Replication Origins:
Basically, replication initiates, whatever may be type of DNA it is, at a specific site or locus called ORIGIN (or Ori) and terminates the replication at a specific site, called Termination site, also called TER sites. The structure, location and the number of Origins and Ter site vary from one system to the other. Origins can be recognized by the site of replicating bubble and fork movement for direction of replication. Every genomic DNA contains an origin site. Lower organisms like E.coli have only one Ori, but higher eukaryotic systems have multiple ORIs’. The size of replicon and the structure and location of Origin vary from one genome to the other. This site is involved in the binding of specific proteins, which results in the unwinding of the DNA into open structure called replication eye or Replication bubble, on to which a variety of proteins bind and initiate replication. Depending upon the kind of origin, the replication from the origin can be bi-directional or unidirectional with some variations in each of the two.
Types: Mutants can delineate the structure and function of ORIs. Understanding of ORIs is very important for amplification of recombinant DNA under in vitro conditions, whether it is for gene cloning or other genetic manipulations.
A DNA containing an Ori site and a Ter site is called a Replicon, the unit of replication, irrespective of the length of the genomic DNA. E.coli DNA consists of one Replicon. In eukaryotes as the size of DNA is thousand times the size of E.coli DNA; it is organized into several nucleo proteinaceous threads called chromosomes. Sheer size of the DNA in eukaryotes (EKs) is so high; each chromosomal DNA contains several replication origins and termination sites, however the Ter sites are not very well defined. Eukaryotic Replicons, have devised replication mechanism with multiple ORIs and TER sites; so as to achieve the completion of replication within a time period.
Replicons, a Comparative Account:
|
Species |
Genome size (bp) |
Replicon numbers |
Size of a Replicon-BP |
Rate of replication (bp)/min |
|
E.coli |
4.6 x 10^6 |
1 |
4.6 x ^6 |
50000 to 60000 |
|
Yeast (n=16) |
1.4 x 10^7 |
500 |
40,000 |
36,000 |
|
Fruit fly (n=4) |
1.4 x 10^8 |
3500 |
40,000 |
2800 |
|
House fly (n=4) |
8.6 x 10^8 |
4000 |
45,000 |
2800 |
|
X.laevis (n=) |
3.1 x 10^9 |
15000 |
200kbp |
2500 |
|
|
|
|
|
|
|
Mus musculus |
2.6 x 10^9 |
25000 |
150kbp |
2200 |
|
Homo sapiens |
3.0 x 10^9 |
25-30 000 |
200kbp |
2800-3000 |
A list of Origins:
|
ORI-Name |
Species |
Replication direction |
Size of the Origin |
|
Ori-C |
E.coli |
Coupled Bidirectional |
250 bp |
|
Ori-Col.E 1 |
Col.E1 plasmid |
Unidirectional, leading strand first initiated |
|
|
Ori Ti |
Ti plasmid from Agrobacterium tumefaciens |
Bidirectional |
|
|
Ori PSc 100 |
PSc plasmid |
|
|
|
Ori Pi |
Pi phage |
Bidirectional |
|
|
Ori Col V |
Plasmid Col.V-k30 |
|
|
|
