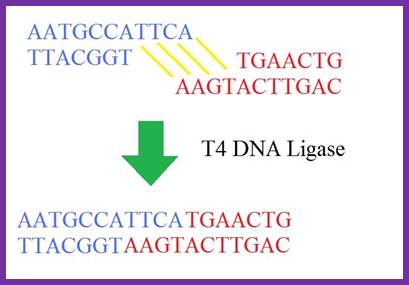
DNA-Ligases:
Ligase is like a molecular stitcher. They are ubiquitous and essential for all cells and perhaps at all times. DNA ligase (polydeoxyribonucleotide synthase) is the enzyme that joins two single stranded DNA fragments by catalyzing the formation of an inter-nucleotide ester bond between phosphate and Deoxyribose. It is active during DNA replication, DNA repair and DNA recombination. There are two forms of DNA ligase: one requires ATP and the other NAD. NAD (+) dependent DNA ligases are present in bacteria, some Entomopox viruses and Mimi virus while ATP-dependent DNA ligases are ubiquitous. Double stranded DNA breaks are common in human cells/fibroblast cells, according to Michael R. Lieber; NCBI.NLH.NIH.GOV, ten breaks per day in fibroblasts. Estimates indicate there are at least ten causes to such breaks; it is astonishing how cells remain functional. Causes- replication across a nick, Ros, Ionizing radiation, malfunctioning of nuclear enzymes and often mechanical stress on DNA
If there is break in one of the double stranded DNA, ligases ligate the broken ends by phosphodiester bond formation between the broken ends. They use the free 5’p of a nucleotide of one segment and the free 3’OH group of the neighboring segment, after recognizing and binding to them they catalyze the reaction to form a covalent bond, thus they seal the nick. For the enzyme to function it requires ds DNA with such gaps or nicks in one of the strands. The nick or the gap should not contain any missing nucleotide in between the ends. A minimal length of the ds strand DNA needed for its function is at least 4 to 8 base pair length and the broken bonds should be side by side. If more than one nucleotide missing they won’t be able to do it. Many DNA Ligases are identified from different sources, which have their own specific characters, molecular weights, requirements and mode of functions.
Prokaryotic Ligases, a Comparison:
|
|
E.coli DNA Ligase |
T4 DNA Ligase |
T7 DNA Ligase |
|
Mol.wt (kd) |
75 |
60 |
41 |
|
Sub units |
Monomer |
Monomer |
Monomer |
|
Cofactors required |
NAD |
ATP |
ATP |
|
Catalyze covalent bond formation between 3’OH and 5’p of nucleotides |
Yes |
Yes |
Yes |
|
Perform sticky end ligation |
Yes, efficient |
Yes efficient |
Yes, efficient |
|
Perform blunt end ligation |
No |
Yes, not efficient |
Yes, |
|
Blunt or sticky ends, require 5’phosphate group and 3’OH group |
Absolutely required |
Absolutely required |
Absolutely required |
|
Can they use 2’OH groups |
No |
No |
No |
|
Can they join ssDNA end to end |
No |
No |
No |
|
Can they ligate RNA strands on a DNA strand |
No |
Yes |
? |
|
Can they ligate DNA strands on a RNA strand |
No |
Yes |
? |
Properties-functions:
- Most of them, whatever may be their sources, are monomers.
- Conformationally they show little elongated shape.
- The ligase in E.coli is NAD dependent for activation.
- Others are ATP dependent ligases.
- Concentration of enzyme is 300 or more per cell.
- During activation, NADH or ATP covalently binds to the enzyme via an amino group lysine in the enzyme, to produce enzyme-NAD/AMP or simply Enzyme-AMP. This is an activated state of enzyme.
- In this state, the enzyme binds to the substrate and traverses the DNA till it encounters the gap or a nick.
- The nick should be between two adjacent nucleotides and the 5’ end should be phosphorylated.
- If the gap is more than one nucleotide and if the base pairing is not correct, Ligases won’t act.
- The bound enzyme transfers the nucleotide phosphate or AMP to the 5’end phosphate group of the nucleotide in the nicked region; the process is called adenylation.
- The enzymes’ covalent bonding to the phosphate group at 5’end activates 3’OH (^-) group, by nucleophilic attack of negatively charged O^-H group on the alpha positioned Phosphate group; it generates a covalent bond, between the two. In this reaction enzyme is silent, yet it is bound to the substrate.
- With the reaction complete, the enzyme dissociates from the substrate.
- Though there are many different Ligases, their basic mechanism of ligation is same.
- T4 DNA ligase is very efficient in ligating sticky ends like NAD dependent E.coli ligase (671 amino acids) encoded by ligA gene.
- While E.coli ligase does not ligate blunt ends, T4 ligase ligates sticky ends efficiently but they also ligate blunt ends but with lower efficiency.
- Adding RNA Ligases to T4 DNA ligase mix in 2:1 or 4:1 ratio can increase its efficiency. RNA Ligases add phosphate groups to the blunt 5’ ends of the DNA, thus enhance ligase activity.
- Efficiency of sticky end ligation and blunt end ligation can be further increases by adding PEG 8000 to the ligating mix to 15%. This result in crowding of reacting molecules to a small volume, thus increase the frequency of enzymes and substrates to collide with each other with higher frequency, which process is called molecular crowding. Increased collision increases the rate of reaction.
