Protein Synthesis Prokaryotic: Mechanism of Synthesis:
In prokaryotes transcription and translation go hand in hand (Coupled); as the 5’ end of the mRNA emerges out of DNA-RNAP complex and the 5’ end is made available, the small ribosomal sub unit binds, then initiator tRNA and initiation factors bind only to associate with large subunit to develop into fully functional ribosome-mRNA complex.
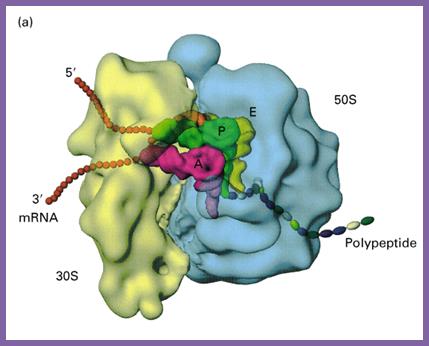
Organization of Ribosome, mRNA and tRNA into a complex for synthesizing a polypeptide chain; http://www.biologyreference.com/
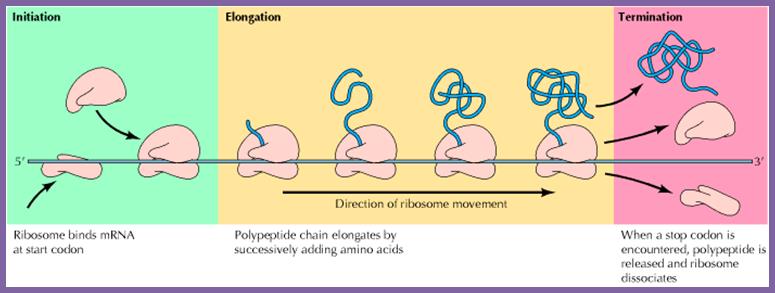
Chain Initiation, elongation and termination; http://biosiva.50webs.org/
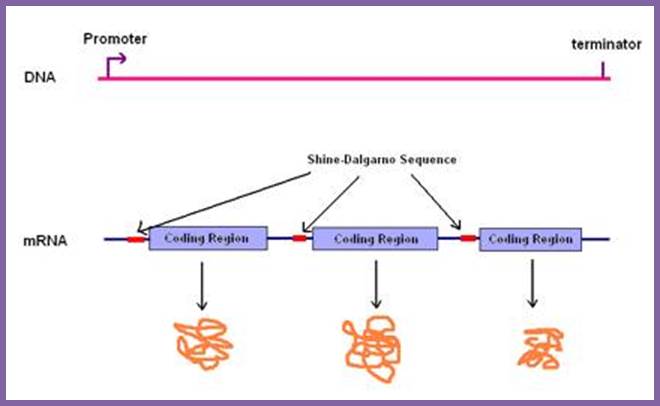
Translation of 3 cistrons interspersed with two intercistronic spaces leads to translation of three cistrons and the release of three polypeptide chains; http://.www.genome.ou.edu
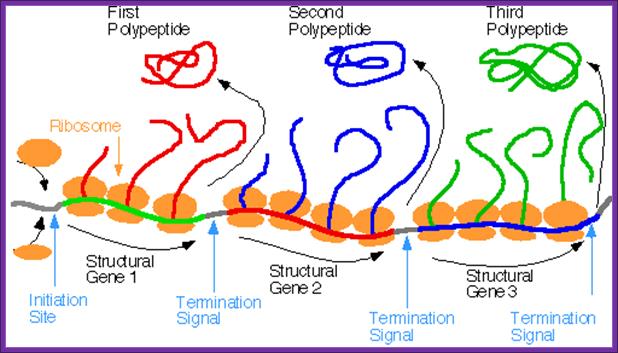
Prokaryotes produce polycistronic mRNA, which on translation each cistron generates one protein; a single mRNA produces different polypeptides/proteins. There are intercistronic spacer which help ribosomes to initiate translation after termination one cistron.www.pixgood.com
Whether it is prokaryotic or eukaryotic system, all mRNAs contain a translation initiation site, a coding region and a termination site all in one frame, it is called open reading frame (ORF). In the case of polycistronic mRNAs each cistron begins with an initiator codon and ends in a terminator codon, in between cistron it has spacer. Very many mRNAs contain IRE (Internal Ribosomal Entry) at 5’UTR or at intercistronic spacers (ex. Some viruses). There are few regulatory sequences at 3’UTR, ex. E.coli SECIS element was located in the 3'-untranslated region (3'-UTR) and adjacent to a sequence complementary to the region downstream of the Sec UGA codon, pubmed, Su D, Li Y, Gladyshev VN.
5’-leader-- SD -AUG----codons--------UAG/UGA/UAA---SECIS---3’
SD = Shine Delgarno sequence at 5’ leader region.
AUG = Initiator codon,
Codons = Code for specific amino acids,
UAG/UGA/UAA Terminator codons: UAA, UAG, UGA = Ochre, Amber and Opal respectively.
S/D sequence; http://themedicalbiochemistrypage.org/
Chelsia A.http://www.studyblue.com/
mRNAs and ribosome binding sequences in Escherichia coli:
|
Gene |
Codes for |
S/D sequence |
Ntd from the start +1 |
|
Lactose utilization enzymes |
5′-A G G A -3′ |
7 |
|
|
galE |
Hexose-1-phosphate Uridyl transferase |
5′-G G A G -3′ |
6 |
|
rplJ |
5′-A G G A G -3′ |
8 |
Archaeal: Initiator factors: IFs: aIF1, AaIF2, aIF5 and aIF6.
Archaeal: Elongation factors: EFs: aEF1 and aEF2,
Archaeal: Ter: factors –aTeFs.
Sometimes redundant AUGs are present in the leader sequence itself (5’UTR), such AUGs are referred to as uAUGs. So they provide false sites for initiation of translation, but they are selected out for initiation because the Shine-Dalgarno (SD) sequence determines correct ORF and provides the positioning of the mRNA on 30s ribosomal subunit. But if there are AUGs before Shine Delgarno or in the middle of the reading frame they are not used as initiator codons. Often chain initiation takes place in 5’UTR and translates the succeeding triplets, but when it reaches the actual initiator codon with SD as an identifying factor; large subunit dissociates only to be reinitiated with proper Initiator f-met tRNA. It is reported that the presence of such uORF significantly reduce the rate of protein expression. However if the uORF contain strong S/D sequence then it can lead to inhibition of translation of downstream ORFs or slow down translation. For example Tryptophan operon polycistronic mRNA contains an uORF which when translated terminates at its Ter sequence. This 5’UTR is used for attenuation mode of transcription of Trp operon. In the case of an eukaryotic Her2 mRNA uORFs have inhibitory effect but the 3’UTR overcome the inhibitory effects.
Such uORFs are present in many operons containing 4-7 cistrons. Ex. Tryptophan operon contains upstream 5’UTR containing uORF which functions as attenuator in regulating Trp operon transcription. Many intercistronic spacers are provided with a minimal S/D sequences. Some multicistronic mRNAs contain Terminator and Initiator as overlapping sequences; in these ribosomes need not reinitiate using S/D sequences to start the second cistron.
ORF is the region defined by (its S/D) initiator codon and successive codon triplets end in a terminator codon. If there are any internal AUGs they are deemed as codon coding for methionine not as initiator codon.
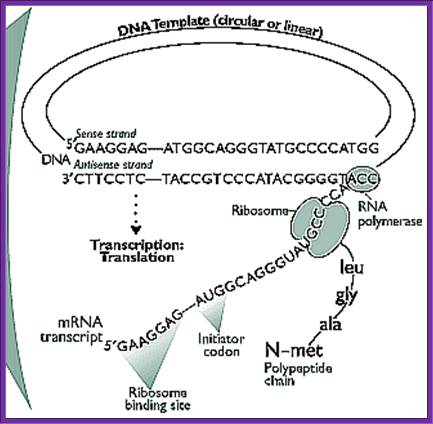
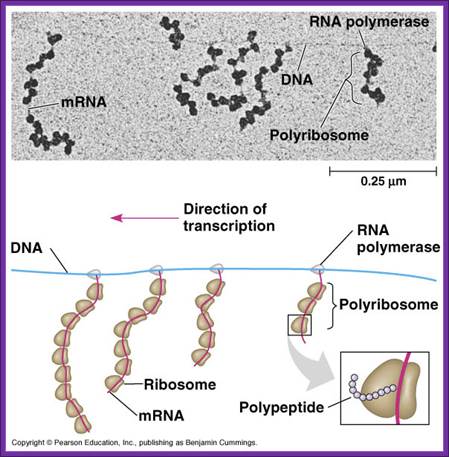
The figure shows transcription and translation events are coupled; http://www.the-scientist.com/ and Gene to protein; http://www.bio.utexas.edu/
Internal ribosome entry site IRES is also common among prokaryotes, and also in eukaryotes ex. Polioviruses (eukaryotic virus).
Chain Initiation:
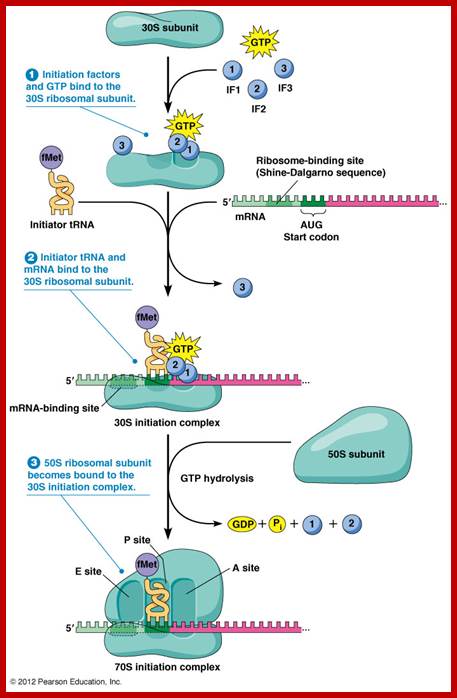
The three initiation factors are involved; they are Initiation Factor-1(IF1) and initiation factor-2 (IF2) and initiation factor-3 (IF3).
During initiation, IF-3 activates the 70 ribosome complex, if it is intact with both subunits joined, where it binds to 30s ribosome near the E-site and initiates dissociation of dimeric ribosomal subunits. When IF-3 associates with 30 S the small subunit gets activated; it prevents the binding of 50s. They are mutually exclusive. This reaction is assisted by IF-1. IF1 prevents binding of amino acyl tRNA to amino acyl tRNA site (A-site). Now the 30S ribosome is in active state.
Binding of IF-3 to 30s induces conformational change into active form. One has to visualize ribosome, as a complex of riboproteins and rRNA, not as rigid structure, but as a highly flexible and the most dynamic structure among all other cell organelles. It reacts in response to the binding of mRNA, tRNA and/or factors during protein synthesis. Position-wise the specific riboproteins and specific RNA sequences play a very important role in translation process.
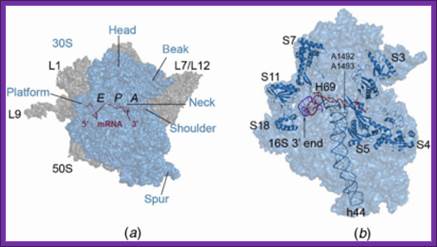
The 30S ribosomal subunit contains two sites for the binding of aa-tRNAs. One called P site (peptidyl site) to which initiator a.a tRNA’ binds first (to begin with); the other is called A- site (amino acyl-tRNA site) to which an aa-tRNA binds for elongation. Binding of any of them induces conformational changes in the structure of ribosome. Besides, the ribosome has another site to the right of A-site; it is called R-site, where the arriving a.a-tRNA is and scrutinized and recruited (?). Similarly one more site exist to the left of the P site called E’ site (exit of tRNA (-a.a). These sites can exist in active or inactive forms.
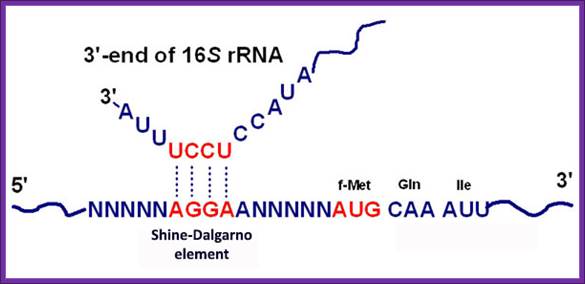
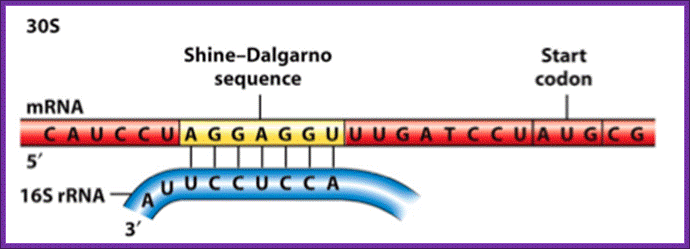
The activated ribosome binds to the leader called 5’ UTR of mRNA and threads through. When it reaches ribosomal binding site called (RBS) 7-8 ntds upstream of the AUG, ribosome binds to a sequence GAGGAAC CCC sequence of mRNA. The sequence acts as a structural motif called Shine-Dalgarno sequence (SD), which base pairs with a sequence found at the 3’ end of 16s ribosomal RNA. The binding site of for mRNA in the small ribosomal subunit is in the cleft region; using Shine Delgarno sequence the AUG codon of the mRNA gets aligned at P site. The S/D sequence determines and provides firm binding of mRNA to ribosomes and also the chain initiator site AUG in line with P site, the beginning of an ORF.
16sRNA: 3’-AUUCCUCCACUAG-5’
mRNAs:
RANAP-b GAGG AAC CCC AUG GUU-3’
Lac-Z GGAA ACA GC AUG ACC.3’
Rec-A GGA GUA AA AUG GCU.3’
Lac-I GGUG GUG AA AUG AAA.3’
QB-A protein GGAG CA AUG CCU.3’
Gal-E GGAG CG AUU AUG AGA.3’
Q B replicase GGA UG AUG UCU.3’
Trp-leader GGG -5- ACC AUG AAA.3’
Rib L10 AGGAG-5-CUA AUG GCU.3’
3’ AUUCCUCCACUAG-5’ 23s rRNA
5’ UAAGGAGG-leader sequence in mRNA
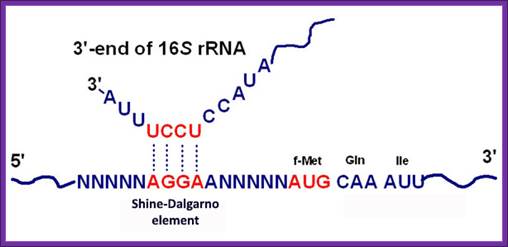
www.themedicalbiochemistrypage.org