Ori-R 300 |
Drug resistant plasmids |
|
|
|
Ori-alpha |
Plasmid alpha, Ori-alpha |
Unidirectional
|
|
|
Ori-y |
Ori-y |
Sequentially bidirectional, one at ori and second at n’pas |
|
|
Ori-F |
Ori-F from F plasmids |
Bidirectional
|
|
|
Ori-R1 |
R1 and R100 |
Unidirectional, lagging strand first initiated |
|
|
Ori-S |
HSV virus |
Sequentially bidirectional, First initiation at ori, then second at n’pas |
|
|
Ori-P |
EBV
|
|
Epstein Barr Viral |
|
Ori-BPV |
BPV
|
|
Ovine Papilloma Viral |
|
Ori Rk2 |
RK2 plasmid |
|
|
|
Ori-ADV |
Adeno virus, at the end of linear DNA |
Unidirectional |
|
|
Ori-Rhi |
Rhizobium plasmid |
|
|
|
Ori-H |
Mitochondria, H stands for Heavy strand replication |
Unidirectional |
|
|
Ori-L |
Mitochondria, for replication of light strand |
Unidirectional |
|
|
Ori T7 |
Phage T7 |
Bidirectional/ Unidirectional |
|
|
Ori lambda |
Phage lambda |
Coupled Bidirectional |
|
|
Ori phi X |
Origin for PhiX174 phage DNA
|
Unidirectional |
Ori (+) located at 4299-4328= ~30 ntds. Ori (-) located at 2220-2300 =~80ntds |
|
Ori M13 |
Phage M13 (filamentous) |
Unidirectional |
Ori (+) located at5750-5850 (contain D and E loops) Ori (-) located at 5600-5750(contain B and C loops) |
Ori sites that Use Both Dna-A and Rep Proteins:
|
Plasmid DNA |
Dna-A box |
A/T (13mers) |
Rep site (18-19 bp) |
A/Y rich (5-11 bp) |
GATC methyl sites |
IHF (histone like factors) |
PSc 101 |
1 |
2 |
5 x18 bp |
None |
1 |
Yes |
|
Pi Ori-R |
2 |
- |
5 x 19 bp |
5 x 7 bp |
5 |
No |
|
F, Ori-S |
2 |
1 |
4 x 19 bp |
4 x 8 bp |
1 |
- |
|
Ori-col-V-K30 |
2 |
- |
5 x 19 bp |
5 x 11 bp |
- |
- |
|
E.Coli |
4 |
3 |
- |
3 x 19 bp |
1 |
Yes |
|
Ori-gamma (R6K) |
1 |
1 |
7 x22 bp |
3 x 10 bp |
1 |
Yes |
Kinds and Components of Origins:
Comparative structural components of different Origins:
E.coli-13----13----13—R1-IHF-M-R2-Fis-R3-R4,R=A=9mer [13mer=GATCTNTTNTTT, 9mer=TGTGAATA],
B.subtilis-----16-----16---16-----A-9----A-9----A-9----A-9 [13mer=GATACCTTANTTTTC],
Pseudomonas---12-----12---12------A-9----A-9----A-9----A-9 [13mer=GAAGAGGTT/AAT/A—T/A],
Lambda [11 AT rich] x 4---------- [19] x 4; TTNTCTTTTGT,
O-lambda binds to 19 mer regions and lambda P-proteins bind AT rich 11bp region
O82 [11 AT rich] x 2-------- [19] x 4; TTGTCTTTTGT, O-82 bind
O80 [6 AT rich] x 3----------- [19] x 4; TCTTGT,
E.coli origin loci:
Replication origin in E.coli DNA is located at 84.3mpu in between genes like gid A and mio C.
--uncB—UncI—gidB-gidA---<OriC>-mioC-asnC<>asnA---dATA
E.coli: -0--13----13----13—<R1-IHF-<M-R2>-<Fis>-<R3-R4>--340bp.
R=A, [3x13mer=GATCTNTTNTTT], [4x9mer=TGTGAATA],
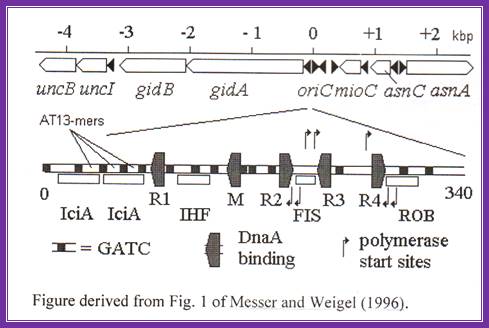
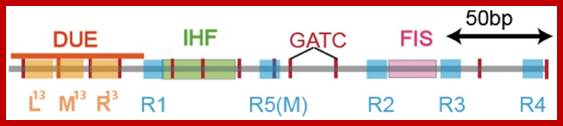
Black shaded rectangles are GATC sequences; R1,R2,R3 and R4 are the DnaA binding sites; IHF-alpha and beta, integration host factor are histone like protein, IciA inhibitors (Inhibitor of chromosome initiation), ROB Right arm binding sites; R3 is weak binding site.
