophosphorylated. We have identified the pyrophosphate-removing hydrolase responsible for that 5'-terminal event, the first such bacterial enzyme ever characterized. That the action of the pyrophosphohydrolase is impeded when the 5' end is structurally sequestered by a stem-loop helps to explain the stabilizing influence of 5'-terminal base pairing on mRNA lifetimes in vivo. Interestingly, this master regulator of 5'-end-dependent mRNA degradation in E. coli not only catalyzes a process functionally reminiscent of eukaryotic mRNA decapping but also bears an evolutionary relationship to the eukaryotic decapping enzyme Dcp2.

Figure . Mechanism of 5'-end-dependent RNA degradation in E.
coli. https://med.nyu.edu/
RNase E is an essential ribonuclease with a key role in the degradation and/or processing of RNA. In E. coli, RNase E is present as a membrane bound multicomponent complex called a "degradosomes". The other major components are polynucleotide phosphorylase (PNPase), the RhlB RNA helicase, and enolase while the DnaK chaperonin protein, GroEL, and polynucleotide phosphate kinase (PPK) are minor components. The ratios of the major components may vary during different stages of cell growth though the RhlB helicase remains at close to a 1:1 ratio.
The C-terminal part of RNase E appears to be responsible for binding these other components. However, these other components are not essential for RNaseE function since a truncated RNase E protein lacking the C-terminal half is sufficient for cell viability and for RNA degradation and processing in vivo in E. coli.
The degradosome has also been shown to contain RNA molecules such as RNA I and fragmented rRNAs which are known to be substrates for RNase E . RNaseE can cleave RNA I at three or four internal sites. More surprising was an observation that degradosomes could also contain the10 Sa/ssrA RNA/tmRNA.
The ssrA RNA is transcribed as a 457-nt precursor which is processed at both 5' and 3' ends to generate the mature tmRNA. Processing at the 5' end requires RNase P; processing at the 3' end requires RNase E, which cuts the precursor at three sites immediately adjacent to the CCA-3'end.
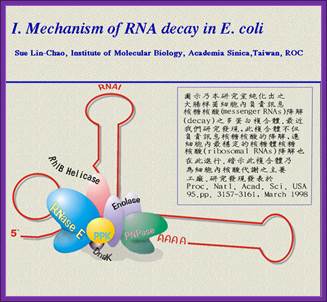
Image from Sue Lin-Chao's web site.
RNAs (sRNAs) and CRISPR RNAs in bacteria:
sRNA binding to a partially complementary mRNA impairs translation initiation, often by occluding the ribosome-binding site. No longer protected by ribosomes, the mRNA becomes vulnerable to attack by RNase E. Antisense sRNA binding to a fully complementary mRNA can create a long, perfectly paired duplex that is susceptible to cleavage by RNase III. A Cmr-associated CRISPR RNA directs endonucleolytic cleavage of a complementary mRNA by one of the six Cmr proteins. Ribosomes have been omitted from this panel because the effect of CRISPR RNAs on translation has not been investigated.
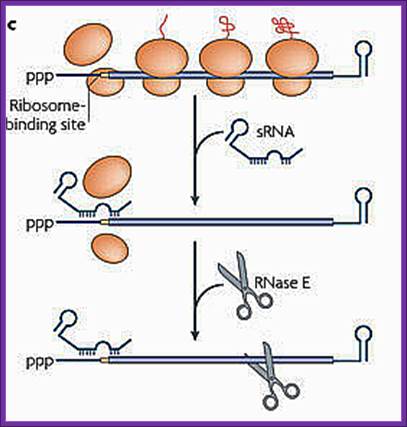
sRNA binding to a partially complementary mRNA impairs translation initiation, often by occluding the ribosome-binding site. No longer protected by ribosomes, the mRNA becomes vulnerable to attack by RNase E.