Shine Delgarno Sequences; the 3’end sequence of rRNA has consistent sequence when compared to S/D sequence
http://www.studyblue.com
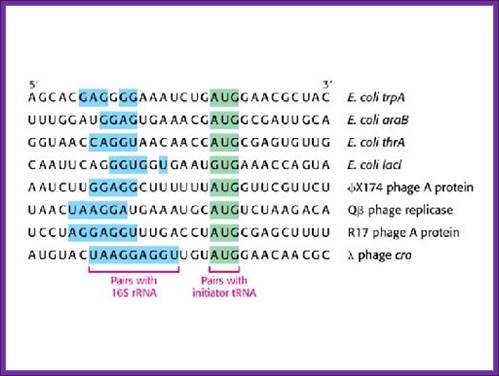
Shine Delgarno sequences of different operons and species; http://www.studyblue.com
The consensus sequence of S/D is ---UAAGGAGGU-- or --GGAGGA--. Positioning of the S/D from the AUG is very important, the distance of 7-8 ntds between them and any drastic deviation from this length hampers initiation.
When the mRNA binds to 30s subunit, the AUG codon is actually positioned at P’ site which is in line with P-site on large ribosomal surface. The mRNA is placed in the cleft, when bound it is in extended form. Between two codons the mRNA is kinked or bent. The three bases in each of this region are in extended conformation so as to base pair with incoming aa-tRNAs.
The initiating factor IF-2 is a GTP binding protein; GTP activates it. The IF-2 has a specific binding site for initiator a.a tRNA (f-met-tRNA) and the IF2-GTP-fmet-tRNA doesn’t bind to P’site and not to ‘A site. This tRNA has 3’ UAC 5’ as an anticodon complementary to initiator codon 5’AUG 3’ in mRNA. The GTP bound IF2 brings f-met tRNA and it is the only amino acyl tRNA that acts as an initiator tRNA. No other tRNA can act as initiator tRNA for its IF2 and tRNA 3-D structure is designed for it. There are other tRNAs, which also bind to methionine, but they act as internal a.a-tRNAs. The initiator tRNA, however, has different but unique structural composition, which differs from the internal met-tRNA or any other tRNAs.
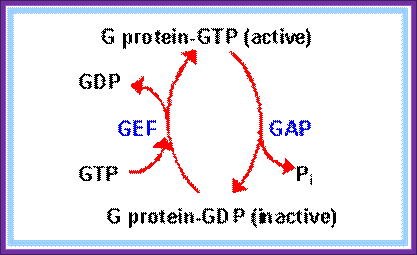
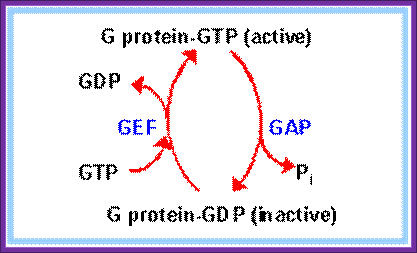
Recycling of GTP and G protein; htpp://www.rpi.edu
The f-met tRNA has unpaired 5’C in amino acyl acceptor stem. The important determinants of initiator tRNA in bacteria are the absence of a Watson–Crick base pair between positions 1 and 72 in the acceptor stem and the presence of six conserved consecutive G:C base pairs in the anticodon stem. But the general aminoacyl tRNAs contain 5’G base paired with C but not in Initiator tRNA. This tRNA has UCC as extra ntds in the D-loop, and there are four GC in anticodon stem-loop.
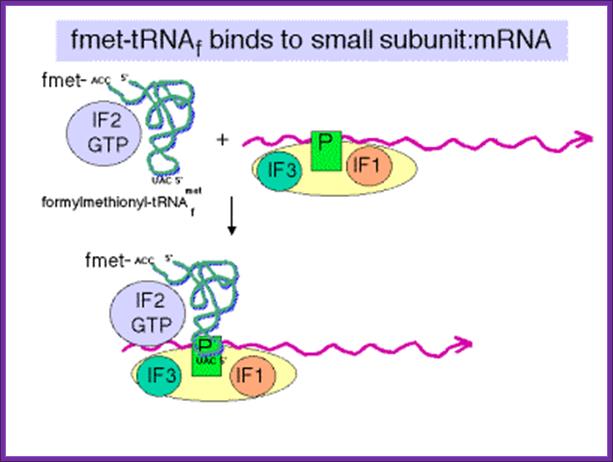
IF2 brings fmet-RNA –f to the partial P site on the small subunit;http://www.personal.psu.edu/
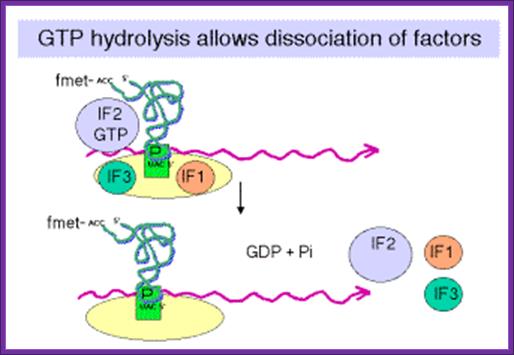
GTP hydrolysis stimulated by IF2 promotes dissociation of IF2, IF1and IF2 and IF3 from the initiation complex and leads to association of 50S subunit. http://www.personal.psu.edu/
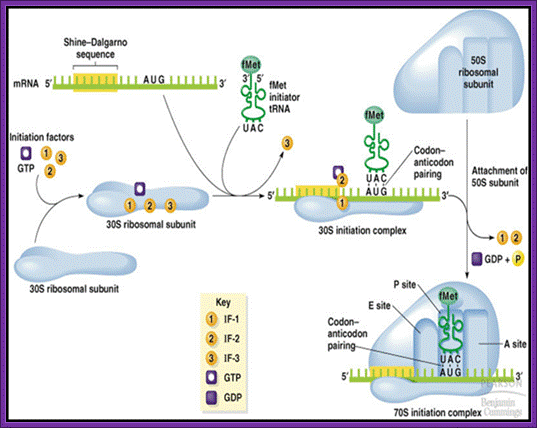
Chain Initiation; http://www.biologyexams4u.com/
Activated, IF-2.GTP-f-met-tRNA complex seeks the activated mRNA.30s. IF3.IF1 ribosomal complex and binds to cleft in such a way, with slight adjustments, positions f-met tRNA on P site in line with AUG codon, a perfect matching indeed. The binding proteins like L27, L2, L14, L15, L16 at the P’ and A’ sites have been determined by affinity labeling. Certain drugs like tetracycline prevent the aminoacyl-tRNA from binding to the ribosomal subunit in prokaryotes. A.P. Czernilofsky et al. (Proc. Natl. Acad. Sci, USA, pp 230–234, 1974; WIKIPEDIA.
The three bases 3’ UAC 5’ in tRNA and 5’AUG3’ of mRNA are stearically out-stretched to meet and properly base pair with each other. The third base redundancy is common so also wobbling of 1st base of tRNA is common, but specific. In this base pairing the first two bases from the 5’end of the codon and the first two bases in tRNA from 3’ end are important. The codon and anticodon base paring fixes them on the template; the third base can be a degenerate nucleotide.
The table below shows the kind of factors involved in protein synthesis in prokaryotes, eukaryotes and archaea. The table shows initiation factors of both prokaryotes and eukaryotes.

The table shows translation factors involved in Prokaryotic , Eukaryotic and Archaea systems


Eukaryotic initiation factors and general functions:
eIF2 binds Met-tRNA to ribosomese’
IF2B activates eIF2 replacing its GDP with GTP’
eIF1 and eIF1A aid in scanning to initiation codone’
IF3 binds to 40S ribosomal subunit, inhibits reassociation with 60S subunite,
IF4 is a cap-binding protein allowing 40S subunit to bind 5’-end of mRNAe,
IF5 encourages association between 60S ribosome subunit and 48S complexes,
IF6 binds to 60S subunit, blocks reassociation with 40S subunit.

Translation initiation. In cap-dependent translation, the small ribosomal subunit (40S) is brought to the 5'-terminal cap structure (represented as a black circle) through the interaction of eukaryotic initiation factor (eIF) 3 (not shown) on the ribosome with the cap-binding complex eIF4F. The ribosome is thought to migrate ('scan') along the 5' untranslated region (5' UTR, shown as a black line), removing any inhibitory stem–loops, until it encounters the first start codon (AUG) in a favourable 'Kozak' context. The codon–anticodon interaction and the initiation factors eIF2, eIF1 and eIF1A all have a role in start codon selection. eIF1/1A and several other initiation factors are omitted from the figure for clarity. During internal ribosome entry site (IRES)-mediated initiation, the ribosomal complex is not assembled at the 5' end of the mRNA, but at an internal site (indicated in red) formed by secondary structures in the 5' UTR. After recognition of the start codon of the ORF (represented as a blue rectangle), a functional ribosome is formed through binding of the large ribosomal subunit (60S). During this process, eIF2-bound GTP is hydrolysed to GDP and initiation factors are released. 'UGA' indicates one of the three possible stop codons of the ORF, UTRs are indicated by black line, and the poly(A)-tail at the 3' end of the mRNA by 'AAAn'. One of the rate-limiting steps in the initiation process is the formation of the ternary complex Met-tRNAi–eIF2–GTP. To enable eIF2 to bind initiator Met-tRNA and participate in the next round of translation, the GDP has to be replaced by GTP. As Met-tRNAi is normally present in abundance, the eIF2B-catalysed GDP–GTP exchange is thought to be rate limiting (bottom). Phosphorylation of eIF2 on its -subunit by eIF2-specific kinases (for example, pancreatic endoplasmic reticulum-resident kinase (PERK)) inhibits eIF2B;
https://www.researchgate.net
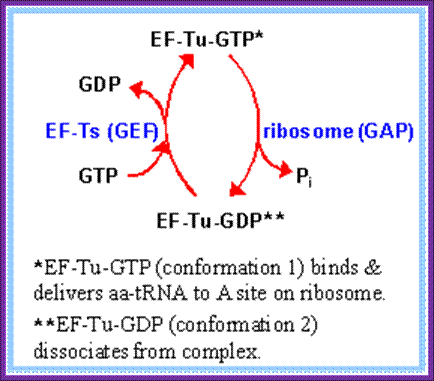
Elongation factorsIn prokaryotes, four are required for translation: EF-Tu, EF-Ts, EF-G and EF-P.
· EF-Tu mediates the entry of the aminoacyl tRNA into a free site of the ribosome.
· EF-Ts serves as the guanine nucleotide exchange factor for EF-Tu, catalyzing the release of GDP from EF-Tu.
· EF-G catalyzes the translocation of the tRNA and mRNA down the ribosome at the end of each round of polypeptide elongation.
· EF-P stimulates peptide formation by catalyzing the first synthesis step between the first amino acid (N-formylmethionine) and the second amino acid.
http://www.mun.ca; https://karimedalla.wordpress.com
Elongation in eukaryotes is carried out with two elongation factors: eEF-1 and eEF-2.
· The first is eEF-1, and has two subunits, α and βγ. α acts as counterpart to prokaryotic EF-Tu, mediating the entry of the aminoacyl tRNA into a free site of the ribosome. βγ acts as counterpart to prokaryotic EF-Ts, serving as the guanine nucleotide exchange factor for α, catalyzing the release of GDP from α.
· The second elongation factor is eEF-2, the counterpart to prokaryotic EF-G, catalyzing the translocation of the tRNA and mRNA down the ribosome at the end of each round of polypeptide elongation.