- T4 DNA ligase can ligate two RNA fragments on a DNA template or they can ligate two adjacent DNA fragments on an RNA template.
T4 DNA Ligase; http://www.dianliwenmi.com
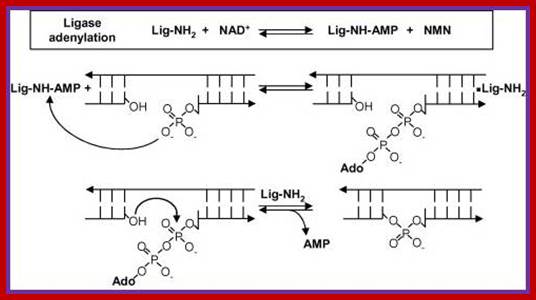
https://chem.libretexts.org/
1. Alignment of conserved sequence elements among Bacterial DNA ligases. Six collinear sequence elements have been identified in DNA ligases and mRNA capping enzymes (motifs I, III, IIIa, IV, V and VI) (Shuman and Schwer, 1995; Aravind and Koonin, 1999; Sriskanda et al., 1999; Doherty and Suh, 2000). For the Bacterial ATP-dependent enzymes, homologous regions were identified by clustal W alignment (http://www.ncbi.nlm.nih.gov/COG/aln/COG1793.aln). Note that the alignment for motif VI is poor in the Bacterial ATP-dependent sequences, and further detailed analysis is required to understand the implications of this. As homology among all NAD+-dependent DNA ligases is high, appropriate sequences are shown for representative examples. Sequences 5–7 are the least homologous of the NAD+-dependent DNA ligases. Sequences 1–3 have been purified and studied biochemically. The consensus sequence derived from all NAD+-dependent DNA ligases is given. All ATP-dependent DNA ligases from currently completed Bacterial genome sequences are shown (sequences 8–19). Of the Bacterial ATP-dependent DNA ligases, only sequence 8 has been purified and confirmed to have DNA end-joining activity. The sequence of T7 DNA ligase is provided for comparison. The number of amino acids not shown is given (-n-). The active-site motif of DNA ligases that contain the adenylated lysine (KXDG) is underlined. Coloured residues are identical in all Bacterial DNA ligases as follows: red, all isozymes; green, NAD+-Adam Wilkinson,Jonathan Day, and Richard Bowater.
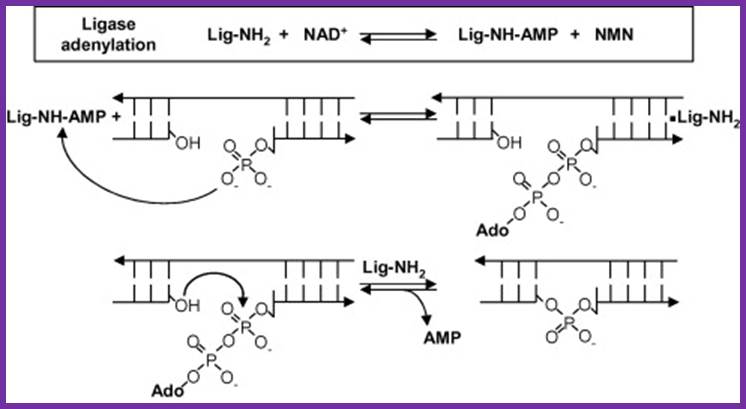
Reaction mechanism of bacterial DNA ligase; The first step is the reversible adenylation of ligase with NAD+ as the adenylyl donor. Ligase then transfers the AMP to the 5′ phosphate end of nicked duplex DNA and finally catalyzes nick closure and the release of AMP. (Figure modified from Ref. [15] with permission).www.lookfordiagnosis.com
This enzyme seals the nick and joints the Okazaki fragments in the lagging strand. It is also involved in the repair of damaged DNA. DNA ligases are extensively used as a joining tool in genetic engineering. http://biogoggles.wordpress.com/
Reaction- Enzyme + ATP-> Enzyme-Amp + PPi,
Enzyme binds to broken region and adds Phosphate group to 5’end P,
Then brings ligation between 3’OH and Phosphate to release AMP
ATP dependent isozymes; blue, ATP-dependent isozymes; (Residues are considered to be identical if they are present in > 90% of the DNA ligases. For the consensus sequence, capital letters denote the sequence of residues that are identical, and small letters denote conserved residues according to the classification of Aravind and Koonin (1999): h, hydrophobic (A, C, F, I, L, M, V, W, Y); s, small (A, C, S, T, D, N, V, G, P); p, polar (D, E, H, K, N, Q, R, S, T).
DNA Ligases: (http://www.tocris.com/pharmacologicalBrowser.php?)
DNA ligases play an integral role in DNA repair and replication through catalyzing the formation of phosphodiester bonds. Two types of DNA ligase have been identified: ATP-dependent DNA ligases (EC 6.5.1.1), and NAD+-dependent DNA ligases (EC 6.5.1.2).