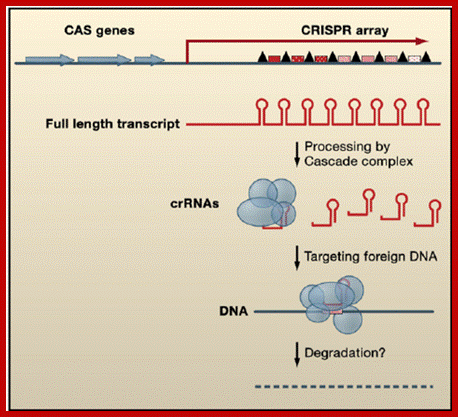
Gene Arrangement and Regulatory Functions of CRISPR RNAs CRISPR arrays are composed of DNA repeats (black triangles) separated by unique spacers (red speckled boxes). CAS genes (blue), which encode proteins that function in CRISPR RNA processing and/or DNA silencing, are located nearby. The CRISPR arrays are initially transcribed as a long RNA, which is subsequently processed by the Cascade complex (blue circles and ovals) to individual repeat-spacer units, called crRNAs. These crRNAs appear to target foreign DNA through an unknown mechanism likely involving other CAS proteins and the degradation of the exogenous DNA. ;https://www.researchgate.net; www.cell.com
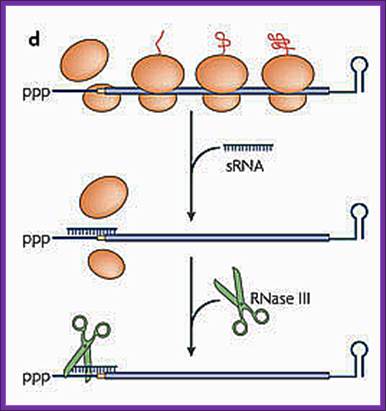
d. Antisense sRNA binding to a fully complementary mRNA can create a long, perfectly paired duplex that is susceptible to cleavage by RNase III.
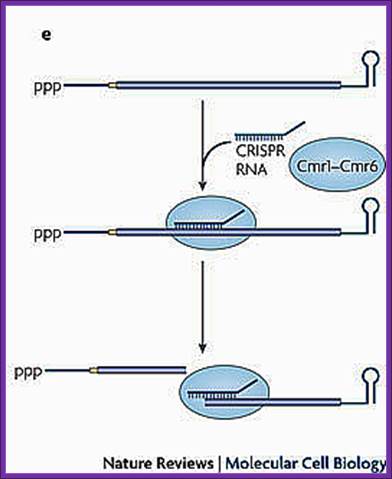
A Cmr-associated CRISPR RNA directs endonucleolytic cleavage of a complementary mRNA by one of the six Cmr proteins. Ribosomes have been omitted from this panel because the effect of CRISPR RNAs on translation has not been investigated.
mRNA Decay:
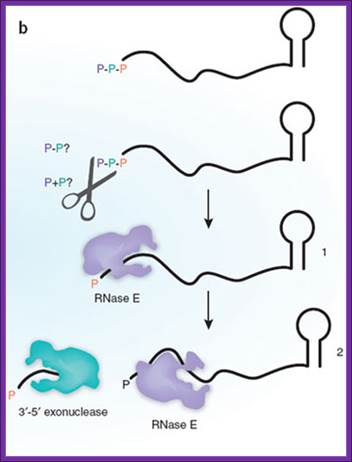
Bacterial mRNAs begin with a 5'-triphosphate and end with a stem-loop structure, and decay involves endonuclease cleavage by RNase E. The preferred substrate for RNase E is RNA with a 5'-monophosphate, a property that is determined by the presence of a monophosphate-binding pocket within the catalytic domain of the enzyme. Celesnik et al.1 describe a previously unknown step in mRNA decay in which pyrophosphate is removed from the 5' end by an unknown enzyme(s) (scissors), in a manner analogous to decapping in eukaryotes, to generate a 5'-monophosphate substrate for cleavage by RNase E. This is the rate-limiting step in decay, and subsequent cleavage by RNase Egenerates an upstream product with a 3'-hydroxyl that is degraded by 3'-5' exonucleases and a downstream product with a 5'-monophosphate. This cycle is then repeated to complete degradation of the mRNA. Daniel R Schoenberg1www.Nature.com,
Bacillus subtilis mRNA decay: new parts in the tool kit
David H. Bechhofer
Focus Article
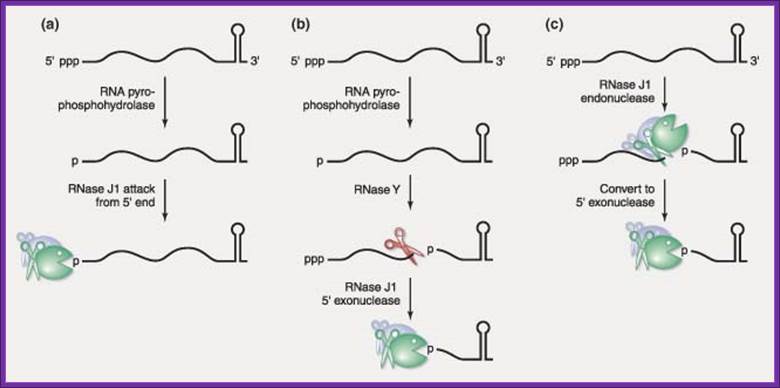
Representatives of two new ribonuclease families have recently been discovered in the gram‐positive model organism, Bacillus subtilis. The RNase J family founding members, RNase J1 and RNase J2, are highly homologous but show differential activities. Although both are broad‐specificity endonucleases, only the essential RNase J1 is a 5′ → 3′ exonuclease—a type of ribonuclease activity that was previously thought not to exist in bacteria. Current data suggest that RNase J1 is highly involved in the turnover of mRNA decay intermediates and may also be involved in the initiation of mRNA decay. A second family of ribonucleases is represented by RNase Y, an endonuclease that exerts a large effect on global mRNA half‐life. The presence of these ribonucleases in B. subtilis predicts significant differences from the well‐established model of mRNA decay in Escherichia coli. WIREs RNA 2010 DOI: 10.1002/wrna.66 .