https://www2.estrellamountain.edu
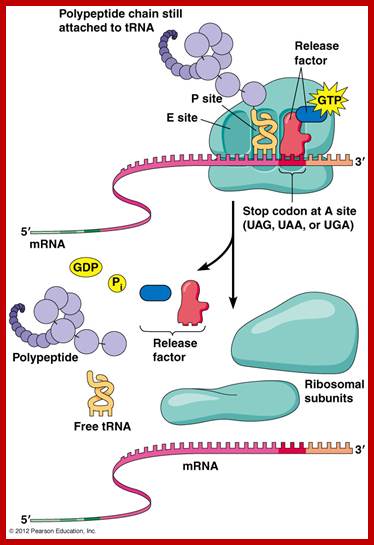
Translation Termination factors:
Prokaryotes:
RF1 recognizes the termination codons UAA and UAG
RF2 recognizes UAA and UGA
RF3 is a GTP-binding protein that leads to the dissociation of RF1/RF2 after peptide release

Translation termination. Click on image to see it on Alila Medical Media website where the image is also available for licensing (together with other related images;
http://classroom.sdmesa.edu
Eukaryotes:
eRF1 recognizes all three termination codons;
eRF3 is a ribosome-dependent GTPase that helps eRF1 release the completed polypeptide
Overall Translation in Eukaryote:

Viruses are fully reliant on the translation machinery of their host cells to produce the polypeptides that are essential for viral replication. Consequently, viruses recruit host ribosomes to translate viral mRNAs, typically using virally encoded functions to seize control of cellular translation factors and the host signalling pathways that regulate their activity. This not only ensures that viral proteins will be produced, but also stifles innate host defences that are aimed at inhibiting the capacity of infected cells for protein synthesis. Remarkably, nearly every step of the translation process can be targeted by virally encoded functions. This Review discusses the diverse strategies that viruses use to subvert host protein synthesis functions and regulate mRNA translation in infected cells. This holds good for eukaryotes also; http://www.nature.com
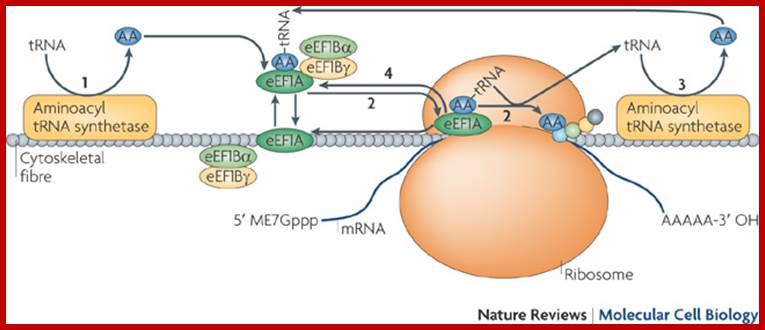
This model shows that aminoacyl (AA; blue)-tRNAs and their metabolites can be efficiently transferred between cytoskeleton-bound components of the protein synthesis apparatus (for example, aminoacyl-tRNA synthetase, eukaryotic elongation factor 1A (eEF1A), eEF1Bα and eEF1Bγ). Although the cytoskeletal fibre shown here is generic, current evidence suggests that filamentous actin (F-actin; also known as microfilaments), in particular, is poised to function in this capacity. Free tRNA molecules are charged as aminoacyl-tRNAs (step 1) and then used as the building blocks of elongating protein chains through eEF1A (step 2; see Supplementary information S1 (box)), and recycled (step 3). eEF1A is also recycled (step 4), an event that is facilitated by eEF1Bα and eEF1Bγ (see Supplementary information S1 (box)). eEF1A occurs as either an aminoacyl-tRNA-bound or a F-actin-bound entity (see Fig. 2). Various perturbations to the cytoskeleton lead to a loss in translational activity, supporting the view that the translational apparatus is not randomly organized in the cell, or randomly bound to a solid-phase (cytoskeletal) entity but, rather, is spatially organized onto the cytoskeleton network in an as yet unknown sophisticated manner. This schematic proposes an expanded role for the cytoskeleton in translation through the provision of solid-state support for the optimal spatial organization of the protein synthesis machinery; Seyun Kim & Pierre A. Coulombe; http://www.nature.com

2-D EF-G sketch
2-D sketch-EF-Tu
The 3-D shape of EF-G, EF-Tu, mimics the shape of tRNAs; this shape provides the complementary surface for the binding of them on to ribosomal surface.
The anticodon stem-loop region of initiator tRNA interacts (non- covalently) with 16s rRNA at G926, m2G966, G1338 and G1401 located at P site.
Release of IF-3/IF1 is very essential for the binding of 50S ribosomal subunit. As long as IF-3 is associated to 30s ribosome, the 50s ribosome does not bind the 30s. Once 50s binds to 30s, IF-2GTP-Fmet-tRNA complex loads on to 50S-30s complex at ‘P site. This induces conformational changes in both subunits. Correct base paring between codon-anticodon is monitored by two ribosomal Adenine residues found at nucleotides 1492-1493.
Correct base pairing activates IF-2‘s GTPase activity leading to the hydrolysis of GTP and release of the same. Whether base pairing is facilitated by the sugar in anti or cis conformation is not clear. This process is called Kinetic selectivity as found in DNA nucleotide assembly. The release of initiation factors from the ribosomal surface reforms the active site on ribosomes. This conformational change is very important. The released IF-2.GDP is recycled through recycling factors.
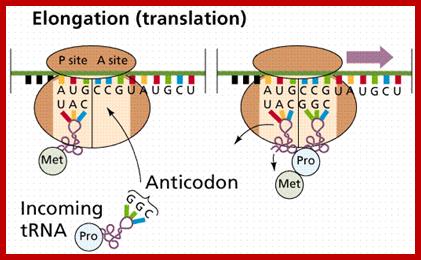
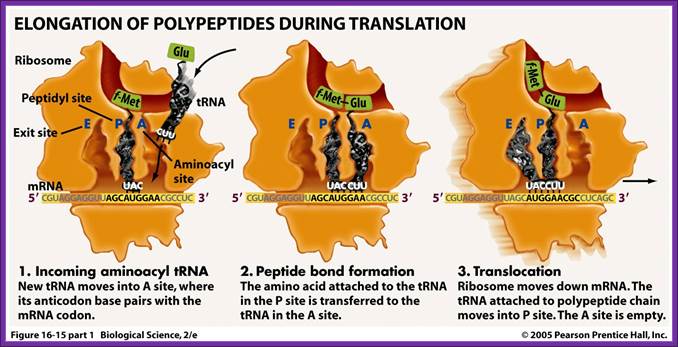
Chain elongation:
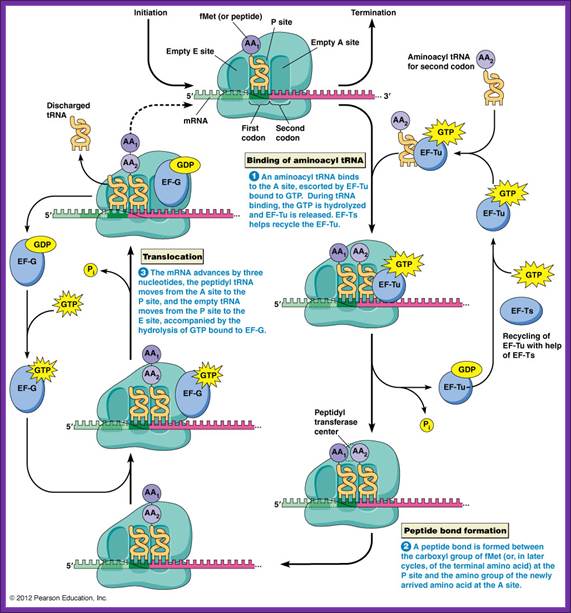
Three special Elongation Factors are required for this phase of protein synthesis: EF-Tu (GTP), EF-Ts and EF-G (GTP).
The Elongation phase of protein synthesis involves a cyclic process whereby a new aminoacyl-tRNA is positioned on the ribosome, the amino acid is transferred to the C-terminus of the growing polypeptide chain, and the whole assembly moves one position along the ribosome.
https://www.rpi.edu
Image from Purves et al., Life: The Science of Biology, 4th Edition,; http://www2.estrellamountain.edu/
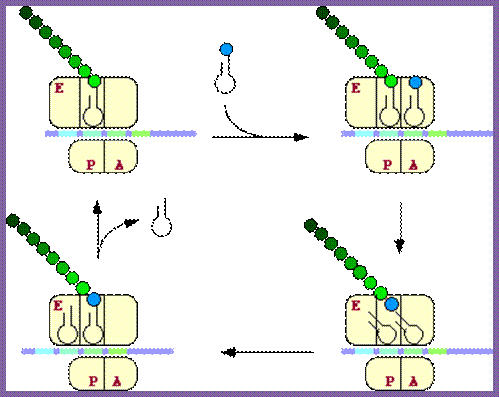
The diagram depicts different steps of chain initiation; http://dwb.unl.edu/