The E. coli origin of replication bears five 9-mer DnaA-binding sites (R1, R2, R3, R4 and R5) as well as three 13-mer binding sites included in an A+T-rich DNA unwinding element (DUE). Although the 9-mers show no differential specificity between DnaA-ATP and DnaA-ADP, the 13-mers specifically recruit DnaA-ATP. In addition to DnaA-binding sites, oriC hosts-specific binding sites for IHF, SeqA and FIS, three proteins that regulate the activity of DnaA. https://www.researchgate.net
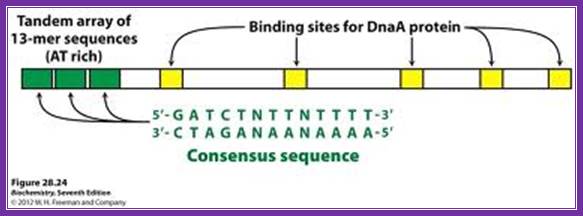
http://oregonstate.edu/

The above figure shows E.coli Ori C components, the binding of DNA to 9-mer sequences lead to melting of the DNA into replication bubble. http://slideplayer.com/
Orisome:
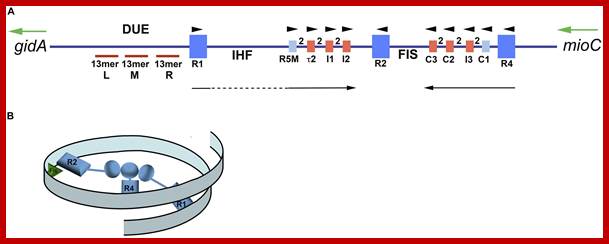
Map of Escherichia coli oriC and conformation of bORC. (A) The oriC region is mapped, showing positions of binding sites for DnaA, IHF, and Fis, as well as the right (R), middle (M), and left (L) 13mer sequences in the DNA unwinding element (DUE). The three high-affinity sites R1, R2, and R4 are designated by royal blue squares, and the low-affinity sites are marked by small light blue or red rectangles. The red rectangles designate sites that preferentially bind DnaA–ATP, while the sites marked by light blue rectangles bind both nucleotide forms of DnaA equivalently. Small arrowheads show orientation of sites, the two between sites indicates the number of bp separating the sites. Arrows under the map indicate growth direction of DnaA oligomers. The dotted line marks that the oligomer does not span the region between R1 and R5M. Two genes, gidA and mioC, flanking oriC are shown, with the green arrows marking the direction of transcription. (B) Proposed looped conformation of bORC in E. coli. OriC DNA (ribbon) is constrained by DnaA (gray-blue figures in center of loop) bound at R1, R2, and R4 sites, as labeled. Interaction among the three bound DnaAs proposed to be via domain I. The green triangle represents Fis bound to its cognate site.; http://www.frontiersin.org
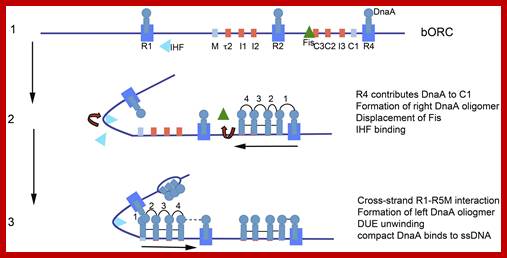
Model of staged orisome assembly. Stage 1 (bORC): after initiation of chromosome replication, DnaA rebinds to high affinity R1, R2, and R4 sites. Fis is also bound at this stage, but IHF is not. Low affinity sites are unoccupied. Stage 2: DnaA bound to R4 recruits DnaA for binding to C1. DnaA then progressively fills the remaining arrayed sites, forming an oligomer in the gap region