The Role of Polyadenylation in mRNA decay in E.coli.
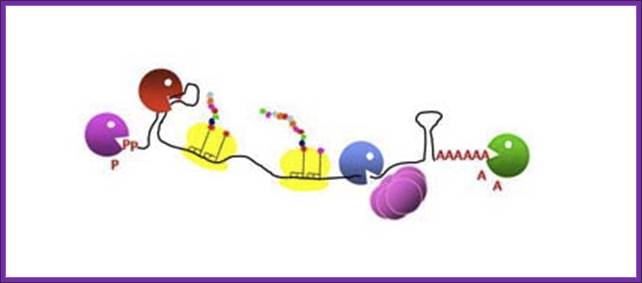

“Poly(A)-dependent degradation of RNA harboring 3' stable secondary structures”. Types of intermolecular RNA–RNA interactions. When an RNA molecule interacts with another RNA molecule at loops by base- pairing hybridization (small bars), loop-loop interactions at stem-loop structures induce three different types of RNA–RNA interactions. Each stem-loop structure is depicted by two parallel bars and an open circle. Models are shown. Type A interaction (dimerization). A loop– loop interaction— i.e ., kissing loop interaction—triggers the hybridization of two RNA molecules to form a long RNA duplex, most likely in concert with dimerizing enzymes ( e.g. , 131). Type B interaction (stabilization). Loop–loop interactions result in the recruitment of a stabilizing protein, such as HuR (60). RNA remains stable until the stabilizing protein detaches or a destabilizing protein attaches. Type C interaction (degradation). A loop–loop interaction results in the recruitment of a destabilizing protein ( e.g ., tristetraprolin) (84) and finally the degradation of the RNA (broken line). The fate of RNA after an RNA–RNA interaction is determined by several trans-acting factors, such as stabilizing and destabilizing proteins, microRNA, and drugs.
;https://www.researchgate.net

Gram-negative mRNA degradation model so also gram positive mRNA are also degraded;http://www.itqb.unl.pt
A) Poly (A) tails synthesized at the 3' ends of RNAs offer a toehold where exonucleases can initiate decay. RNase II removes the poly (A) tails before to be arrested by 3' hairpins. It generates tail-less molecules and therefore protects the RNA from exonucleolytic decay. Similarly, PNPase, alone or in association with other degradosome components including the RhlB RNA helicase, can remove the tail and stop few nucleotides downstream of the secondary structure. A cofactor referred to as EIF could promote PNPase stalling at secondary structures. PNPase can also slightly nibble the bottom of the hairpin before dissociating from the RNA. Then, the RNA is readenylated by PAP I thus offering to PNPase the opportunity to nibble the descending strand of the hairpin a little further and finally to degrade completely the RNA into nucleotides. Unstructured RNA fragments are degraded by PNPase and RNase II whose activity is facilitated by PAP I. The oligo ribonuclease degrades short oligo ribonucleotides released by PNPase and RNase II into mononucleotides.71 B) Degradosome is represented by a circle containing an endonucleolytic shaded domain (RNase E) indicated by “endo” and an exonucleolytic hatched domain (PNPase and RhlB) indicated by “exo”. The model postulates that this particle interacts simultaneously with an internal processing site, shown by a star, through RNase E and with the 3' poly(A) tail through PNPase and RhlB and that this interaction favors degradation mediated by RNase E. C) In contrast, RNA fragments resulting from endonucleolytic cleavages harbor 5' monophosphorylated extremities which promote processing by RNase E or degradosome. The model proposes that degradosome interacting simultaneously with the 5' end of the RNA fragment and an internal processing site looses its affinity for poly(A) tails and that this explains why RNA fragments can be degraded simultaneously by RNase E and poly(A)-dependent ribonucleases. PNPase, free or associated with degradosome, carrying out the exonucleolytic degradation of the RNA is indicated by a small hatched circle.