The ATP-dependent DNA ligases are divided into four classes: DNA ligase I, II, III and IV. DNA ligase I links Okazaki fragments to form a continuous strand of DNA; DNA ligase II is an alternatively spliced form of DNA ligase III, found only in non-dividing cells; DNA ligase III is involved in base excision repair; and DNA ligase IV is involved in the repair of DNA double-strand breaks by non-homologous end joining (NHEJ).

DNA ligase I- ligation of Okazaki fragments, Lig2 ligation of alternatively splicing of RNA, Lig3 DNA repair protein XRRCC1, and Lig4 complexes with XRCC4 repair nonhomologous end joining of dsDNA breaks. Joining of Okazaki fragments http://faculty.samford.edu; https://www.youtube.com
A List of Eukaryotic Ligases:
|
Properties |
Ligase-1 |
Ligase-2 |
|
Molecular mass (kd) |
85-125 |
68 |
|
ATPs requirement |
Yes, require 2ATPs |
Yes, require 4 ATPs |
|
Blunt end ligation |
Yes |
No |
|
Sticky end ligation |
Yes |
Yes |
|
Location |
Nucleus |
Nucleus |
|
Fractional activity |
90% |
<10% |
|
Overall function |
Replication |
Repair |
Eukaryotic Ligases (Mammalian):
|
Name |
Mol.wt |
Subunits |
Cofactor |
Ligase-I |
85-125 |
Monomer |
ATP |
|
Ligase-II |
68 |
Monomer |
ATP |
Properties:
- Structure, mode of function and mechanism of catalysis of Eukaryotic ligases is same as that of prokaryotic Ligases.
- Two types of enzymes are present; both are located in the nucleus.
- But type I is mostly involved in replication and found in large numbers in proliferating cells or during cell division.
- On the contrary, DNA-ligase type II was found to be very active during DNA repair or recombination events.
- The enzymes are heat labile.
- Both of them are serologically unrelated, it means their gene and their amino acid sequences are different.
RNA Ligase:
RNA Ligases are also located in nuclei and their activity is high when transcriptional activity is at its peak. In bacterial hosts, T4 Phage produces an RNA ligase 43 KD (gp 63). RNA Ligases extracted from wheat germ and yeast cells do exhibit different activities. They join 3’ OH of one piece of RNA to 5’P of another piece of RNA in 5’ to 3’ end-to-end manner. Enzymes require first two to three nucleotides of the RNA fragments to recognize the ends and bind to perform catalytic activity. Most of the RNA ligases found in eukaryotic systems are involved in RNA splicing. They can also phosphorylate 5’ends and can cleave cAMP leaving Phosphate group at 2’end.
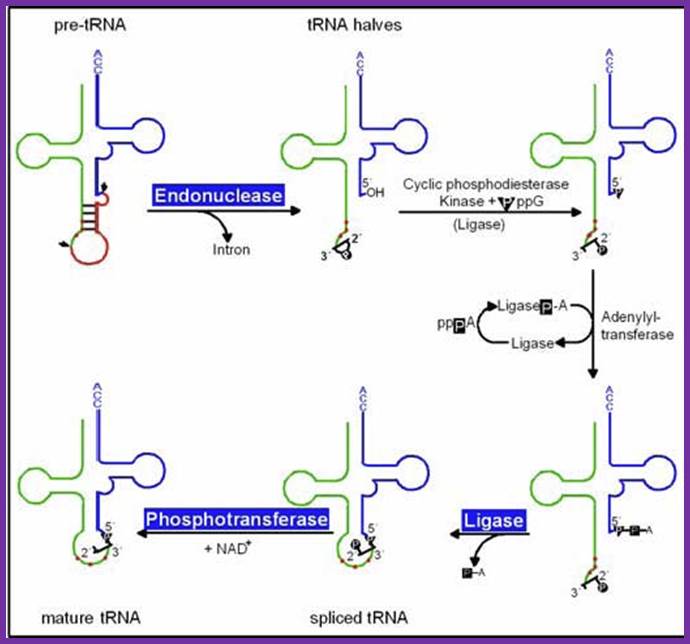
pre-tRNAs introns are spliced by two enzymes: First, an endonuclease cleaves the phosphodiester bonds at the 3´ and 5´ splice sites, producing paired tRNA halves with 2´,3´-cyclic phosphate and 5´ OH ends as well as a linear intron. Secondly, the halves are joined by an ATP/GTP-dependent RNA ligase to generate mature tRNA, Hildburg Beier
Structural Biochemistry/T4 DNA Ligase
From Wiki books, the open-content textbooks collection-< Structural Biochemistry
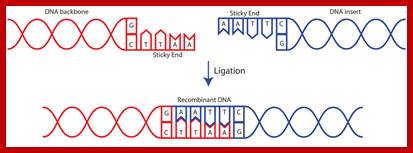
Process of ligation of two strands of DNA. Usually, scientists select two different enzymes for adding an insert into a vector (one enzyme on the 5' end and a different enzyme on the 3' end). This ensures that the insert will be added in the correct orientation and prevents the vector from ligating to itself during the ligation process. If the sticky ends on either side of the vector are compatible with each other, the vector is much more likely to ligate to itself rather than to the desired insert. If you are in this situation, it is important to treat the digested vector backbone with a phosphatase before performing the ligation reaction (phosphatase removes the 5' phosphate and therefore prevents the ligase from being able to fuse the two ends of the vector together) https://www.addgene.org
DNA ligase is a special type of ligase that can bring about the bond between the two DNA strands that have a broken ends in both complementary strands. DNA ligase has applications in both recombination and DNA repair. The enzymes are used extensively in molecular biology laboratories for genetic recombination experiments. DNA ligases are very useful tools in generating recombinant DNA. For example, DNA fragments are cut with restriction enzymes and then recombined with DNA ligase.