Peptide bond formation by the ribosome. The three lines
between the mRNAs and the tRNA indicate base pairing between the codon of the
mRNA and the anticodon of the tRNA. http://www.biologyreference.com/
http://www2.estrellamountain.edu/
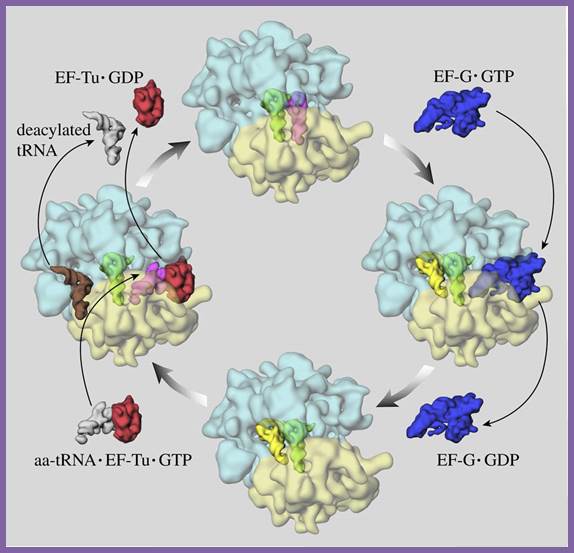
Colors:
The large ribosome subunit is cyan, the small ribosome subunit pale yellow,
EF-Tu red, and EF-G blue. tRNAs are gray (free or complexed with EF-Tu),
magenta (binding at A site), green (in P site), yellow or brown (in the process
of exiting).
Dr. Frank's laboratory group at the Wadsworth Center, New York State Department
of Health, used cryo-EM and 3D image reconstruction to determine ribosomal
structures and positions of EF-Tu and EF-G. Structures of EF-Tu and EF-G are
based on separate X-ray crystallographic studies; https://www.rpi.edu/
Both subunits of ribosomes have active sites, one called peptidyl site (P) and other site is aminoacyl-tRNA site called ‘A’ site i.e incoming aminoacyl tRNA site. The cleft region of small ribosomal subunit, has binding sites for initiator tRNA-IF-2-GTP at P’ site and EF-tu-GTP amino acyl-tRNA complex at A’ site. It also acts as a platform for mRNA binding. The small subunit also contains one more site called R-site to the right of ‘A’ site, which increases accuracy of translation by monitoring incoming aminoacyl-tRNA-IF2Tu-GTP. The incoming aminoacyl-tRNA-IF-2-GTP complex binds to R-site, once it screens it flips to A site; ATP is required at this step?. The A site is continuously bombarded with a.a tRNAs; only that tRNA’s anticodon matches with the codon in mRNA, base pairs and binds.
The large ribosomal subunit also contains another site called E site to the left of P’ site, which provides space for the exit of unloaded or (-a.a) tRNA. The large ribosomal subunit also contains what is called ‘factor binding site’ at which EF2G-GTP binds.
Chain elongation progresses in two distinctive but processive steps. The GTP by binding to elongation factor EF-Tu activates the factor and it now binds to an aa.tRNA, which have unpaired bases between C at 5’terminal to A on 3’ stem. Some tRNAs which are fully base paired at 5’ends with acceptor stems, open when the EF-Tu-GTP complex binds to a.a-tRNA.
The EF-Tu-GTP complex has N-terminal GTP binding domain. Hydrolysis of GTP induces dramatic structural conformational changes. The factor has a cleft between domains 1 and 2 with several positive charged side chains, which actually facilitates the binding of aminoacyl tRNA. The changes in this factor also induce some significant changes in the ribosomal complex when they bind to it.
These factors bind to any amino-acylated tRNAs and they are found in large numbers. Almost ~200,000 a.a. tRNAs, found in cytoplasm are sequestered by the above said factors.
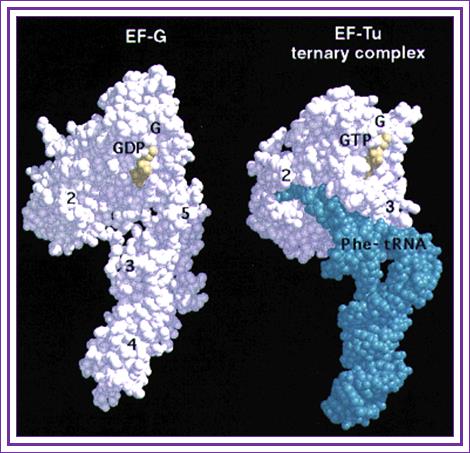
The 3-D model depicts EF-G and EF-Tu have more or less same shape and they occupy the same site but mutually exclusive; http://www/cell.com
These aa.tRNA.EF-Tu.GTP factors now diffuse on to ribosomal ‘A’ sites; interact with codon positioned at A site. The EF-Tu GTP always binds 50s ribosomal subunit. The region between adjacent codons in the mRNA, at P and A sites, is kinked by 45°. In this recognition process the projected codon and anticodon nucleotides, held in geometrically and stearically directed positions, interact with each other for complementarity; it is only the tRNA whose bases with proper complementary base-pair are retained. Two adjacent Adenine residues in 16s rRNA also monitors this proper base paring, by what is called kinetic selectivity. If the base pairing after diffusing on to the site fails, they are expelled from the surface with hydrolysis of GTP.
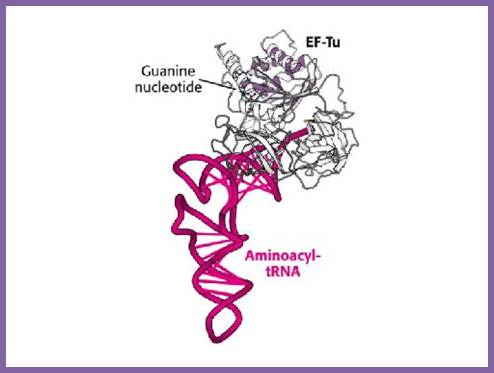
This diagram shows the ribbon model of EF-Tu bound to aminoacyl tRNA loaded with an amino acid residue; www.qizlet.com
If the base pairing is perfect to the rule, it activates the EF-Tu to hydrolyze GTP. This leads to the dissociation of the EF-Tu-GDP from ribosomal surface and further readjustments takes place at the region of active site, through conformational changes; such as positioning of aa.tRNA in it nearest neighborhood of f-met-tRNA in such a way the distance between the two is approximately < 5 to 7 Å, which is the distance required for any covalent bond formation and also the aminoacyl end of tRNA now directed towards 'P' Site.
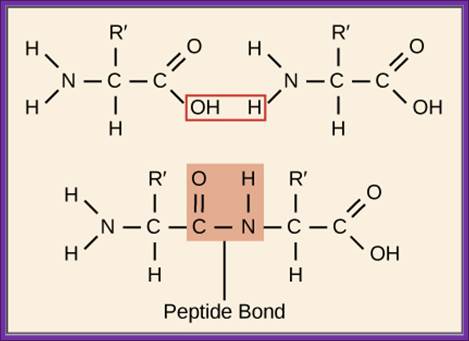
USDAVIS chemWIKI
Chain elongation steps; www.uic.edu
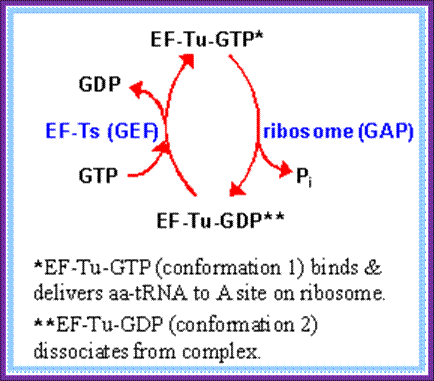
Then EF-Tu is regenerated by GTP exchange factor EF-Ts (GEF)
https://www.rpi.edu
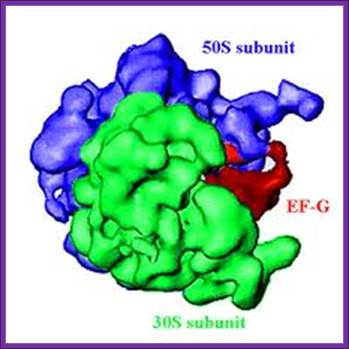
Position of EF-G binding site on Large ribosome; Bmb.leeds,ac UK
Crystal structure of EF-Tu-GTP-aa-tRNA shows structural similarity with that of EF-G, they also show functional similarities. The 3 domains at the C-terminal region of EF-G-GTP resemble tRNA and positions in same way as that of tRNA ternary complex.
Both bind to ribosomal surface with overlapping sequence, and they bind only when they are bound by GTP. Both have GTPase activity when bound to ribosomes. After hydrolysis of GTP, both factors assume different conformation and have very low affinity for ribosomes in this form. EF-G with hydrolysis of GTP drives translocation, where ribosomes under go flip-flop conformational change. EF-G s-IV th domain and anticodon domains have similarities. The IV th domain is required for GTP hydrolysis and it is also required for the release of EF-G-GDP complex from ribosomal surface. Both EF-Tu and EF-G hydrolysis of GTP induce conformational changes in the ribosomal structure, so ribosomes become active form for their respective functions.
Principles of Cell Biology; Dept. of Biology, Memorial University of Newfoundland; http://www.mun.ca/biology;

Side view of the large ribosomal unit showing an exit tunnel though which newly synthesisized polypeptide chain tyraverses through; www.cipsm.de; plant cell biol.org
Dissociation of EF-Tu-GDP provides the space for the binding of EF-G-GTP. The position of EF-Tu and EF-G are overlapping so, their binding is mutually exclusive. EF-G, which is present in about 20 000 copies per cell, is 691 amino acids long and contain five domains. This provides energy for chain translocation, so it can be considered as Translocation factor. The activity of this factor can be inhibited by Fusidic acid, which stabilizes EF-G-GDP complex and such complexes remain bound to the surface of ribosomes. The two N-terminal domains (191) resemble N-terminal region of EF-Tu. The COO- terminal region of EF-G also has sequence similarities with certain motifs of riboproteins; hence it is believed that this protein reacts with 23s ribosomes.
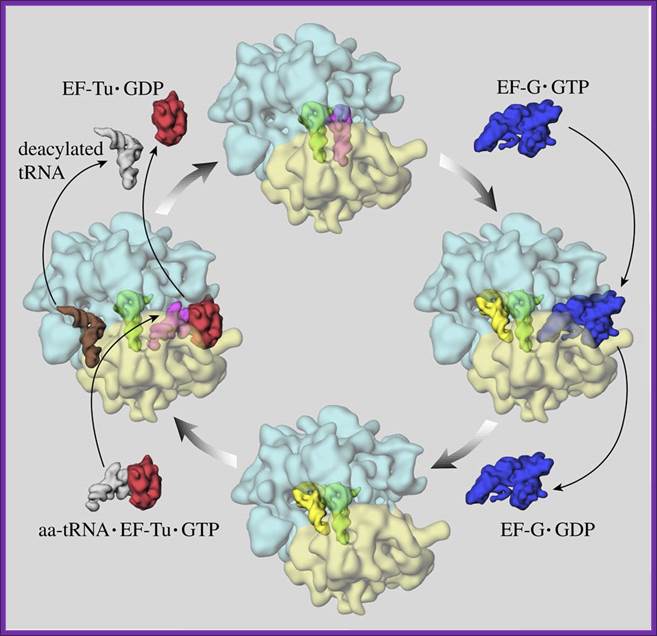
Mechanism of protein synthesis by ribosomes; EF TU-GTP and EFG-GTP-GDP recycle; EF-G’GTP binding site abutted to A’ site on ribosomal surface; this site often referred to as ‘Factor Binding Site’, the factors such as EFTu, EF.G with their GTP bind to these sites. http://www.ks.uiuc.edu/
‘Elongation cycle of protein synthesis; The ribosome is shown in top view, with the small subunit (transparent yellow) below the large subunit (transparent blue). (i) The ribosome in the pre-trans locational state with tRNAs in the A (magenta) and P (green) sites. After spontaneous peptidyl transfer, the nascent peptide is covalently attached to the A-site tRNA. (ii) The elongation factor EF-G in complex with GTP (blue) has bound to the ribosome to facilitate the translocation of tRNAs to the P (green) and E (yellow) sites. The translocation is induced by GTP hydrolysis accompanied by large transient conformational changes in the EF-G and the ribosome. (iii) The release of the EF-G after GTP hydrolysis leaves the ribosome in the post-translocational state. It is now ready to accept a new amino acyl-tRNA (white) presented to the ribosome by the ternary complex, comprising, in addition to the new amino acyl-tRNA, the elongation factor EF-Tu and a GTP (red). (iv) The ribosome with a bound ternary complex (magenta and red) in place. It has been suggested that when the ternary complex binds to the ribosome, the E-site tRNA moves further away from the P site to the so-called E2 site (orange). The snapshot shown here is part of the decoding step, where the amino acyl-tRNA whose anticodon matches the next codon in the mRNA is selected to enter the A site, accompanied by GTP hydrolysis and conformational changes. In case of a match, EF-Tu with the hydrolyzed GTP and the E-site tRNA leave the ribosome, leaving it in the pre-translocational state (i). This figure is adapted from Frank, J. in Conformational Proteomics of Macromolecular Architectures. World Scientific Publishing Comp. Singapore, 2004. Watch a movie of this process”.
Large ribosomal subunit contains a protein that acts as an enzyme, called peptidyl transferase, mostly located in between the two sites. The enzyme brings about peptide bond formation. However it is now thought that the large ribosomal RNA by itself at space between tRNAs can act as an enzyme in bringing about peptide bond formation.
Partial digestion of ribosomal surface with protease, does not inhibit peptide bond formation, on the other hand digestion of rRNA inhibit the process.
- Ribosomes obtained from Thermophilous aquaticus are mildly treated with phenol, SDS and proteases, when allowed them to synthesize protein, they still showed translational activity, which strongly suggests that 23S-rRNA has the ability to perform peptide bond.
- Peptide bond formation between the aa.tRNA on P site and aa.tRNA on the A site is greatly facilitated and favored by the close vicinity of the two ends of aa.tRNA on the ribosomal surface, which is about 2-4Å angstroms. The carbonyl bond between an amino acid and the tRNA at the 3’ of A is already an energy rich bond. This carbonyl bond may be found at 3’OH or 2’OH; it does not matter at the time of peptide bond formation. When two amino acids are brought closer to each other by ribosome conformational change, carbonyl bond from tRNA of the P site is transferred to the NH2 group of the tRNA at A site. This produces a C-N bond or peptide bond. This event doesn’t require any input of energy.
Peptidyl transferase that performs peptide formation is a part of 23S rRNA, which acts like a Ribozyme unlike other ribozymes, it performs peptide bond formation. The aa-tRNAs at P and A sites are bound to large ribosomal surface through base pairing between 23S rRNA and acceptor CCA3’. Base pairing to rRNA facilitates hydrogen atoms from alpha amino group of aa-tRNA at A-site are drawn by the RNA nucleotides, thus rendering the N atom of the alpha amino group as a strong nucleophile. This nucleophile attacks the carbonyl group aa-tRNA at peptidyl site, resulting in a C-N bond formation. A similar action is also performed by the proteins in peptide bond formation.
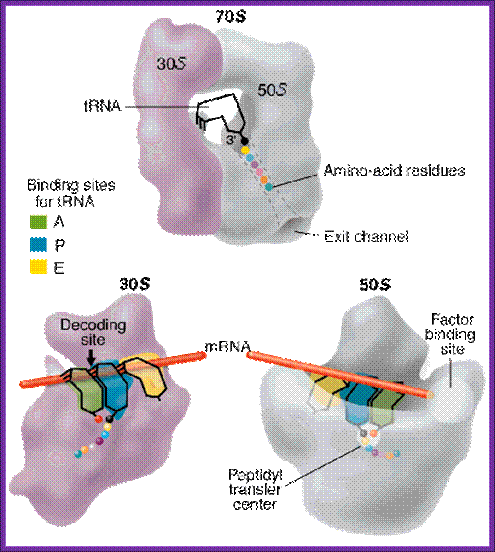
Different factors’ binding sites are shown. The mRNA between two bound tRNA is bent. http://dc445.4shared.com/