The first indication that poly(A) tails destabilize RNA came from the discovery that PAP I controls the stability of the small RNA (RNA I) which regulates the replication of ColE1 plasmids (see above). Further study led to the conclusion that PNPase does not bind RNA I, whose 3' end is sequestered in a secondary structure and that poly(A) tails provide sites where PNPase can bind and initiate the exonucleolytic degradation of RNA I. Polyadenylation has since been shown to be involved in the degradation of other RNA species and of RNA in general9 and it is admitted that the mechanism of degradation of RNA I can be extended to the poly(A)-dependent degradation of any RNA with a 3' secondary structure. The current idea is that PNPase can carry out the complete processive degradation of RNAs containing weak secondary structures but is blocked when it encounters stable hairpins which cause dissociation of the ribonuclease from its substrate. It has been proposed that an Exonucleolytic Impeding Factor, referred to as EIF, might provoke PNPase stalling at secondary structures.85 When blocked at secondary structures, PNPase releases RNAs devoid of a 3' single-stranded stretch of nucleotides that cannot be bound by exo ribonucleases). The current model of RNA decay postulates that these tail-less RNAs are readenylated thus allowing PNPase to reinitiate exonucleolytic decay. Again, PNPase can generate tail-less RNA or, possibly, continue to degrade the RNA upstream of the tail and remove few nucleotides at the bottom of the hairpin before to dissociate from the RNA .
Decay of Prokaryotic mRNAs:
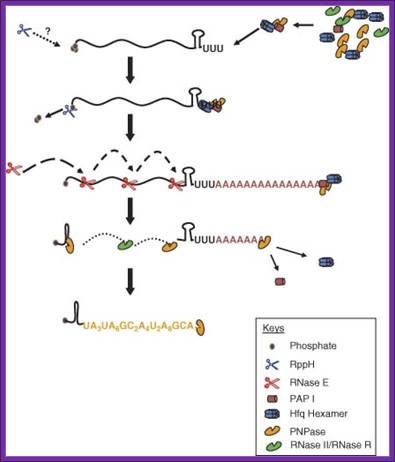
In E.coli poly-U mediated degradation; https://www.researchgate.net
Although the first poly (A) polymerase (PAP) was discovered in Escherichia coli in 1962, the study of polyadenylation in bacteria was largely ignored for the next 30 years. However, with the identification of the structural gene for E. coli PAP I in 1992, it became possible to analyze polyadenylation using both biochemical and genetic approaches. Subsequently, it has been shown that polyadenylation plays a multifunctional role in prokaryotic RNA metabolism. Although the bulk of our current understanding of prokaryotic polyadenylation comes from studies on E. coli, recent limited experiments with Cyanobacteria, organelles, and Archaea have widened our view on the diversity, complexity, and universality of the polyadenylation process. For example, the identification of polynucleotide phosphorylase (PNPase), a reversible phosphorolytic enzyme that is highly conserved in bacteria, as an additional PAP in E. coli caught everyone by surprise.