T4 DNA ligase: It is an enzyme that
is encoded by the T4. In a reaction where the DNA molecules are being joined
together at the 3'-hydroxy and 5'-phosophate termini, the ligase is used as a
catalyst. Given that there are no missing nucleotides in the repair reaction,
the ligase can also catalyze the covalent joining of two segments to one uninterrupted
strand in a DNA duplex. In order to accomplish this catalytic activity, ATP and
Mg2+ is required. DNA that lacks the required phosphate residues can
still be made to ligate through phosphorylation with T4 polynucleotide kinase.
Additionally, an exchange reaction of phosphate between pyrophosphate and ATP
can also be catalyzed through use of the ligase.
Characteristics of T4 DNA Ligase from E. coli lysogenic NM989:
The T4 DNA ligase is a single polypeptide with a molecular weight of 68,000 daltons. In order to obtain the maximum activity, a pH of 7.5-8.0 is desired. At pH levels of 6.9 and 8.3, the enzyme exhibits 40% and 65% of its full capabilities, respectively. As previously mentioned, Mg2+ is necessary for T4 DNA ligase to be effective, and the optimal concentration of Mg2+ is 10mM. Sulfhydryl reagents (DTT, 2-mercaptoethanol) are also essential in order to utilize the enzyme. If there is NaCl present, concentrations over 200 mM will stop all enzymatic reactions from occurring. For intermolecular ligation, especially when the substrate DNA consists of large DNA molecules PEG (concentrations of 1-10%) appears to stimulate the enzymatic activity.
Application:
T4 DNA ligase is mostly used in the joining of DNA molecules with compatible cohesive termini or blunt-ended, double-stranded DNA to one another, or to synthetic linkers. The reaction that involves blunt-ended DNA is slower than the other reactions, but the rate of ligation can be accelerated by adding 150 to 200 mM of NaCl along with a low concentration of PEG. If the 5' phosphate is absent in the DNA, and then phosphorylation is necessary before ligation can be performed. Phosphorylation is achieved through utilization T4 polynucleotide kinase with ATP. If the DNA fragments being joined together have protruding 5' termini that are not compatible with one another, it is still able to join the two fragments together by partial filling of the recessed 3' termini in controlled reactions using the Klenow fragment of E. Coli DNA polymerase I.
Ligase will also work with blunt end although higher enzyme concentrations and different reaction conditions are required.
Mammalian ligases;
Some forms of DNA ligases present in bacteria (usually larger) may require NAD
to act as a co-factor whereas other forms of DNA ligases (usually present in
E.Coli, and usually smaller) may require ATP to react. Also, a number of other
structures present in the DNA ligase are the AMP and lysine which are both
important in the ligation process since they create an intermediate enzyme.
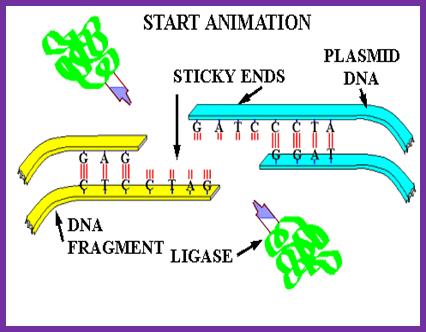
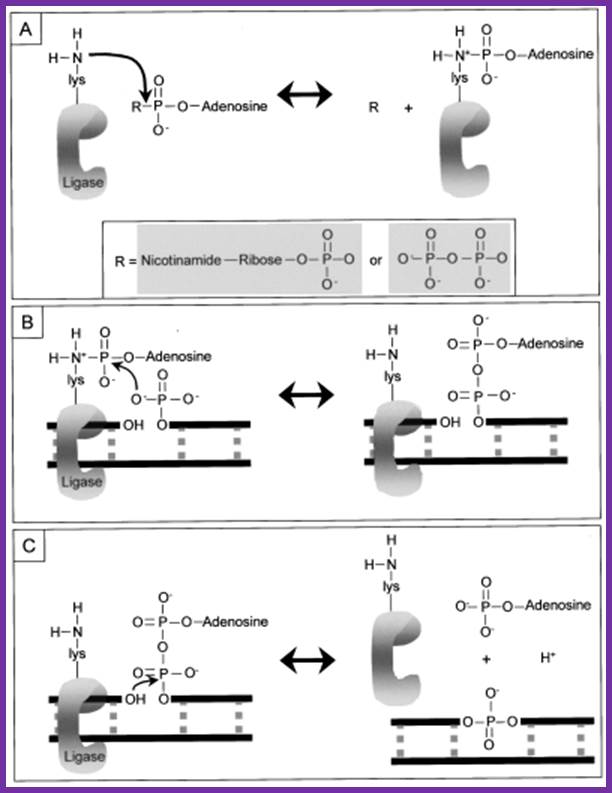
The above figure illustrates the ligation reaction pathway for DNA ligase and it’s interaction with the DNA strands during the process.