Figure: Possible reaction mechanism for the peptidyl transferase activity present in the large ribosomal subunit; observe the role of Adenine in peptide bond formation.www.quizlet.com
The overall reaction is catalyzed by an active site in the 23S rRNA. In the first step of the proposed mechanism, the N3 of the active-site adenine abstracts a proton from the amino acid attached to the tRNA at the ribosome's A-site, allowing its amino nitrogen to attack the carboxyl group at the end of the growing peptide chain. In the next step this protonated adenine donates its hydrogen to the oxygen linked to the peptidyl-tRNA, causing this tRNA's release from the peptide chain. This leaves a polypeptide chain that is one amino acid longer than the starting reactants. The entire reaction cycle would then repeat with the next aminoacyl tRNA that enters the A-site. (P. Nissen et al., Science 289:920–930, 2000.)
The Role of 23S Ribosomal RNA Residue ‘A’-2451 in Peptide Bond Synthesis; Revealed by Atomic Mutagenesis:
Kathrin Lang1, Matthias Erlacher2, Daniel N. Wilson3, 4, Ronald Micura and Norbert Polacek.
Summary of the peptide bond formation reaction:
Peptide bond
formation is a fundamental reaction in biology, catalyzed by the ribosomal
peptidyl-transferase ribozyme. Although all active-sites of 23S ribosomal RNA
nucleotides are universally conserved, atomic mutagenesis suggests that these
nucleobases do not carry functional groups directly involved in peptide bond
formation. Instead, a single ribose 2![]() -hydroxyl
group at A2451 was identified to be of pivotal importance. Here, we altered the
chemical characteristics by replacing its 2
-hydroxyl
group at A2451 was identified to be of pivotal importance. Here, we altered the
chemical characteristics by replacing its 2![]() -hydroxyl
with selected functional groups and demonstrate that hydrogen donor capability
is essential for transpeptidation. Authors mentioned below propose that the
A2451-2
-hydroxyl
with selected functional groups and demonstrate that hydrogen donor capability
is essential for transpeptidation. Authors mentioned below propose that the
A2451-2![]() -hydroxyl
directly hydrogen bonds to the P-site tRNA-A76 ribose. This promotes an
effective A76 ribose C2
-hydroxyl
directly hydrogen bonds to the P-site tRNA-A76 ribose. This promotes an
effective A76 ribose C2![]() -endo
conformation to support amide synthesis via a proton shuttle mechanism.
Simultaneously, the direct interaction of A2451 with A76 renders the
intramolecular transesterification of the peptide from the 3
-endo
conformation to support amide synthesis via a proton shuttle mechanism.
Simultaneously, the direct interaction of A2451 with A76 renders the
intramolecular transesterification of the peptide from the 3![]() - to 2
- to 2![]() -oxygen
unfeasible, thus promoting effective peptide bond synthesis.
-oxygen
unfeasible, thus promoting effective peptide bond synthesis.
Important Contribution to Catalysis of Peptide Bond Formation by a Single Ionizing Group within the Ribosome; Vladimir I Katunin1, Gregory W Muth2, Scott A Strobel2, Wolfgang Wintermeyer3 and Marina V Rodnina.
The
catalytic mechanism of peptide bond formation on the ribosome is not known. The
crystal structure of 50S ribosomal subunits shows that the catalytic center
consists of RNA only and suggests potential catalytic residues. Here we report
rapid kinetics of the peptidyl transferase reaction with puromycin at rates up
to 50 s![]() 1. The
rate-pH profile of the reaction reveals that protonation of a single ribosomal
residue (pKa = 7.5), in addition to protonation of the nucleophilic
amino group, strongly inhibits the reaction (>100-fold). The A2451U mutation
within the peptidyl transferase center has about the same inhibitory effect.
These results suggest a contribution to overall catalysis of general acid-base
and/or conformational catalysis involving an ionizing group at the active site.
1. The
rate-pH profile of the reaction reveals that protonation of a single ribosomal
residue (pKa = 7.5), in addition to protonation of the nucleophilic
amino group, strongly inhibits the reaction (>100-fold). The A2451U mutation
within the peptidyl transferase center has about the same inhibitory effect.
These results suggest a contribution to overall catalysis of general acid-base
and/or conformational catalysis involving an ionizing group at the active site.
The Role of 23S Ribosomal RNA Residue A2451 in Peptide Bond Synthesis Revealed by Atomic Mutagenesis: Kathrin Lang; Matthias Erlacher; Daniel N. Wilson; Ronald Micura; Norbert Polacek :Chemistry & Biology; online article 24.04.2008
Peptide bond formation is a fundamental reaction in biology, catalyzed by the ribosomal peptidyl-transferase ribozyme. Although all active-site 23S ribosomal RNA nucleotides are universally conserved, atomic mutagenesis suggests that these nucleobases do not carry functional groups directly involved in peptide bond formation. Instead, a single ribose 2’0-hydroxyl group at A2451 was identified to be of pivotal importance. Here, the above authors altered the chemical characteristics by replacing its 2’-0-hydroxyl with selected functional groups and demonstrate that hydrogen donor capability is essential for transpeptidation. We propose that the A2451-2’0-hydroxyl directly hydrogen bonds to the P-site tRNA-A76 ribose. This promotes an effective A76 ribose C20-endo conformation to support amide synthesis via a proton shuttle mechanism. Simultaneously, the direct interaction of A2451 with A76 renders the intramolecular transesterification of the peptide from the 3’0- to 2’0-oxygen unfeasible, thus promoting effective peptide bond synthesis.
- The released EF-Tu-GDP is recycled by another cofactor called EF-Ts. The EF-Ts interacts with EF-Tu-GDP, where the GDP is released from EF-Tu complex and EF-Ts-EF-Tu complex in turn reacts with GTP and now GTP binds to EF-Tu and EF-Ts is released. The EF-Ts acts like a GTP exchange factor or GEF.
This recycling process can be subjected to regulation. The EF-Tu has proof reading property. Kirromycin binds to EF-Tu, which in turn binds to A’ site. On hydrolysis, with Kirromycin that is bound is not released from the A site and prevents the peptide bond formation and creates a ‘stalled state’.

EM picture of polyribosome
“Note: The information from Rodina regard to peptide bond formation.
“Peptide bond formation is the fundamental reaction of ribosome. The ribosome's active site – the peptidyl transferase center – is composed of rRNA, and thus the ribosome is the largest known RNA catalyst. The ribosome accelerates peptide bond formation by 107-fold relative to the uncatalyzed reaction. Recent progress of structural, biochemical and computational approaches has provided a fairly detailed picture of the catalytic mechanisms employed by the ribosome. Energetically, catalysis is entirely entropic, indicating an important role of solvent reorganization, substrate positioning, and/or orientation of the reacting groups within the active site. The ribosome provides a pre-organized network of electrostatic interactions that stabilize the transition state and facilitate proton shuttling involving ribose hydroxyl groups of tRNA. The catalytic mechanism employed by the ribosome suggests how ancient RNA-world enzymes may have functioned. (Published Online August 8 2006)” Correspondence: Institute of Physical Biochemistry, University of Witten/Herdecke, Stockumer Str. 10, 58448”.

