Protein Synthesis – Regulation at Recoding Level:
A single transcript can generate:
- More than one kind of polypeptide can be generated by alternative splicing of the same mRNA in different tissues.
- More than one polypeptide chains can be generated by editing the same in different tissues.
- More than one protein can be generated by frame shift reading in different tissues.
- A single protein can be cut and joined to generate more than one protein.
- Frame shift reading signals in an ORF are encoded in mRNA sequences and structure, such structural elements are called recoding signals.
- In retroviruses such as Rous Sarcoma Virus, infection leads to the production of double stranded complementary DNA with Long terminal sequences, by means of which the ds DNA integrates into the host genome, in non-site specific manner.
- Though the insert is most of the times not active, it gets activated under certain stimulus. In such situations, using 3’ LTR region as promoter, a single large transcript can be generated.
- By splicing, it can generate two kinds of mRNA. One codes for ‘gag’ and ‘pol’. And the other codes for ‘env’ proteins.
- In the first case most of the time translation of the transcript generates gag protein only. Rarely, but infrequently it produces a polypeptide chain containing both gag and pol proteins. This is achieved by shifting the reading frame. At the junction of gag and pol cistrons; the mRNA has the following sequence.
Phe leu gly
--- UUU. AUU. AGG G----3’
-----UUUAU. UAG.G--------3’
· If this reading frame is read without any change, it generates gag and pol fused protein, but most of the times, the mRNA at this site produces a secondary structure in such a way, when the ribosome reaches phenyl alanine (UUU) codon and reads leu (AUU) and when it is on AGG, the ribosome at this codon, slides back by one nucleotide and read it as UAG, which is a terminator codon. Thus translation terminates and produces only gag protein.
· The above said process exist in many systems such as retroviruses HIV, and HTLV viruses.
· This kind of shift in the reading frame occurs by shifting the reading frame by one nucleotide backwards or it can shift one nucleotide forward and can generate different codon may be sensible or nonsense codons.
· Shifting the reading frame one nucleotide backwards or forwards leads to the new a.a sequence in the protein, so also its function.
· In some eukaryotes, the transcript of a particular gene, for example Apolipoprotein, produced in liver is a full-length protein, but the Apolipoprotein produced in intestine is a truncated one. This is by editing.
· In this case the mRNA for Apolipoprotein produced in liver is read unhindered, but the same transcript produced in intestine it is edited at a particular codon so as to produce a terminator codon, so the synthesis of the protein progresses up to that region, then it is terminated. So the original meaning of the message is recoded to generate a truncated protein.
· In phi-X174, the single stranded DNA genome is 5386 ntds long. But it has endowed with sequences that can generate 10 proteins, not as polycistronic, but the mRNA produced is recoded at different sites to generate two proteins from the same ORF or some time it can generate more than two proteins from the same reading frame.
· Here the reading frame is organized in such a way; the end of one reading frame overlaps the start of the next reading frame. The sequences are organized in a manner that the terminator codon of the first reading frame overlaps the first nucleotide of initiator codon of the second reading frame.
· In this viral genome, the sequences are organized in such a way that genes F, G. H and J are arranged in non-overlapping fashion. But the genes A and A* are organized so as to generate A as a full-length product and A* as product of frame shift translation.
Glu Ser Lys Gly Gly Lys TER
5’XAT GAG TCG AAA ATT---II GCT GGT GGA AAA TGA GAA AAT TCA A
A TG AGA AAA TTC AAT
Met Arg Lys
Met Ser Arg Lys
ATG AGT CGA AAA TT.. IIGCT GGC GGA AAA TGA GAAA AATT CAA
· In this kind of overlapping organization of genes, it generates different polypeptide chains only at translation level.
· The sequences are organized so as ribosome while translating reads the ORF according to the dictates of the structure of mRNA and the sequence context. For example in ‘A*’ is located within the ‘A’ gene. The B gene is located within the ‘A’ gene but at the fag end of the ‘A’ gene.
· The protein ‘B’ is generated by shifting the reading frame by one nucleotide backwards (-1) in ‘A’ reading frame.
· The gene ‘K’ is located at the end of the ‘A’ and so it overlaps both ‘A’ and ‘C’ gene. The ‘K’ is generated by shifting the reading frame within A by shifting forward by one nucleotide.
· The ‘C’ gene itself over laps the ‘A’ gene at its end and it initiates translation by shifting the reading frame backwards by one nucleotide to generate an initiator codon.
Recoding:
Three examples of recoding events: (A) Antizyme frame shifting. The +1 shift at the last codon (UCC) before the termination codon of ORF1 of human antizyme 1 is stimulated by polyamines and by a 5′ mRNA element and a 3′ pseudo knot. (B) Gene 60 bypassing. Fifty nucleotides between codons 47 and 48 of phage T4 gene 60 coding sequence are bypassed by half the ribosomes in response to matched takeoff and landing site codons, a stop codon directly after the take-off site in a stem–loop structure and a nascent peptide signal that acts within the ribosome. (C) Prokaryotic selenocysteine insertion—redefinition. UGA codons in prokaryotes that specify selenocysteine are directly followed by a stem structure whose apical loop is bound by a selenocysteine tRNA specific elongation factor SELB, resulting in a tethered amino acylated tRNA poised for the oncoming ribosome. These figures are adapted from; Atkins et al.