T7 DNA ligase Structure;
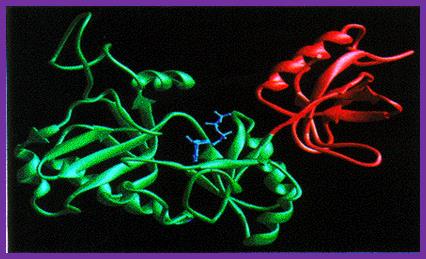
T7 DNA Ligases; http://www.biochem.umd.edu
The T7 DNA ligase structure consists of two distinct domains with the ATP binding site formed by the larger N-terminal domain. In the complex with ATP, the adenine ring is buried in a pocket in the enzyme with the alpha-amino group on the ATP close to the side-chain of lysine 34, with which it forms a covalent bond during the first stage of the reaction.
The conformation of the ATP suggests that, in the complex with AMP-DNA, the adenine residue would be ‘flipped-out’ of the DNA duplex, preventing it from interfering with base-pairing of the DNA in the region of the nick-site.
DNA ligases close nicks in the phosphodiester backbone of DNA. Biologically, DNA ligases are essential for the joining of Okazaki fragments during replication, and for completing short-patch DNA synthesis occurring in DNA repair process. There are two classes of DNA ligases. The first uses NAD+ as a cofactor and only found in bacteria. The second uses ATP as a cofactor and found in eukaryotes, viruses and bacteriophages. The smallest known ATP-dependent DNA ligase is the one from the bacteriophage T7 (at 41KdA). Eukaryotic DNA ligases may be much larger (human DNA ligase I is > 100KDA) but they all appear to share some common sequences and probably structural motifs.
DNA Ligase Mechanism:
The reaction occurs in three stages in all DNA ligases:
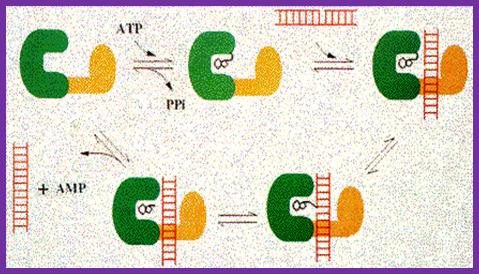 1.
Formation of a covalent enzyme-AMP intermediate linked to a lysine side-chain
in the enzyme. 2. Transfer of the AMP nucleotide
to the 5’ phosphate of the nicked DNA strand. 3.
Attack on the AMP-DNA bond by the 3’-OH of the nicked DNA sealing the phosphate
backbone and resealing AMP.
1.
Formation of a covalent enzyme-AMP intermediate linked to a lysine side-chain
in the enzyme. 2. Transfer of the AMP nucleotide
to the 5’ phosphate of the nicked DNA strand. 3.
Attack on the AMP-DNA bond by the 3’-OH of the nicked DNA sealing the phosphate
backbone and resealing AMP.
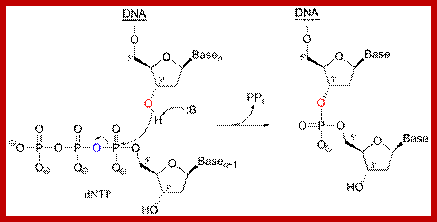
The above figure illustrates the three reaction stages of ligase reaction; http://www.biochem.ucl.ac.uk/bsm/xtal/teach/repl/ligase.html
“The joining of DNA fragments with protruding 5' termini that are not compatible (for instance, restriction digestion of DNA with Xba I and Hind III) can be accomplished by partial filling of the recessed 3' termini in controlled reactions using the Klenow fragment of E. coli DNA polymerase I. it’s an interaction with the DNA strands during the process.
DNA ligation of double strand breaks:
The
process of nonhomologous end-joining in eukaryotes and prokaryotes is similar,
but remarkably, prokaryotes accomplish these multiple steps with only two
proteins.
The current working model for end-joining in eukaryotes holds that the Ku70−Ku80 heterodimer encircles a broken end, then recruits DNA-PKcs and perhaps other factors such as the end-modifying enzyme terminal transferase. Activated DNA-PKcs recruits Artemis and the Mre11 complex to process ends (for example, removing 3' or 5' flaps); once the DNA ends are trimmed, single-strand gaps resulting either directly from the initial breakage or from the trimming process need to be filled in by polymerases such as Pol Mu. XRCC4 may bind directly to DNA or be recruited by Ku; ligation is carried out by DNA ligase IV, which binds to XRCC4; additional unidentified factors also seem to be necessary for ligation. In contrast to this veritable army of factors for eukaryotic NHEJ, only two proteins—albeit with quite distinct functional domains—are needed for mycobacterial NHEJ.