Polysome formation steps and protein synthesis
Peptide bond formation leads to EF-G hydrolysis of GTP to GDP and Pi and the release of the EF-G factor are not linked to any other events. However GTP hydrolysis leads to translocation of peptidyl tRNA from P’ site to A’ site, during which the deacylated tRNA is displaced to E’ site. The energy released is used for physical movement.
Another factor 4.5s RNA, a 114-base long RNA, which is more or less double stranded, once thought to be involved in protein synthesis. This factor is present in about ~3000 copies per cell. Mutants in 4.5s RNA gene, grow very slow. Mutants in EF-G suppress 4.5 RNAs action. The 4.5s RNA interacts with EF-G; how it acts? It is now known that 4.5 dsRNA, is known to be involved in protein secretion. Its action is similar to eukaryotic 7s RNA.
The 4.5s RNA has domains similar to 7sL RNA, which plays an important role in transferring proteins destined to ER. The 4.5s RNA acts as Srp in transport of protein in E.coli.
Translocation of ribosomes on mRNA involves physical movement of ribosomal structures in relation to mRNAs, which requires energy, and it is actually provided by the hydrolysis of GTP by EF-G itself.
Translocation perse involves regulation, but stepwise movement of ribosomal complex on mRNA by 3 nucleotides i.e. one codon at each step, towards the 3’ end of the mRNA. Whether the mRNA is pulled in 5’ direction or ribosome moves in 5’ to 3’ direction of the mRNA (Click-Clock), it is all the same.
But movement of ribosome requires a motor protein or helicase like protein (requires energy provided by the hydrolysis of EF-G-GTP), which is suspected to be one of the riboproteins, because only motor proteins have the ability to move on polynucleotide strand. During the movement base pairing of tRNA to coding bases is very important. In certain frame shift reading, the tRNA containing 4 nucleotides (anticodon) pair with 4 nucleotides of the codons, when it is translocated it translocates by four nucleotides, rather than three nucleotides (frame shift).
Translocation requires GTP energy, which is provided by the hydrolysis of GTP by EF-G factor. Once hydrolysis is over the EF-G-GDP dissociates from the ribosome.
According to one view, 50s subunit with bound peptidyl tRNA at A’ site and deacylated tRNA at P site together move by three nucleotides towards the next codon i.e. towards 3’ direction of mRNA. Then 30s subunits slide along to join 50s in proper orientation. The process by which 30s ribosome bound with tRNAs and mRNA and 50S ribosome bound with tRNAs in their respective sites act as small machine process called ‘Ratcheting’. Animation of the same provides an excellent view.
According to another view the 50s ribosome in relation to 30s, move in such a way, the peptidyl tRNA end moves towards the P site, yet the anticodon end is still base paired at A site, thus both anticodons ends are still bound to P and A site respectively and the their a.a ends are displaced in such a way, the peptidyl end of the tRNA of the A site is now in P site and the free 3’ end of the discharged tRNA is now displaced towards E site, yet the anticodon ends are still in their respective positions i.e. at P and A sites. It is then the other subunit moves forward, in such a way, the tRNA free from amino acid, is transferred to the E site and the tRNA with peptidyl chain positions at P site and makes the A site free.
The two sites, with respect to the tRNA binding affinity, exhibit allosteric properties. When the discharged or deacylated tRNA is shifted to E site it binds with high affinity and the A site has very low affinity to the deacylated tRNA, but when a charged aa-tRNA contacts A site, it becomes high affinity site, and the exit site becomes a low affinity site, so the tRNA is discharged from the E site. And the ribosomal surface is again ready for one more set of reaction for another peptide formation.
This process of positioning of the aa.tRNA onto the A site, formation of peptide bond and translocation and readjustments and activation of each of the ribosomal active sites processively progress.
The rate of assembly varies, but a single ribosome can decode or move at the rate of 100 to 200 codons in just 10 to 20 seconds. This, however, is not the situation, as the 5’, end of the mRNA is projected out of the RNAP-DNA complex during transcription, the end is complexed with the ribosomes for translational initiation and starts synthesis. As this progresses by about 20 –23 codon, the free 5’ end is again gets associated with another ribosomal complex, by that time the first ribosome might have already moved by another 20—23 codons. This way, during translation, as and when the 5’ end is made available, it can be associated with the respective ribosomal factors to reinitiate another translation event.
The N-terminal part of the protein chain is threaded through the tunnel or a narrow groove in the large ribosomal subunit. The end of the chain comes out through exit point. The length of the tunnel is about 100 Aº and it can hold 35 to 45 amino acids. It is through the exit site the ribosomes bind plasma membranes and thread secretary proteins through. The ribosomal tunnel is aqueous-flooded with water.
Depending on the needs and inherent factors, the rate of translation can be accelerated or decelerated. In an active translational system a single mRNA can be associated with more than 8 to 10 ribosomes at any given time, where initiation, elongation and termination processes are going on simultaneously. Such mRNAs with multiple ribosomes operating are called polyribosome or polysome; and the rate of synthesis is high.
The rate of translation though range from 15 -20 amino acids per second, some genes do control the rate. Mutants ‘ram’ and ‘tuf’ and ‘Ar’, show faster rate of protein synthesis; in this accelerated protein synthesis proof reading is a casualty. In mutant strain str.A, which is streptomycin resistant, the rate is half the normal, but translation is extremely accurate. Instead of one ribosome translating one mRNA at a time, many ribosomes translating simultaneously, is another way of increasing the rate of translation.
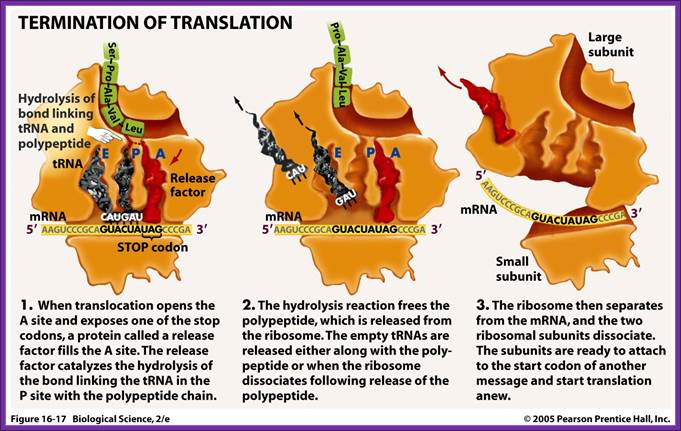
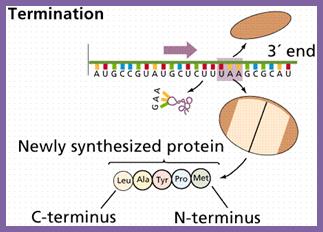
Chain Termination:
Once the chain initiation sets into elongation mode, decoding takes place, one codon after another codon processively, till it comes to one of the three termination codons i.e. UAA, UAG or UGA which are located at the end of ORF. These were discovered as mutants, such as UAA = Ochre, UAG = Amber and UGA = Opal. A point mutation that changes the codon coding for an amino acid into a different amino acid, then it is called ‘missense mutation’. If the mutation in coding sequence changes into noncoding terminator codon then it is called ‘nonsense mutation’.
When the translating ribosomal machinery reaches any one of the above said codons at A’ site, it stops. This is due to the fact that there are no tRNAs having anticodons which can base pair with ‘ter’ codons; it is sine quo non device, so ribosomes halt or stall at this position. This is an incredible molecular device to end mRNA translation.
The empty ‘A’ site is recognized by chain releasing factors like RF1 or RF2 and RF-3. RF1 and RF2 belong to class-I termination factors and RF3 is class II factor. RF-1 recognizes UAA and UAG and RF-2 recognize UAA and UGA termination codons and bind to sequence on the ribosomal surface.
The termination factors RF1 and RF2 have structural shape similar to that of tRNA with domain-1 having SPF (Serine, Proline and Phenylalanine) motif recognizes the ter-codon and the domain 2 at the distal end that is similar to the domain of aminoacyl acceptor end of tRNA contain an amino acid sequence GGQ (Glycine, Glycine, Glutamine). The GGQ ‘glutamine’ carboxyl end with an amino and oxygen group holds a molecule of water, thus provides water for the lysis of the bond between aminoacyl end of tRNA and peptide chain.
Factors RF1 and RF2 bind to empty A-site on ribosome. The number of RF factors is about ~600 per cell. When they properly bind recognizing their respective TER codons, they activate ribosomal peptidyl transferase, which uses the water molecule provided by the Glutamine, transfers one Hydroxyl group to the carbonyl group at peptidyl end and the polypeptide is released from the ‘P’site, where the empty tRNA is still bound.
Then the RF3-GTP, (RF3 is a GTP binding protein), having domain similar to EF-G and EF-Tu, binds to the large ribosomal surface at a site called ‘factor binding site’. The bound RF3-GTP interacts with RF-1 or RF-2 and also interacts with the last tRNA at P’ site now deacetylated. With the hydrolysis of GTP, it releases RFs from the ribosomal surface.
![]()
![]()
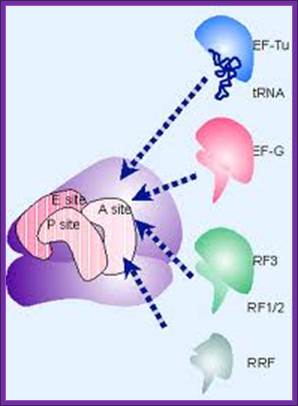
Molecular mimicry enables the elongation factor Tu-tRNA complex, the translocation factor EF-G, and the release factors RF1/2-RF3 to bind to the same ribosomal site. RF1 and RF2 recognize the termination codons and activate the ribosome to hydrolyze the peptidyl tRNA. Cleavage of polypeptide from tRNA takes place by a reaction analogous to the usual peptidyl transfer, except that the acceptor is H2O instead of aminoacyl-tRNA. Then RF1 or RF2 is released from the ribosome by RF-3, which is a GTP-binding protein related to EF-G (1161,1162). RF3 resembles the GTP-binding domains of EF-Tu and EF-G, and RF1/2 resemble the C-terminal of EF-G, which mimics tRNA. This suggests that the action of RF3 on RF1/2 utilize the same site that is used by the elongation factors. Figure 6.27 illustrates the basic idea that these factors all have the same general shape and bind to the ribosome successively at the same site (basically the A site or a region extensively overlapping with it); genes.atspace.org; http://molecularstudy.blogspot.in/
Role of RfsS(A):
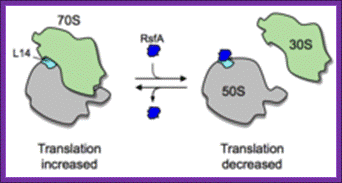
Mechanism of ribosomal subunit dissociation by RsfS(A): The RsfS-A binding to ribosomal L14 blocks joining of small and large ribosomal subunits during stationary phase. This factor is found in all bacterial systems and mitochondria and chloroplast; looks they are homologues; WIKI. www.en.wikipedia
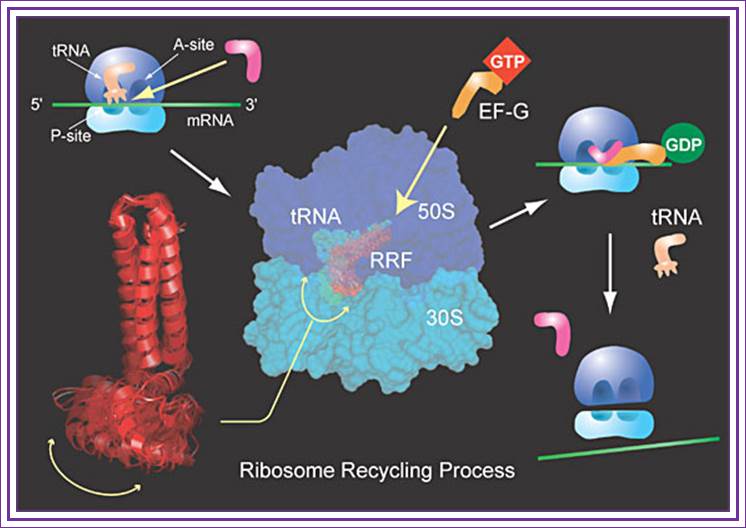
Role of RRF:
Dissociation of ribosomal subunits and the release of tRNA from P-site is actuated by a factor called Ribosomal Recycling Factor (RRF). RRF recruits EF-G-GDP to the ribosomal surface at factor binding site and gets activated by the exchange of GDP to GTP, then the hydrolysis of GTP leads to the release of tRNA from the P-site and RRF locates itself on to P-site and EF-G on to A site. The RRF3 that is bound to the ribosome interact with 30S ribosome and releases RRF-EF-G and RRF itself from the ribosomal surface. The small subunit of ribosome that is released from large subunit is still intact with mRNA. This ribosome bound mRNA can be activated only when IF-3 interacts with 30s subunit and activates it for recycle.
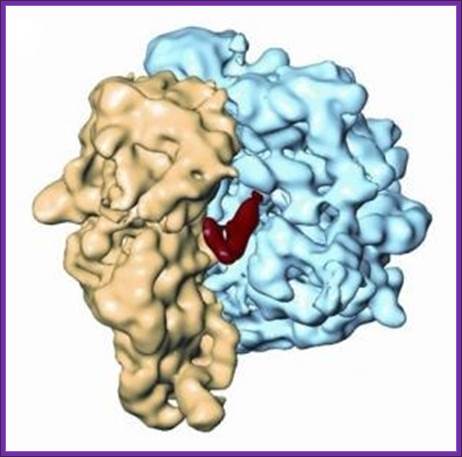
Representation of the three-dimensional cryo-electron micrograph structure of the ribosome-RRF complex. The ribosome recycling factor (RRF) appears in red; the small ribosomal subunit in yellow; and the large ribosomal subunit in blue; Akira Kaji, Rajendra Agrawal, and Joachim Frank/Proceedings of the National Academy of Sciences
In short, the recycling process goes like this: RRF, along with EF-G, binds to the ribosome. This promotes the release of tRNAs by the movement of RRF, similar to tRNA movement. “This is the first example of a functional mimic of tRNA by a protein,” adds Kaji. After the tRNAs leave, RRF, EF-G, and mRNA also detach from the ribosome. The released ribosome is now empty and free to start a new session of translating mRNA into protein. Where RRF binds is near the key ribosomal spot holding mRNA. “Since the main function of RRF is to release mRNA, this makes sense in terms of function,” explains Kaji.
The UGA based termination can also be regulated for this terminator codon is the leakiest one that can lead to continuation of protein synthesis; some times in different reading frame from that point i.e. UGA.
|
Chain Release factors: |
||
|
RF-1 |
Recognizes the termination codons 5′-UAA-3′ and 5′-UAG-3′ |
|
|
RF-2 |
Recognizes 5′-UAA-3′ and 5′-UGA-3′ |
|
|
RF-3 |
Stimulates dissociation of RF1 and RF2 from the ribosome after termination; GDP, GTP binding factors |
|
|
RRF |
Responsible for disassociating the ribosome subunits after translation termination. EGF-GTP binding factor. |
|
Contours of two ribosomal subunits, the small 30 S (yellow) and the large 50 S (red), associated in the full 70 S ribosome, with designations of some morphological features.Left, the so-called overlap projection when the 30 S subunit is facing the viewer and covers part of the 50 S subunit; right, the lateral projection viewed from the side of the L7/L12 stalk. CP, central protuberance.
Chain termination reaction. Refer to http://www.dnatube.com/video/11/Visualization-of-tRNA-Movements-on-70S-Bacterial-Ribosome. This figure is not accurate for RRF.
Few authors’ view on chain release and ribosome recycling:
Following the release of the polypeptide chain, a series of reactions is required to enable ribosomes to recycle and initiate the synthesis of a new protein. In E. coli three proteins are required for this process, in addition to other proteins involved more specifically in the process of translation initiation. The three proteins are RRF (Ribosome Recycling Factor, discovered by A. Kaji in the late 1960s), EF-G the same factor that catalyses translocation during the elongation cycle) and initiation factor IF3. The function of RRF requires that RF1 (or RF2) should no longer be bound to the ribosome. The work of Reza Karimi showed that the role of RRF and EF-G is to dissociate the ribosomal subunits once the A-site has been liberated by the departure of RF1 or RF2. This reaction requires GTP hydrolysis. After the departure of the 50S subunit the mRNA and deacylated tRNA remains bound to the 30S subunit. IF3 destabilizes this complex allowing the deacylated tRNA to be replaced by fMet-tRNA in readiness for translation initiation. The three factors RRF, EF-G et RF3 implicated in ribosome recycling are also able to to catalyze the dissociation of peptidyl-tRNA at very early stages of mRNA translation (Heurgué-Hamard et al., 1998); Dinçbas et al., 1999 ; Heurgué-Hamard et al., 2000). (Pavlov et al., 1997; (Karimi et al., 1999).
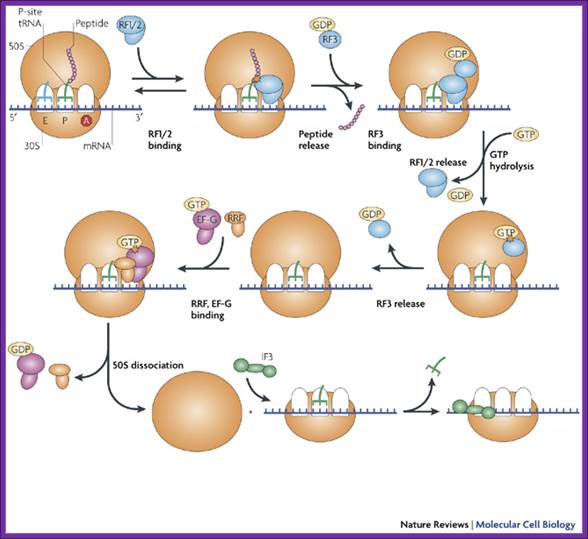
There are two hypothesis one of them is- The diagram shows chain termination factors and the events; binding of Rf1/2 to ‘A site, then RF3-GDP binds upstream of Rf1/2; this releases peptide chain. Then the binding of GTP releases Rf1/2 and later RF3-GDP is also released. The binding of RRF and GTP leads to the release of subunits and mRNA; IF3 is required for the activation of 30s. In the above figure RRF-EF.G dissociate large subunit from the small subunit, but small subunit still retains mRNA. www.nature.com
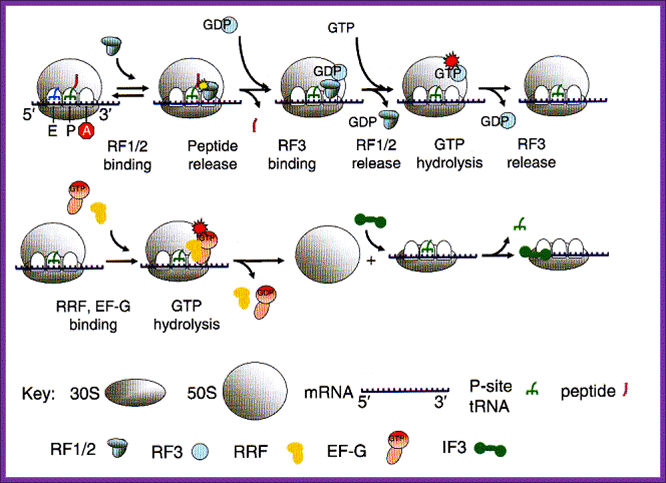
Overview of the process of translation termination. See text for detailed explanation. Note: RF1/2 means either RF1 or RF2. The red stop sign with a white A indicates that a stop codon is positioned in the A site of the ribosome. Other symbols are explained in the Key below; The process of releasing the ribosome after protein synthesis has terminated is fundamental because the gene for RRF is essential. After release of the peptide chain, the ribosome is left with mRNA and a deacylated tRNA in the P site figure above. This complex needs to be disassembled to prepare the ribosome for a new round of protein synthesis. Ribosome recycling factor (RRF) along with EF-G is required for this process. RRF and EF-G lead to the dissociation of ribosomes into subunits upon GTP hydrolysis. Subsequently, initiation factor IF3 is required for removal of the deacylated tRNA from the 30S subunit. Oregonstate.edu.
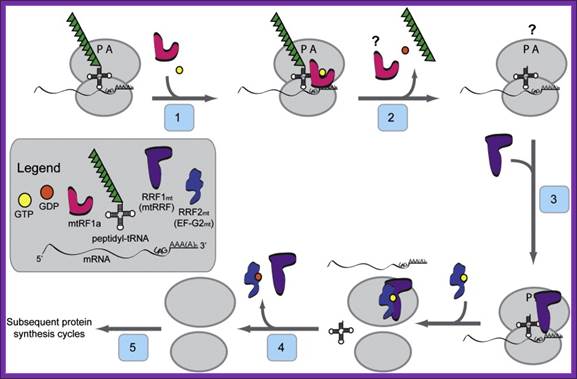
Model for the initiation phase of mitochondrial translation: In the current model for the initiation of protein synthesis, mitochondrial initiation factor 3 (IF3mt) actively dissociates 55S ribosomes, forming a transient [IF3mt:55S] complex (Step 1) and leading to the formation of an IF3mt:28S complex (Step 2). Mitochondrial initiation factor 2 (IF2mt) bound to GTP binds to the small subunit (Step 3), followed by the fMet-tRNA and mRNA (Step 4), and although the exact order of binding is not clear. The mRNA feeds into the mRNA entrance gate and the 5′ end pauses at the P-site of the ribosome for inspection of its 5′ start codon. In the presence of the correct start codon and fMet-tRNA, the mRNA is locked in place by codon: anticodon interactions to form the 28S initiation complex. If fMet-tRNA binds in the absence of mRNA, or if the mRNA does not contain a proper start codon, the inspection step fails. The failed inspection causes the mRNA to continue sliding through the ribosome to exit. Once the 28S initiation complex has formed, the large subunit joins, and along with the hydrolysis of GTP to GDP, the initiation factors exit (Step 5) leaving a 55S:fMet-tRNA:mRNA complex that is ready for the elongation phase of protein synthesis.
Recycling of post-TCs (ternary Complex) requires elongation factor EF-G, the ribosome recycling factor RRF and initiation factor IF3. EF-G and RRF dissociate post-TCs into free 50S subunits and 30S subunits bound to mRNA and P-site deacylated tRNA in a GTP-dependent manner, after which IF3 induces tRNA release from 30S subunits, and mRNA then dissociates spontaneously. RRF binds to post-TCs in the ratcheted state, interacting with segments of 23S rRNA that are constituents of B2a and B3 inter-subunit bridges. It has been proposed that EF-G•GDP binds RRF-associated post-TCs and exchanges GDP for GTP; EF-G•GTP then induces rotation of RRF's head domain, which after GTP hydrolysis splits ribosomes into subunits, (reviewed in Petry et al., 2008, (Gao et al., 2005).

