Other DNA polymerases:
DNA-Pol of B. subtilis:
- B.subtilis is a sporulating bacterium.
- They induce spore formation when growth conditions are not optimal.
- Formation of endospores is a fascinating phenomenon, and regulated by a sequential (temporal) mode of gene expression.
- The bacteria produce three DNA polymerases, Pol-I, pol-II and pol-III, which are more or less similar to E.coli enzymes.
- They are coded for by genes pol-A, pol-B and pol-C respectively.
- Pol II and I show remarkable similarities to E.coli, in their a.a sequence, structure and function.
- But the pol-C product i.e. alpha subunit (similar to that of E.coli Pol-IIIs alpha subunit) has remarkable characters.
- It encompasses 5->3 polymerase activity (like alpha subunit).
- It has 35 exonuclease activity of theta subunit of E.coli enzyme.
- In B.subtilis, unlike E.coli both functional activities are combined in the same polypeptide chain, which is a unique combination of gene segments, though such polypeptide chains are found separately in E.coli DNA Pol-I.
- Its enzyme activity is inhibited by aryl- Azopyrimidines, while this drug does not inhibit E.coli enzyme.
- The alpha subunit of B.subtilis is 163 KD and the 35 exonuclease activity is located at N-terminal region of the protein and its polymerase activity is at the other end of the polypeptide chain.
DNA Pol from Thermophilic Bacteria:
Sulfobolus acidocaldarius:
DNA POL -Solfobolus. Acidocaldarius; http://www.rcsb.org
- It is an archive bacteria, grows in sulphur hot springs (85^o C and pH 2.0).
- The DNA pol is made up of a single polypeptide chain of 100 KD mass.
- It shows optimal activity at 70^C.
- It is also stable at 80^o C for more than 35 to 45 minutes.
- This enzyme is capable of synthesizing a new strand at 100^o C up to 100 ntds.
- However , most of the thermophilic DNA Pols lack 3-->5' proof reading activities under in vitro conditions. This cannot be true under in vivo conditions for the bacterial systems grow and multiply under such severe conditions with out generating abnormalities. Perhaps they may have some accessory factors that may elude isolation during purification process.
Thermus aquaticus DNA- Polymerase:
- It is a eubacteria.
- The enzyme is 94 KD.
- It is stable at 95^oC for 45 minutes.
- It has 5ΰ3 polymerization activity with a processivity of 100 or little more ntds, which is also the rate of synthesis.
- Under in vitro conditions, it lacks 3>5 exonuclease activity and 5>3 exonuclease activity.
- It is a surprising feature.
- Its N-terminal part and the Catalytic cleft have strong similarities with that of E.coli- Pol. I.
DNA polymerase;Thermophilus aquaticus; www.hindawi.com
Halophytic DNA-Polymerases:
- Halobacterium halobium has several polymerase enzymes, two of which resemble eukaryotic polymerases in having multi subunits, with one of the subunit resembling alpha subunit.
- The alpha like subunit is sensitive to Amphidicolin and N-ethylmaleimide.
- The enzymes have 3->5 exonuclease activity.
- Its another, small Beta like subunit, that has polymerase activity and resistant to the above drugs.
- Surprisingly the enzyme has both 3->5 and 5->3 exonuclease activities.
T4-DNA polymerase:
- After infection, the phage DNA replicates ten times faster than the host DNA. The phage gene product called gp 43 methylates its own Cytidine ntds, in sequence specific manner, within the host, thus enabling the faster rate of replication of the phage DNA. How?
- The T4 phage enzymes are similar to T2 and T6 phages. The enzyme binding to 5 end of its mRNA autogenously regulates the gene 43.
- The enzyme is made up of a single polypeptide chain similar to E.coli DNA pol -I.
- It requires primed ssDNA as the template, but it cannot use nicked DNA.
- Turn over of the enzyme activity is 400 ntds per sec. Actually it functions as a Holozyme with some accessory proteins. The core complex, with gp44, gp62, and gp45, acts as holozyme.
- With gp 41, which acts like helicase, it efficiently replicates nicked DNA also. The gp 61 is a primase. With accessory proteins, like gp 43, 62, 45, and 32, the rate of replication is 40 fold faster to that of gp 43 alone.
- This is because one of the accessory proteins, provide clamp structure and can progress through the strongest possible hairpin structure. The processivity is good, and it can progress on the template up to 20000 or more with out dissociating from the template.
- Interestingly it has 3ΰ5 exonuclease activity and it can generate 5 ss overhangs from blunt ends. Though the enzyme lacks 5ΰ3 exonuclease activity, its N- terminal region is similar to that of E.coli pol Rnase-H.
T7 DNA polymerase:
· The polymerase is product of phages gp5 gene.
· The protein is a single polypeptide chain of Mol.wt 79.7 KD.
· In host cells, it is complexed with hosts accessory subunit of 11.7 KD called thioredoxins; they are associated in 1:1 ratio.