Bacterial/archaeal/organellar polyadenylation
Advanced Review; Bijoy K. Mohanty,Sidney R. Kushner
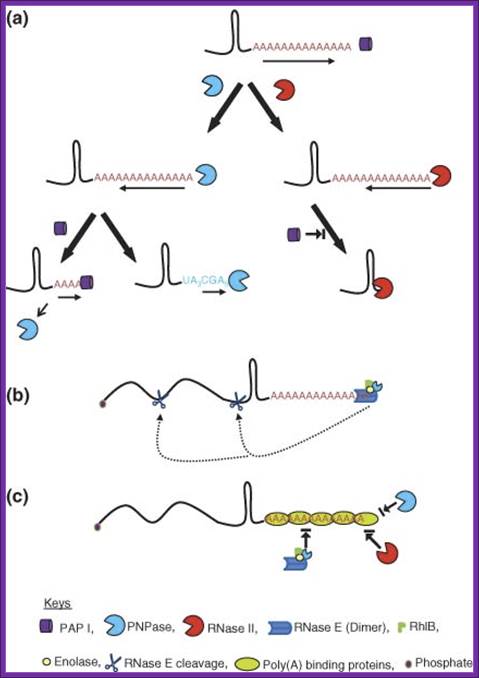
PNPase is involved in 3’ degradation; Polyadenylation-assisted RNA decay in E. coli. (a) Addition of poly(A) tails by PAP I to the 3′ ends of an RNA substrate provides the single-stranded binding site for both PNPase and RNase II. Although PNPase catalyzes both 3′ → 5′ phosphorolytic degradation in the presence of inorganic phosphate and 5′ → 3′ polymerization in the presence of NDPs, RNase II can only degrade RNA hydrolytically in the 3′ → 5′ direction. Both the ribonucleases pause upon encountering a G/C-rich secondary structure. PNPase either dissociates relatively quickly or reverses its activity to polymerize polynucleotide tails. Dissociation of PNPase may initiate multiple rounds of polymerization by PAP I. In contrast, RNase II remains bound to the base of the secondary structure thereby effectively blocking the binding of either PAP I or PNPase. (b) RNase E alone or as part of the multiprotein complex called the degradosome can bind A/U-rich poly(A) and polynucleotide tails to initiate degradation of a potential substrate through endonucleolytic cleavage. A full-length polyadenylated RNA substrate may be degraded very fast125 by direct or internal entry116 resulting in very few steady-state polyadenylated RNA species. This type of RNase E entry to an RNA substrate has yet to be experimentally demonstrated. (c) Potential poly(A)-binding proteins can block RNA decay in E. coli. Proteins such as CspE, Hfq, and ribosomal protein S1 could bind to poly(A) or polynucleotide tails blocking endonucleolytic access by the RNase E-based degradosome through its PNPase moiety or direct exonucleolytic degradation by exoribonucleases such as RNase II, RNase R, and PNPase. https://www.researchgate.net
In fact, PNPase has now been shown to be the source of post‐transcriptional RNA modifications in a wide range of cells of prokaryotic origin including those that lack a eubacterial PAP homolog. Accordingly, the past few years have witnessed increased interest in the mechanism and role of post‐transcriptional modifications in all species of prokaryotic origin. However, the fact that many of the poly(A) tails are very short and unstable as well as the presence of polynucleotide tails has posed significant technical challenges to the scientific community trying to unravel the mystery of polyadenylation in prokaryotes. This review discusses the current state of knowledge regarding polyadenylation and its functions in bacteria, organelles, and Archaea. Copyright © 2010 John Wiley & Sons, Ltd.
Facilitation of 3′ exonucleolytic degradation of bacterial mRNA decay by polyadenylation: All things must pass: contrasts and commonalities in eukaryotic and bacterial RNA decay- Joel G. Belasco:Nature Reviews Molecular Cell Biology 11, 467-478 (July
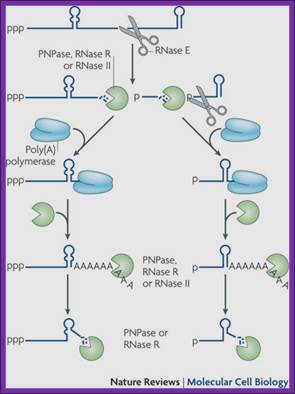
Endonucleolytic cleavage of mRNA by RNase E generates multiple fragments, one of which ends with the original 3′-terminal stem-loop. The others undergo 3′ exonucleolytic attack by polynucleotide phosphorylase (PNPase), RNase R and/or RNase II until an upstream stem-loop is encountered, which interrupts further degradation owing to the preference of these ribonucleases for unpaired 3′ ends. The resulting decay intermediates are then polyadenylated by poly (A) polymerase, thereby enabling the exonucleases to re-engage. The repeated addition of single-stranded poly (A) tails to the 3′ ends of these intermediates provides many opportunities for PNPase and RNase R to overcome structural impediments to exonucleolytic degradation, and eventually they succeed. The ability of PNPase to digest base-paired RNA is enhanced by its association with the RNA helicase RhlB, whereas RNase R requires no such assistance. By contrast, RNase II can degrade poly(A) and other types of unstructured RNA but not structured RNA. Ribosomes and coding regions have been omitted from this figure for simplicity
Add on notes- Certain critical materials taken from different authors has been used for students to refer and update further.