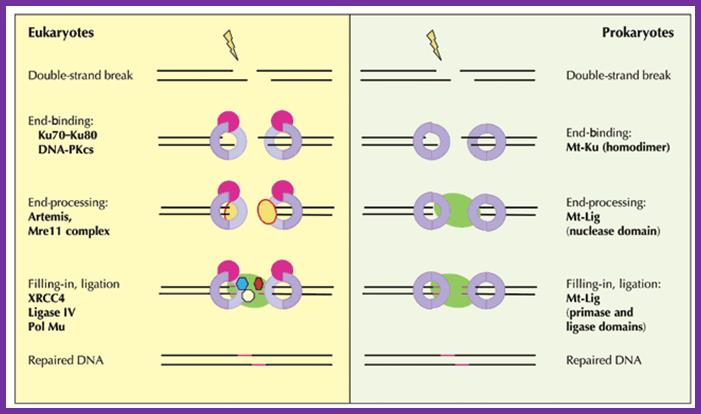
Geoffrey R Weller, Vicky L Brandt & David B Roth; http://www.nature.com/
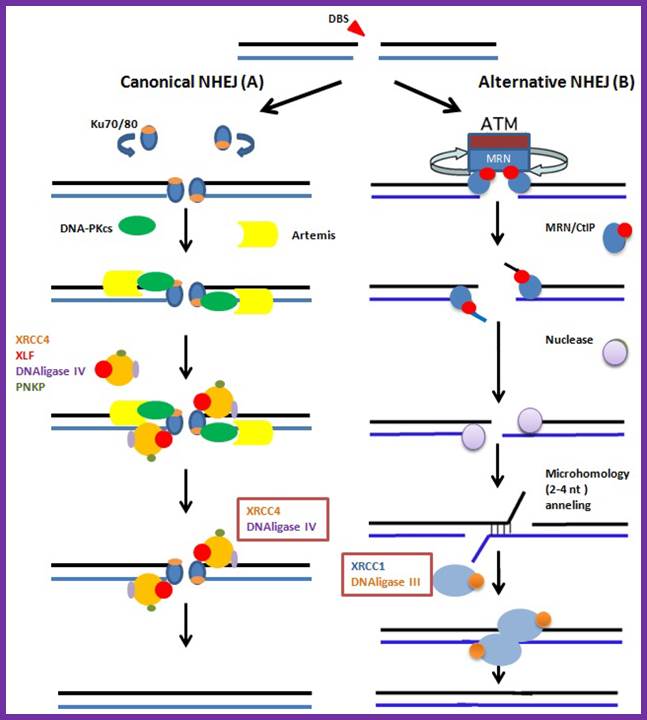
The canonical and NHEJ-alternative pathways. (A) The canonical NHEJ pathway, involving KU and XRCC4, can seal double-strand ends, even distal and non-fully complementary ends, in a conservative fashion; (B) In the alternative pathway, the main event is extended deletion at the junction, generally associated with the use of internal microhomologies distant from the ends. And XRCC1 and DNA ligase-III are used for strand ends ligation. Clara González-Marín 1, Jaime Gosálvez 2 and Rosa Roy;http://www.mdpi.com/
DNA Ligation and Bacterial Transformation: Ligation Reactions. (by MIT Open Course Ware.) Note: There are few overlapping notes emphasize certain specific points.
Introduction:
Today you will ligate your linearized M13KO7 backbone with your oligonucleotide insert by mixing the two in the presence of ATP and an enzyme, T4 DNA ligase. During the ligation reactions, hydrogen bonds will form between the overhangs on the fragments, and then the ligase will repair the phosphate backbone, creating a stable circular plasmid;
DNA Ligation
Hopefully, your ligation reactions will generate your desired construct, namely the M13KO7 backbone carrying a short added sequence in the gene for p3. Alternative ligation products may arise, including a simple reclosing of the singly cut M13KO7 backbone. We will remove these easy to form but undesired products with a trick called a "kill cut," described below.
You will transform today's ligation reactions into bacteria. During 'transformation,' a single plasmid from the ligation mixture enters a single bacterium and, once inside, replicates and expresses the genes it encodes. One of the genes on the M13KO7 genome leads to kanamycin-resistance. Thus, a transformed bacterium will grow on agar medium containing kanamycin. Untransformed cells will die before they can form a colony on the agar surface.
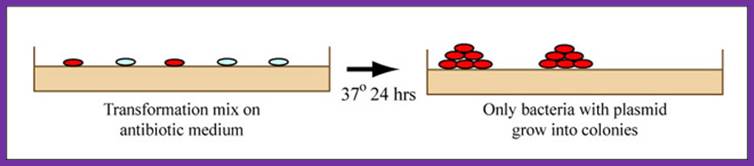
Growing colonies on medium, showing that only bacteria with plasmid incorporated grow into colonies. (Figure by MIT Open CourseWare.)
Bacterial Transformation:
Most bacteria do not usually exist in a 'transformation ready' state, but the bacteria can be made permeable to the plasmid DNA, and cells that are capable of transformation are referred to as 'competent.' Competent cells are extremely fragile and should be handled gently, specifically kept cold and not vortexed. The transformation procedure is efficient enough for most lab purposes, with efficiencies as high as 109 transformed cells per microgram of DNA, but it is important to realize that even with high efficiency cells only 1 DNA molecule in about 10,000 is successfully transformed.