between R2 and R4. The DnaA oligomer displaces Fis, and loss of Fis allows IHF to bind to its cognate site. Stage 3: the bend induced by IHF binding allows DnaA, recruited by R1, to bind to R5M, and form a cross-strand DnaA interaction. A DnaA oligomer then progressively grows toward R2, bound to arrayed low affinity sites, and anchored by R2. In this configuration, oriC DNA is unwound in the DUE, and DnaA in the form of a compact filament binds to the ssDNA. http://journal.frontiersin.org
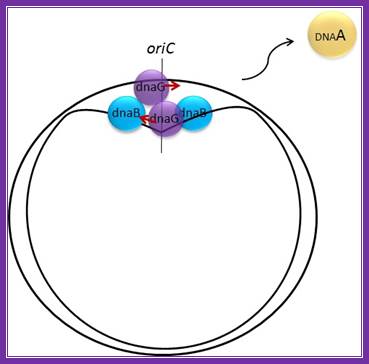
E.coli- Pre-primosomes; https://www.flickr.com
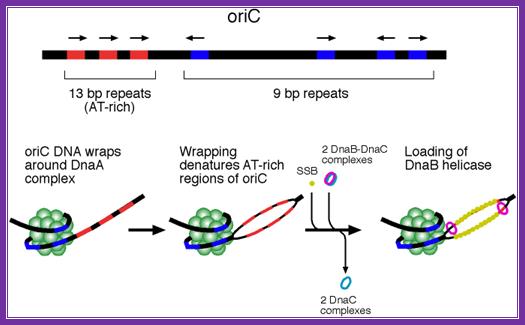
http://oregonstate.edu/
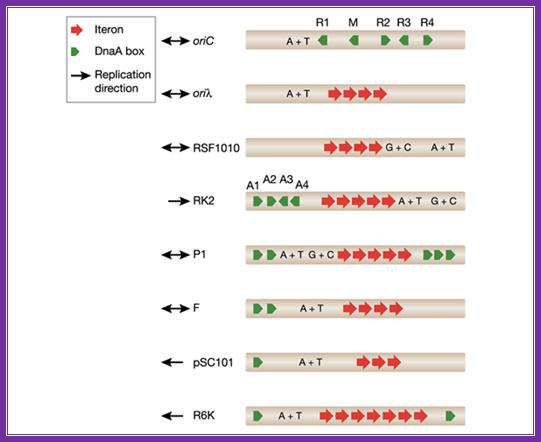
The diagram shows origin components of different DNAs from different plasmids and viruses; http://embor.embopress.org/
Properties of some Plasmid DNA and their Origins:
|
Plasmid |
Ori |
Size of the plasmid (Kbp) |
Copy no.s/cell |
Required proteins |
Host range |
PSC 101 |
|
9 |
4-6 |
Rep-A (38kd) |
Limited |
|
F |
Ori-S |
90 |
1-2 |
Rep-E (29kd) |
Limited |
|
Col V K30 |
Ori V |
90 |
1-20 |
|
Limited |
|
R 300 B |
|
9 |
10-20 |
|
Limited |
|
R1, R100 |
|
90 |
1-2 |
Rep-A (33kd) |
Limited |
|
R6K |
Ori-R |
38 |
1-11 |
36KD |
Limited |
|
RK2 |
|
56 |
5-8 |
Trf-A (32-43kd) |
Broad host range |
|
Col-E1 |
Col E1 ori |
6kbp |
10-20 (100) |
|
|
|
Ti |
Ori-T |
150-500 |
1-2 |
|
|
|
Rhi |
Ori-rhi |
40-50 |
1-2 |
|
|
Replication Termination Regions in E.coli:
When replication progresses; it has to end at some region. In the case of E.coli replication progresses in bidirectional and the replication forks meet at a common point called termination site or TER region. This is about 36 kbp and consists of six 23bp long consensus sequences; they are located almost opposite to origin region and oriented in opposite orientations, the are Left- < E <D <A : : : >C > B > F, the sequences- AATTAGTATGTTGTAACTAAAGT. This region is recognized by TER utilizing substance called TUS protein of 36Kd and they bind and prevent the movement of Helicase into double stranded daughter DNA molecule, thus prevent the second round replication.
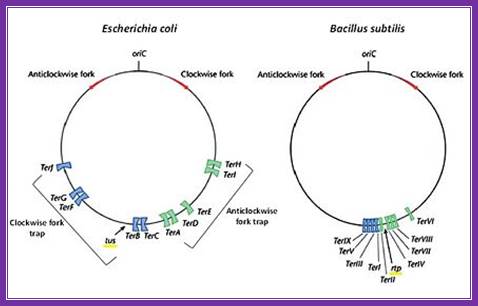
The circular chromosomes of E. coli (left) and B. subtilis (right) showing their respective origin of replication (Ori C), direction of the two replication forks (red arrows) and their subsequent fork traps (blue and green). Modified from; The diagram shows both Origin sites and TER sites; http://proteopedia.org/