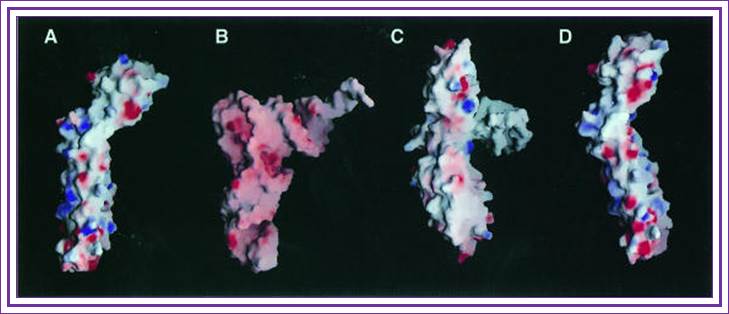
The above three ribbon models show the shapes of the RF1, RF2 and RRF; they almost look similar with their domains. Further examples of molecular mimicry among translation factors. Structures of eukarytotic Release Factor 1 (eRF1), prokaryotic Release Factor 2 (RF2), and prokaryotic Ribosome Recycling Factor (RRF) are shown below. The domains that are thought to mimic tRNA are shown in magenta.
Crystal structure of the Ribosome Recycling Factor (RRF) from Escherichia coli: Kyeong Kyu Kim, Kyeongsik Min, and Se Won Suh.
Fig. Structural
comparisons of RRF with tRNA and EF-G. RRF superposed on (A)
tRNA, (B) yeast tRNAPhe (Suddath et al., 1974) and (C)
EF-G (Czworkowski et al., 1994), and (D)
RRF superposed on EF-G are drawn by surface charge distribution with the
program GRASP (Nicholls et al., 1991).
The red and blue colors represent negatively and positively charged surfaces,
respectively. For the superposition of RRF on tRNA (A), the coil domain of RRF
is superimposed on the anticodon arm of tRNA and the α/β domain is
oriented toward the acceptor arm in tRNA. The position of EF-G in (C) is
determined by overlapping domains I and II of EF-G with the EF-Tu![]() GDP
GDP![]() tRNA complex (Nissen et al., 1995). For the
superposition of RRF on EF-G (D), the coil and α/β domains of RRF are
overlapped with domains IV and V of EF-G, respectively
tRNA complex (Nissen et al., 1995). For the
superposition of RRF on EF-G (D), the coil and α/β domains of RRF are
overlapped with domains IV and V of EF-G, respectively

Figure: Comparison of different tRNA binding sites with the possible conformations of RRF on the ribosome. RRF overlaps with both A- and P-site tRNA binding sites. Binding of RRF is therefore concurrent or subsequent to translocation of P-site tRNA to the P/E-hybrid state.
The ribosome deciphers and translates the genetic code in all organisms. Ribosome recycling is the final stage of translation and involves the concerted action of the ribosome recycling factor (RRF) and elongation factor G (EF-G) to disassemble the post-termination complex for the next round of translation. Although RRF was discovered in the early 1970s, the exact mechanism by which RRF mediates ribosome recycling still remains to be fully elucidated
RRF is universally conserved in bacteria, but not present in archaea or eukaryotes (with the exception of chloroplast and mitochondrial RRFs, they are similar to that of E.coli). Deletion of frr, the gene encoding RRF, is lethal to E. coli cells and, in the absence of RRF, ribosomes remain bound to the mRNA and initiate spontaneous translation downstream of the stop codon. The cellular importance and kingdom distribution of RRF make bacterial ribosome recycling an attractive target for drug design; however, such an undertaking requires an atomic understanding of the ribosomal binding site of RRF.
Despite having a shape remarkably similar to tRNA, recent studies using hydroxyl radical probing and cryo-EM have revealed that RRF adopts a significantly different orientation on the ribosome to that of a tRNA. In this orientation domain I of RRF interacts predominantly with the large subunit, rather than with the small subunit as predicted by tRNA mimicry, which is in agreement with the observation that domain I alone (RRF-DI) can bind to the 50S subunit with an affinity comparable to that of RRF. Though biochemical methods and cryo-EM provide a good model for the location of RRF on the ribosome, the precise mechanism of termination remained open due to the rather limited spatial resolution of these methods.
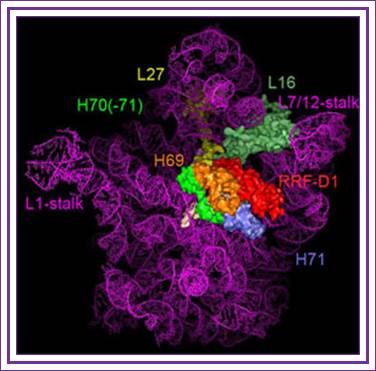
The binding site of RRF-D1 domain;
RRF-Ribosome binding model:
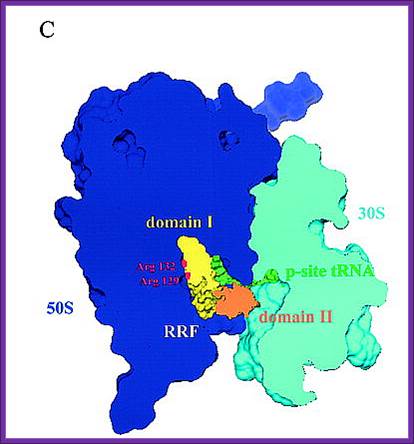
A new concept of mimicry between RRF and tRNA is shown. Domain I of RRF corresponds to the acceptor arm of tRNA. B, the models of RRF and ribosome complex was constructed according to Selmer's proposal. Diagram C, a new model for the A-site bound RRF. This arrangement is consistent with biological findings that domain I of RRF interacts not with the 30 S subunit but with the 50 S subunit.
Ribosome
recycling is the final stage of translation and involves the concerted action
of the ribosome recycling factor (RRF) and elongation factor G (EF-G) to
disassemble the post-termination complex for the next round of translation.
In collaboration with the group of Prof. Kobayashi
, authors have determined the X-ray crystal structure of domain I of RRF bound
to the D. radiodurans large 50S ribosomal subunit at 3.3 Å resolutions.
Their results provide a structural description of the interactions that RRF makes with components of the large subunit and rationalizes how the dual action of RRF and EF-G act to disassemble the post-termination complex during ribosome recycling.
Ribosome recycling factor (RRF) consisting of 20.3 Kd (185 amino acid residues) is a translational factor for protein biosynthesis. It disassembles the post-termination complex (PoTC) which is composed of (50+30) 70S ribosomal subunits, deacylated tRNA and mRNA in concert with elongation factor G (EF-G) in a GTP- dependent manner. X-ray and NMR analyses on RRFs from thermophilic bacteria showed that they display a tRNA-like L-shaped conformation with two domains. Since then, it has been accepted that domain I, consisting of a three-helix bundle, corresponds to the anticodon arm of tRNA, and domain II consisting of a β/α/β sandwich structure, corresponds to the acceptor arm. In this study, authors obtained a RRF from a mesophilic bacterium, Vibrio parahaemolyticus, by gene cloning and carried out an X-ray analysis on it at 2.2 Å resolutions. Analysis of the relative orientations between the two domains in the structures of various RRFs including this RRF revealed that domain II rotates about the long axis of the helix bundle of domain I. The peptide fragment (RRF-DI) corresponding to domain I of RRF was shown to bind to 70 S ribosome and the 50 S subunit with an affinity similar to that of wild-type RRF. But it does not bind to the 30S subunit. These finding made us to propose a new model where domain I corresponds to the acceptor arm of tRNA and domain II corresponds to the anticodon arm. This is just the reverse of a model that is now widely accepted. However, the new model is in better agreement with published biological findings.
Ribosome recycling is more or less same in cell organelle like mitochondria and chloroplast. Both are derived from prokaryotes. From the cDNA analysis and from the amino-terminal sequencing of the isolated proteins, the mature RRFHCP was deduced to have a Mr of 21,838 with 193a.a. Its biosynthesis is limited to photosynthetic tissues. Further, human mtRRF is able to associate with Escherichia coli ribosomes in vitro and can associate with mito-ribosomes in vivo. Mitochondrial and chloroplast RRF are coded by nuclear genes. Proteins translated are transported in to the organelles.
1. Ribosome Remodeling Factor (RMF): RMF is one of the factors thought to be responsible for the translational inhibition observed upon entry of bacterial cells into stationary growth phase. It is 50 a.a protein binds to PTC Peptidyl Transferase Center region and dimerize ribosomes. RMF binds to the 50S subunit of the 70S ribosome and has been shown to inactivate translation by inducing dimerization of the 70S ribosomes (100S formation). Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp at stationary or stress condition; Izutsu K, Wada A and Wada C.
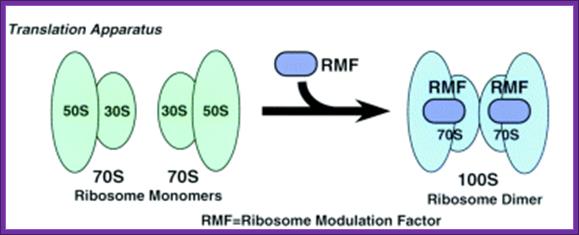
The expression of ribosome modulation factor (RMF) is induced during stationary phase in Escherichia coli. RMF participates in the dimerization of 70S ribosomes to form the 100S ribosome, which is the translationally inactive form of the ribosome. (Toshiko Aiso1etal).