RNA Ligases:
T4 RNA ligase: It catalyzes the ATP-dependent covalent joining of single-stranded 5-phosphoryl termini of DNA or RNA to single-stranded 3’-hydroxyl termini of DNA or RNA. T4 RNA ligase 2 also catalyzes the joining of a 3-hydroxyl terminus of RNA to a 5’-phosphorylated RNA or DNA; unlike T4 RNA ligase 1, this enzyme prefers double-stranded substrates. A truncated form of T4 RNA ligase 2 requires a pre-adenylated substrate for ligation. This unit describes specific reaction conditions, as well as applications such as radioactive labeling of the 3 termini of RNA, circularizing oligodeoxyribonucleotides and oligoribonucleotides, ligating oligomers and nicks, creating hybrid and chimeric DNA/RNA molecules, and miRNA cloning. Curr. Protoc. Mol. Biol. 84:3.15.1-3.15.4. © 2008 by John Wiley & Sons, Inc.
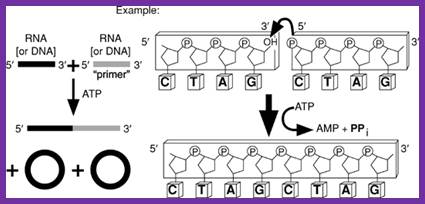
T4 RNA ligase catalyzes the ATP-dependent covlent joining single stranded RNA with 5’p with 3’OH of the broken ends of RNA or two pieces of RNA segments; http://www.lookfordiagnosis.com
mRNA Intron’s ligation:
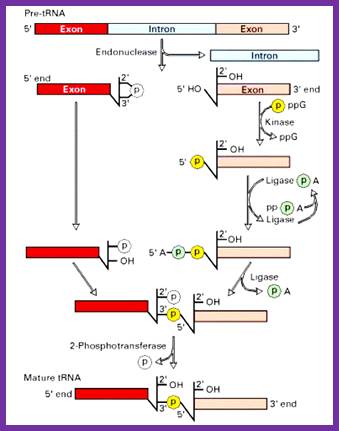
Splicing reactions requires four enzymes namely t-RNA specific endonuclease, cyclic phosphodiesterase, t-RNA specific ligase and 2-phosphotransferase.
By the action of endonuclease, introns removed. Following, excision of introns, a 2’-3’ cyclic phosphomonoester bond forms on the cleaved end of the 5’-exon. The multi step reaction joining the two exon requires two nucleoside triphosphates: a GTP, which contributes the phosphate group for the 3’à5’ linkage in the finished t-RNA molecule and an ATP, which forms an activated ligase—AMP intermediate. The 2’ phosphate on the 5’-exon is removed in the final step. http://www.biosivva.50webs.org
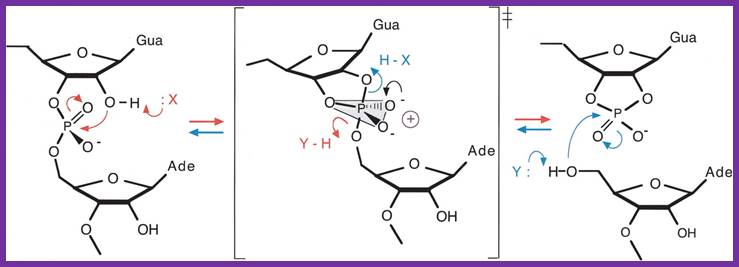

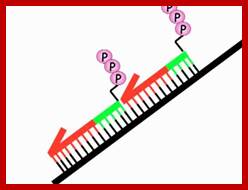
The ends of either single stranded DNA RNA can be joined by T4 RNA ligase by the mechanism shown (bottom figure)
The Varkud satellite ribozymes; The reaction mechanism for cleavage and ligation by the VS ribozyme. In the cleavage reaction (right, red) the 3′-phosphorus is attacked by the 2′-oxgen, with departure of the 5′-oxygen to leave a cyclic 2′3′-phosphate. In the ligation reaction (left, blue) the attacking nucleophile is the 5′-oxygen, with departure of the 3′-oxygen to form the internucleotide phosphodiester linkage. The reaction could be accelerated by general acid-base catalysis, whereby the attacking nucleophile is deprotonated by a base, and the leaving oxyanion is protonated by an acid. Note that a given group must fulfill opposite functions in the two reactions. The transition state for the reaction is a doubly charged oxyphosphorane, which could be stabilized in various ways, including the juxtaposition of positive charge. In the transition state, the 2′-oxygen, 3′-phosphorus, and 5′-oxygen are colinear, and the local RNA structure could be exploited to align the reactants in an optimal manner for reaction. http://rnajournal.cshlp.org/;http://www.ufrgs.br/
Molecular Tools-for DNA Ligation:
This web page was an assignment for an undergraduate course at Davidson College. General Information;
Ligases are enzymes that seal breaks in the phosphate-sugar backbone of DNA and RNA. There are two main classes of DNA ligases, those that use NAD+ as a cofactor (only in bacteria), and those that use ATP as a cofactor (eukaryotes and viruses) (Kahn, 2003). Mammalian cells have four types of DNA ligase, which together accomplish three main functions: joining Okazaki fragments, sealing repairs, and sealing recombination fragments (Purves et al., 2001).
DNA ligase I: connects Okazaki fragments of the lagging strand in DNA replication, and can also seal some repair and recombination fragments (Wei et al., 1995).
DNA ligase II: an alternatively spliced form of DNA ligase III that is only expressed in non-dividing cells.