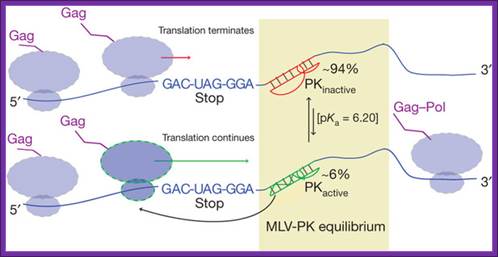
Gene expression via translational recoding is regulated by a
dynamic equilibrium (PKactive ![]() PKinactive) between an active, read-through
permissive conformation and an inactive, non-permissive conformation the
distribution of which determines the Gag:Gag–Pol ratio. Ribosomes that
encounter the PKactiveconformation continue translating through the
stop codon. Structural and functional data indicate that PKactive interacts with the ribosome before the
stop codon is in the decoding centre owing to the shortened nature of the
linker. Brian Houck-Loomis, et al; http://www.nature.com/
PKinactive) between an active, read-through
permissive conformation and an inactive, non-permissive conformation the
distribution of which determines the Gag:Gag–Pol ratio. Ribosomes that
encounter the PKactiveconformation continue translating through the
stop codon. Structural and functional data indicate that PKactive interacts with the ribosome before the
stop codon is in the decoding centre owing to the shortened nature of the
linker. Brian Houck-Loomis, et al; http://www.nature.com/
Frame shifting by (+)1: At defined shift sites in mRNA, ribosomes can be programmed to efficiently change to one of the two alternative reading frames for gene expression purposes. Commonly, there are stimulatory signals in mRNA distinct from the shift site that greatly increases the level of frame shifting at the shift site. As a result, ribosomes initiating at the same start codon produce two different protein products, one being the product of standard decoding and the other the product of a recoding event. Depending on the configuration of the open reading frames (ORFs) in an mRNA relative to the site of frame shifting, two outcomes are possible. If the stop codon of the new ORF (ORF2) is 3' of the termination codon of the original ORF (ORF1), the frame shifting product is longer (sometimes much longer) than the product resulting from standard decoding. Alternatively, if the stop codon of ORF2 is 5' of the end of ORF1, the frame shift protein product is shorter. Many more examples are known where the frame shift product is longer but that may be because of inherent biases associated with the discovery of frame shifting examples.
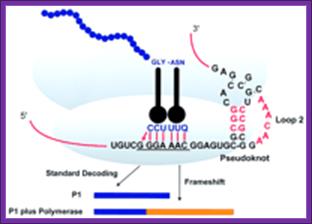
Frame shifting by (-)1: From: Recoding: Site- or mRNA-Specific Alteration of Genetic Readout Utilized for Gene Expression; S. L. Alam, J. F. Atkins, and R. F. Gesteland;http://www.pnas.org/
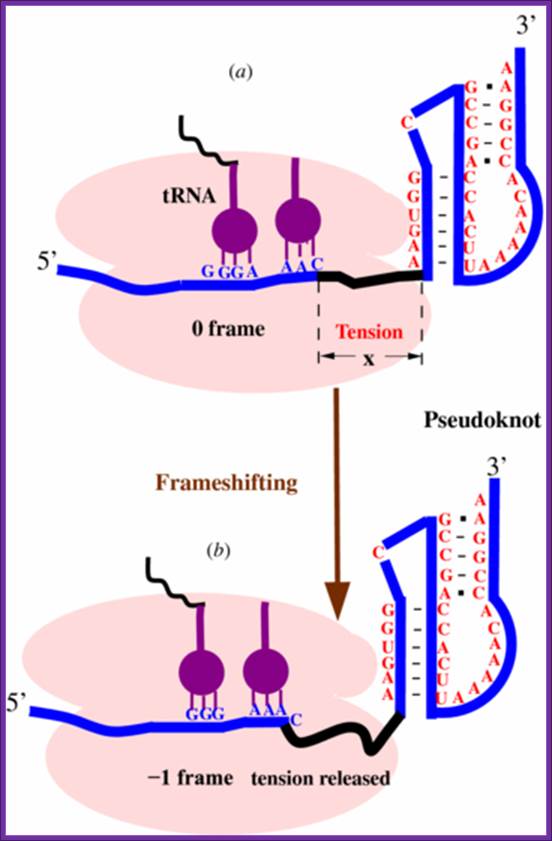
Predicting ribosomal frameshifting efficiency. Song Cao and Shi-Jie Chen http://iopscience.iop.org/.