Recently, Dr. Hideji Yoshida et al have determined the binding site of RMF on the D. radiodurans large ribosomal subunit at 3.5 Å revealing that RMF binds in close proximity to the PTF centre, explaining how RMF can inactivate translation on the 70S ribosome.
The role of ribosome modulation factor (RMF) in protecting heat-stressed Escherichia coli cells was identified by the observation that cultures of a mutant strain lacking functional RMF (HMY15) were highly heat sensitive in stationary phase compared to those of the parent strain (W3110). No difference in heat sensitivity was observed between these strains in exponential phase, during which RMF is not synthesized. Studies by differential scanning colorimetry demonstrated that the ribosomes of stationary-phase cultures of the mutant strain had lower thermal stability than those of the parent strain in stationary phase, or exponential-phase ribosomes. More rapid breakdown of ribosomes in the mutant strain during heating was confirmed by rRNA analysis and sucrose density gradient centrifugation. Analyses of ribosome composition showed that the 100S dimers dissociated more rapidly during heating than 70S particles. While ribosome dimerization is a consequence of the conformational changes caused by RMF binding, it may not therefore be essential for RMF-mediated ribosome stabilization.
The ribosome modulation factor (RMF) binding site on the 100S ribosome of Escherichia coli: Yoshida H, Maki Y, Kato H, Fujisawa H, Izutsu K, Wada C, Wada A.
During the stationary growth phase, Escherichia coli 70S ribosomes are converted to 100S ribosomes, and translational activity is lost. This conversion is caused by the binding of the ribosome modulation factor (RMF) to 70S ribosomes. In order to elucidate the mechanisms by which 100S ribosomes form and translational inactivation occurs, the shape of the 100S ribosome and the RMF ribosomal binding site were investigated by electron microscopy and protein-protein cross-linking, respectively. We the above authors show, that (1) the 100S ribosome is formed by the dimerization of two 70S ribosomes mediated by face-to-face contacts between their constituent 30S subunits, and (ii) RMF binds near the ribosomal proteins S13, L13, and L2. The positions of these proteins indicate that the RMF binding site is near the peptidyl transferase center or the P site (peptidyl-tRNA binding site). These observations are consistent with the translational inactivation of the ribosome by RMF binding. After the "Recycling" stage, ribosomes can readily proceed to the "Initiation" stage during exponential growth, but during stationary phase, the majorities of 70S ribosomes is stored as 100S ribosomes and are translationally inactive. We suggest that this conversion of 70S to 100S ribosomes represents a newly identified stage of the ribosomal cycle in stationary phase cells, and we have termed it the "Hibernation" stage.
Translation inhibitors:
Translation inhibitors are any and they act at different levels of translation such as initiation, elongation, proof reading peptide transfer and termination.
Tetracyclins: block at A site prevent a.atRNA binding
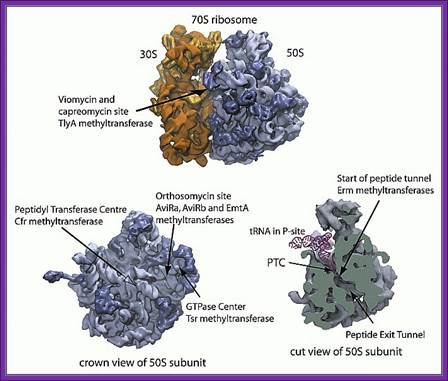
Translation inhibitor sites are depicted
Linezolid- acts at initiation stage.
Amioglycosides- interfere with proof reading.
Chloramphenicol- Blocks peptidyl transfer- acts on bacteria and Mitochondria.
Macrolides- bind to 50s and prevent ribosomal translocation.
Fusic acid- acts on elongation factor- prevents turnover.
Macrolides- cause premature termination by dissociating peptidyl tRNA from the ribosome.
Puromycin- mimics tRNA tyr, it binds to ribosome at A site and involves in peptide bond formation, but fails to elongate.
Streptogramins - Cause premature chain release.
Streptomycin- causes misreading and produces aberrant proteins.
Chain Termination of broken mRNAs or mRNAs without Ter codon:
In mRNAs, for some reasons, like deletions or additions TER codon can change into a coding sequence or coding sequence into TER codons or an mRNA with no terminator codons may continue to translate till the end and perhaps stall at some point.
Stalled translation due to lack of stop codon is often rescued by a chimeric RNA; it resembles tRNA and mRNA, what is called TmRNA (tRNA-mRNA like).
While ribosomal machinery is translating, if it finds an end of mRNA without a TER codon, it halts. In such situations the P site is loaded and the A-site is empty. In such cases ribosomes associate with tm-RNAs (ssRNAs), which are 363 or 457 ntds long. Such tmRNAs are coded by ssR gene in E.coli. This tm-RNA binds to stalled ribosome to a site where an mRNA normally is located.
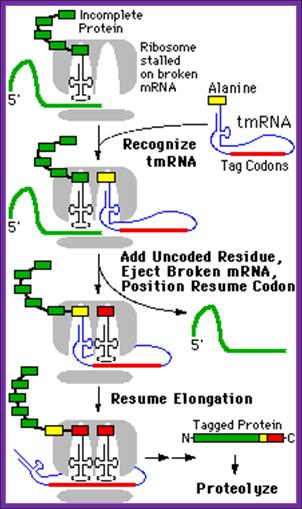
The above diagram illustrates the mechanism of translational termination of mRNA which does not contain TER codons
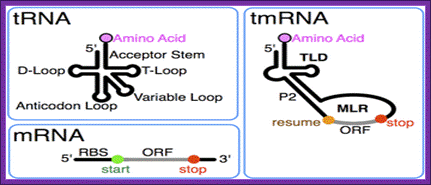
The tmRNA combines the features of mRNA and tRNA; http://en.wikipedia.org/

OR


Tm RNA model
This RNA has both tRNA structural domains at 3’ end and mRNA structural feature at the other end. The 3’ of tRNA like structure gets loaded with alanine, then it is placed on A-site in association with EF-Tu.GTP. This leads to peptide bond formation, thus the stalled protein is extended.
The tm RNA at the other end with specific 5’>3’codon sequences is threaded into small ribosome in line with A-site as if it is an mRNA. The truncated mRNA bound to small ribosomal subunit now is threaded out. The tmRNA is also associated with few proteins like SmB1 and SmB2. Once the mRNA like sequence is placed in line with A-site one more peptide bond is formed; thus the stalled ribosome on an mRNA moves on in 5’-3’ direction and translates approximately 10 codons; at the end of which a TER codon is located in the Tm RNA, at which translation stops and terminates. Thus it produces a fused protein with terminal a.a sequence of A N T N Y A L A A sequence. As this protein is nonfunctional, it is then presented to specific proteases and ultimately it is degraded.
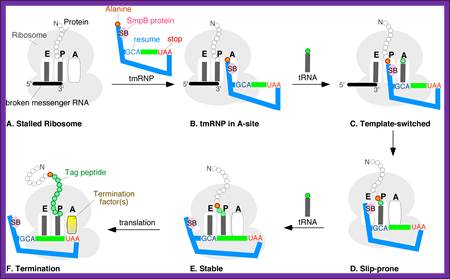
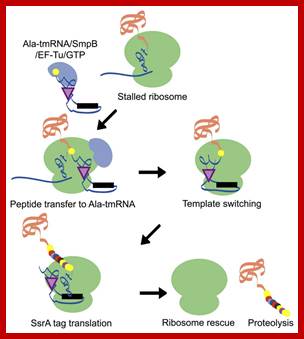
A schematic model of the trans-translation system. A complex comprised of Ala-tmRNA, SmpB, EF-Tu, and GTP enters the A site of the stalled ribosome and receives the nascent polypeptide from the peptidyl-tRNA in the ribosomal P site. The template is then switched to the ORF region of tmRNA and the SsrA tag is synthesized and attached to the C-terminus of the nascent polypeptide. Translation ceases at the termination codon in the ORF region of tmRNA and the ribosome is rescued. The SsrA-tagged nascent polypeptide is identified by the proteolysis system and degraded.; http://www.frontiersin.org
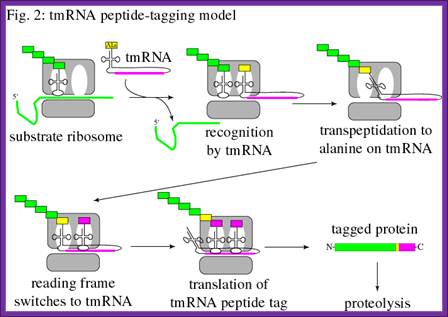
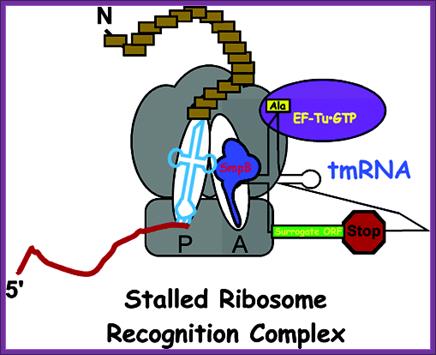
The mechanism of ssr action is shown in the fig.Another model for translation of Tm RNA When ribosome stalls, the ssrA RNA charged with alanine is brought and binds at A site of the ribosome; it is facilitated by SsrB. The peptidyl transferase brings about the bond formation between the existing a.a and alanine, thus the protein chain is elongated, till the end of ANDENYLAA. The last two a.a A.A marks the protein fror degradation. http://dwb.unl.edu/