DNA ligase III: works with protein XRCC1, which is a DNA repair protein. DNA ligase III is a primary agent in sealing base excision-repairs and recombination fragments.
DNA ligase IV: works with protein XRCC4, which is another DNA repair protein. DNA ligase IV is also important in sealing base excision-repairs and recombination fragments, especially during development (Schär et al., 1997).
Mechanism
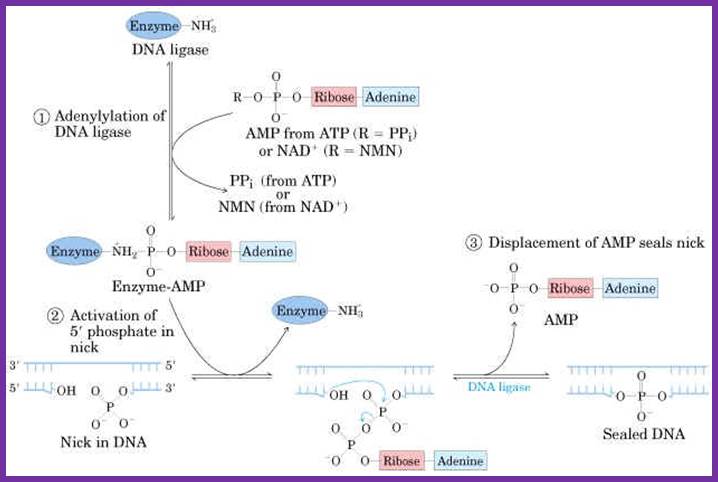
The following diagrams and descriptions are of the three main steps of ligation among all types of DNA ligases. Notice how ligase follows the general properties of enzymes, solely providing a docking area for the reaction between the AMP and DNA.
1. Adenylation of DNA ligase:
The side chain of lysine 34 in ligase forms a bond with ATP, where ATP kicks off two phosphate groups to become an AMP-ligase complex (Kahn, 2003).
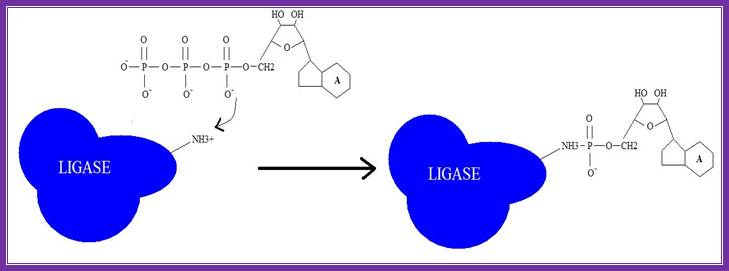
Adenylation of DNA ligase by ATP. http://good-stuff.info
2. Activation of 5’ phosphate:
The monophosphate of the AMP-ligase complex forms a bond with the 5’ phosphate of the broken strand. This bonding activates the 5’ phosphate group for the next step.
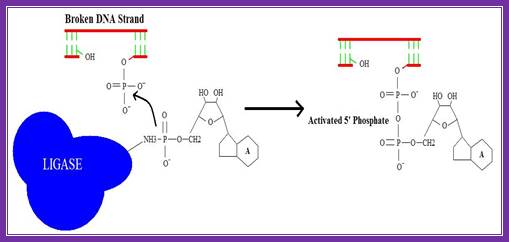
Figure . Activation of 5’ phosphate mediated by AMP-ligase complex.
3. Displacement of AMP connects the broken strand:
Now that the 5’ phosphate has been activated by the AMP-ligase complex, the 3’ hydroxyl group attacks the 5’ phosphate and forms a new bond, releasing the AMP. Ligase is crucial for holding the complex together in the necessary orientation.
The Overall Reaction:
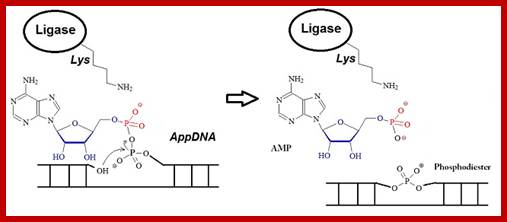
Rapid progress has been made in recent years in high-throughput next-generation sequencing of small RNAs, which has increased the demand for 5’-adenylated DNA linkers or adapters. ;http://www.biosyn.com

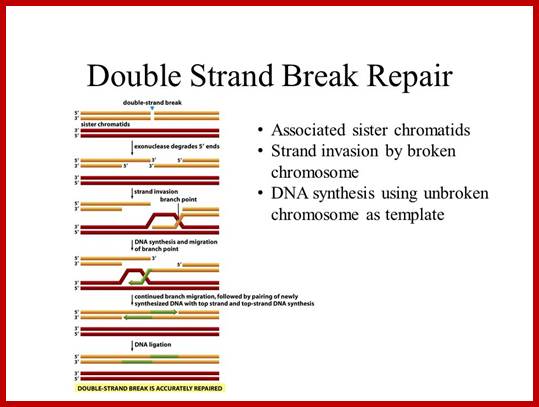
Overall reaction of the ligation of a broken DNA strand to form a new phosphodiester bond between the adjacent phosphate and sugar. http://slideplayer.com