Ribosomal Hopping or Bypassing:
Ribosomal hopping can be seen as an extreme case of translational frameshifting. Therefore, its mechanism can be viewed as a variation of P-site slippage resulting in +1 frameshifting. Most of our knowledge about ribosomal hopping comes from work on bacteriophage T4 gene 60, which encodes a subunit of phage topoisomerase. At the end of gene 60 ORF1, up to half of all translating ribosomes bypass 50 nucleotides and then resume normal translation on a second downstream ORF2. Three stages define this remarkable event. During the first stage (take-off ), the tRNA dissociates from the P-site GGA codon.
In the next stage (scanning), the ribosome traverses the coding gap checking for a matching P-site codon. In the last stage (landing), the P-site tRNA-mRNA pairing is re-established and regular translocation resumes. In translational bypassing (hopping), a fraction of the translating ribosomes “skip” a portion of the mRNA without inserting any amino acid. Regular decoding is resumed some distance downstream. Gag-pol- In most viruses and retro transposable elements, the recoding event serves to link structural (e.g., retroviral Gag) and catalytic polypeptides (e.g., retroviral Pol or Pro-Pol) (reviewed in refs. 2 and 3). Standard decoding of retroviral genomic RNA typically produces only Gag protein whose ORF is located 5' on the mRNA but through recoding, Gag-Pol, or Gag-Pro-Pol, is produced as a single polypeptide.
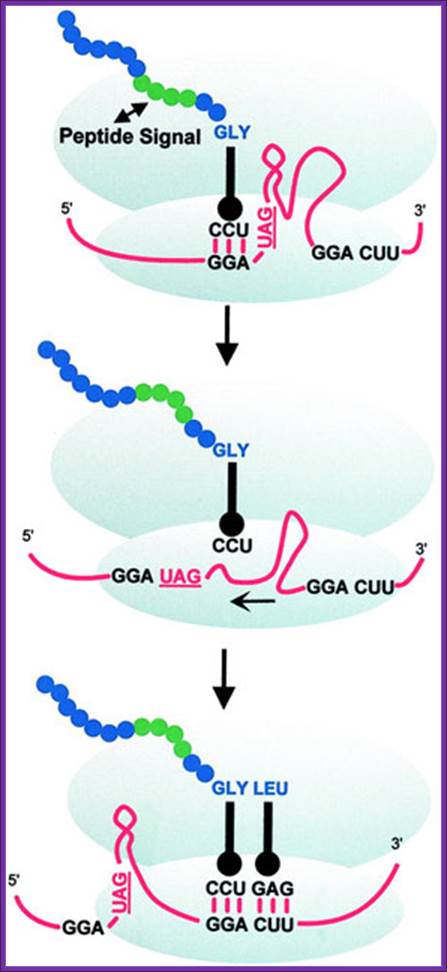
During translation ribosomes jump nearly 50-60 nts of mRNAs; Alan J. Herr, et al; http://emboj.embopress.org/
Redefinition: Selenocysteine incorporation (Sec):
Selenium is an essential trace element that is incorporated into proteins as selenocysteine (Sec), the twenty-first amino acid. Sec is encoded by a UGA codon in the selenoprotein mRNA. The decoding of UGA as Sec requires the reprogramming of translation because UGA is normally read as a stop codon. The translation of selenoprotein mRNAs requires cis-acting sequences in the mRNA and novel trans-acting factors dedicated to Sec incorporation.
Selenoprotein synthesis in vivo is highly selenium-dependent, and there is a hierarchy of selenoprotein expression in mammals when selenium is limiting. This review describes emerging themes from studies on the mechanism, kinetics, and efficiency of Sec insertion in prokaryotes. Recent developments that provide mechanistic insight into how the eukaryotic ribosome distinguishes between UGA/Sec and UGA/stop codons are discussed. The efficiency and regulation of mammalian selenoprotein synthesis are considered in the context of current models for Sec insertion.
Insertion of Selenocysteine; tRNASec-dependent amino acid transformations leading to Sec in eukaryotes as elucidated by using recombinantly produced mammalian components ; http://www.pnas.org/
In redefinition, codon meaning is changed in an mRNA specific manner (as opposed to reassignments of the “universal” genetic code that are species or organelle-specific). Specification of an amino acid by a “stop” codon results in the production of protein product, which is longer than the product of standard decoding. Most known cases of redefinition fall into this category, perhaps only due to the fact that the products of standard decoding and recoding, in this case, have usually very different sizes and therefore can easily be detected and distinguished by standard laboratory techniques. Encoding of selenocysteine by special UGA codons allows incorporation of this 21st amino acid thus extending the capabilities of the genetic code. Encoding an alternative amino acid by a sense codon could, in theory, result in the production of two proteins with the same number of amino acids but differing biochemical activities. Perhaps because of the technical difficulties of identifying such cases, no examples of this kind have been discovered.