· Many phages like F2, QB and ms2 and other RNA phages are also associated with host components.
· Structurally the T7 DNA polymerase has similarities with Large Fragment (Klenow) of E.coli DNA pol-I and alpha subunit of DNA pol-III.
· The T 7 polymerase with theoredoxin exhibits high processivity and high fidelity.
· The gp5 gene product perse shows 3ΰ5 exonuclease activity on ssDNA as well as on dsDNA.
· With thioredoxins it exhibits full polymerase activity.
· This enzyme lacks 5 exonuclease activity.
· The gp6 gene is located downstream of the gp5.
· T he gp6 protein shows 5->3 exonuclease activity.
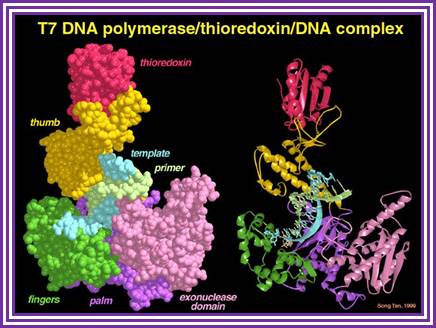
T7DNA pol/Trx/DNA; Doublie, Tabor, Long, Richardson & Ellenberger, Nature 391:251-258, 1998www.personal.psu.edu
· Structurally it shows strong homology with the N-terminal segment of DNA pol-I.
· The combined mass of gp 5 and gp6 is almost same as that of DNA pol I.
· The combination of gp5, gp6 and thioredoxin has 5-3 polymerase, 53 exonuclease and 35 exonuclease activity.
· It is a unique combination of features, which suggest, how recombination of independent gene segments, during evolution, can give rise to genes whose products have multiple activities.
· The reverse is also true. It means that a single gene with different structural and functional coding domains can be recombined in different ways to be distributed to different genes or they work as independent genes. These are some of the steps or modes by which natural selection and evolution operates at molecular level.
· Another gene product, gp 4 has very unique functions; it has helicase as well as primase activities.
· This protein produces just a tetra nucleotide RNA primer.
· The T7 DNA polymerase, without EDTA gets modified during purification process.
· This leads to loss of its 3ΰ5 exonuclease activity. This loss is due to the selective oxidation and modification of an amino acid residue in the vicinity of iron binding site in the protein.
· Iron ion is very essential for 3->5 exonuclease activity.
· Commercially available enzymes lack 35 exonuclease activity, and they dont discriminate dNTPs from normal ddNTPS (di Deoxy NTPs). The enzyme even tolerates d ITPs in the place of dGTPs.
· Substituting Mg2+ with Mn 2+ further enhances the lack of discrimination between dNTPs from that of dideoxy NTPs.
· Because of the above features T7 DNA polymerase is extensively used in DNA sequencing experiments.
· It is also used in a variety of genetic engineering techniques.
· Presently this enzyme is available as genetically modified product and sold by by many commercial Biotechnology firms as Sequenase.
Pol IV
· In E. coli, DNA polymerase IV (Pol 4) is an error-prone DNA polymerase involved in non-targeted mutagenesis. Pol IV is a Family Y polymerase expressed by the dinB gene that is switched on via SOS induction caused by stalled polymerases at the replication fork. During SOS induction, Pol IV production is increased 10-fold and one of the functions during this time is to interfere with Pol III holoenzyme processivity. This creates a checkpoint, stops replication, and allows time to repair DNA lesions via the appropriate repair pathway. Another function of Pol IV is to perform tanslesion synthesis at the stalled replication fork like, for example, bypassing N2-deoxyguanine adducts at a faster rate than traversing undamaged DNA. Cells lacking dinB gene have a higher rate of mutagenesis caused by DNA damaging agents. http://en.wikipedia.org/wiki/DNA_polymerase
Pol V
DNA polymerase V (Pol V) is a Y-family DNA polymerase that is involved in SOS response and translesion synthesis DNA repair mechanisms. Transcription of Pol V via the umuDC genes is highly regulated to only produce Pol V when damaged DNA is present in the cell generating an SOS response. Stalled polymerases cause RecA to bind to the ssDNA which causes the LexA protein to auto digest. LexA then loses is ability to repress the transcription of the umuDC operon. The same RecA-ssDNA nucleoprotein posttranslationally modifies the UmuD protein into UmuD' protein. UmuD and UmuD' form a heterodimer that interacts with UmuC which in turn activates umuC's polymerase catalytic activity on damaged DNA. http://en.wikipedia.org/wiki/DNA_polymerase
Motifs and homologies among polymerases:
Amino acid sequences of certain segments of polymerases show significant homologies among several DNA-Pols, RNA dependent DNA- polymerases, DNA and RNA dependent RNA-polymerases. The active sites, where the template bind and nucleotides bind have more or less conserved in both DNA and RNA polymerases, because they have same mode of action and perform similar functions.