Replication of ΦX174 Phage DNA:
Understanding of ssDNA replication is very fascinating. A large number of DNA viruses have single stranded genomes. Ex. Phi X 174 family of phages such as PhiX-S13, phi-A, phi-C, phi K, Phi R, 6SR, BR2, G4, st-2, alpha-3 are similar in nature, but the last three show different mode of replication.
PhiX174 virus has been extensively studied. It is an isometric virus, with 20 triangular unit faces, and 12 vertices (pointed regions where five faces meet). Each of the triangular surfaces is made up of 3 subunits of gp-F (gp means gene products). Every vertices contain 5 gp-G protein subunits and in the center of the five gp-G one spike protein called gp-H is present and it is like a fiber.
The phage infects through its spike protein binding to a lipopolysaccharide receptor present on the host cell surface. The exact mechanism of entry is not known (we always say that we know lot of things about the Viruses, but when comes to specifics, all the time we keep on saying we don’t know).
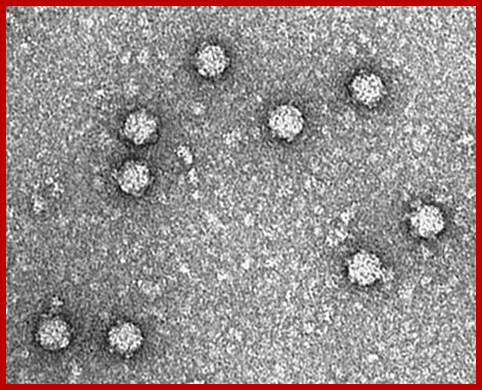
Electron micrographs of PhiX viral particles; https://www.biochem.wisc.edu

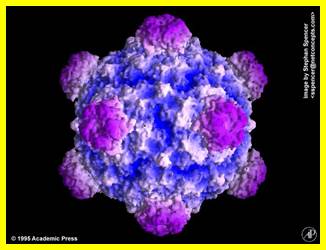
Simple representation of PhiX Phage; http://www.virology.wisc.edu/
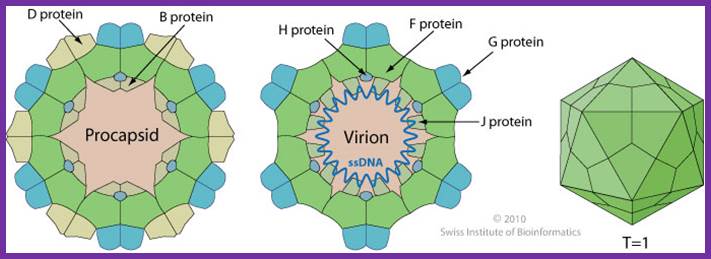
Non-enveloped, round, icosahedral symmetry (T=1), about 30 nm in diameter. The capsid consists of 12 pentagonal trumpet-shaped pentamers. The virion is composed of 60 copies each of the F, G, and J proteins, and 12 copies of the H protein. There are 12 spikes which are each composed of 5 G and one H proteins. http://viralzone.expasy.org/
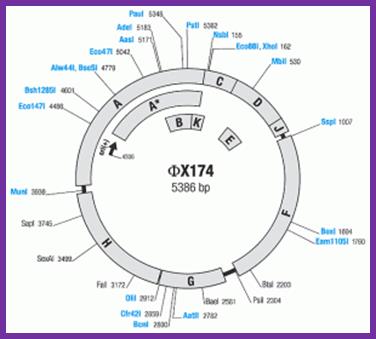
Size of the genome is 5386 nucleotides. It is single stranded and circularly modulated DNA. It is encoded with 10 genes but generates 11 proteins. This genome exemplifies, how a reading frame, if organized in a specific designed way, with minimum length of DNA, one can generate more proteins. This is accomplished by what is called overlapping reading frame. In this, the reading frame of a gene is organized in such a way if one gene ends in a particular position; the succeeding gene starts with few nucleotides overlapping the terminal region of the first gene. This is what, classically called overlapping genes, where reading of two genes is overlapped in their sequence.
2 nd Gene
5’----ATGAAA-1 st gene------------UAATGGAG
5’---ATG------------------------------------ATGAAA
|
Gene, Gene size |
Protein (Kd) |
Function |
|
A=3981-136=1539 |
60 |
Role in (+) strand synthesis |
|
A*=4497-136=1022, A* overlaps A |
37 |
Shuts of Host DNA replication |
|
B = 5075-51=362, B overlaps A |
20 |
Capsid morphogenesis |
|
K = 51-221=170 Overlaps A, |
8 |
Stimulates phage production and increase phage burst size |
|
C =133-393=263 |
6 |
DNA maturation |
|
D = 390-848=342 |
14 |
Capsid morphogenesis and assembly |
|
E = 568-843=270 |
10 |
Host cell lyses |
|
J = 848-964=116 |
4 |
DNA condensation and packing |
|
F = 1001-2284=1283 |
50 |
Major coat protein |
|
G = 2395-2922=527 |
20 |
Major Pentamer spike protein |
|
H = 2931-3917=986 |
37 |
Minor spike protein-central |
|
Ori(-)=IgS+1283-2395=111 |
|
|
|
Ori(+)=4305-4306 |
|
|
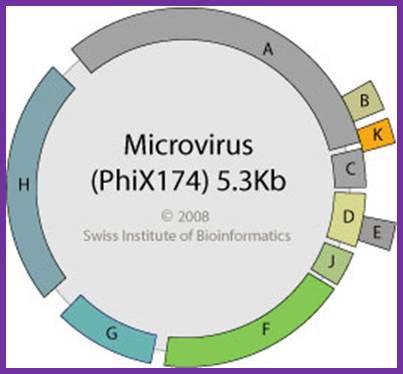
http://viralzone.expasy.org/
There are four distinct intergenic regions (IR), one between D and J (1001 to 1070), second between F and G (2200 to 2300, the numbers are from 0 map unit position), the third is in between G and H and the fourth is between H and A. The IR between F and G is used as origin for (-) strand synthesis. The origin for (+) strand synthesis is at left end of A gene, located approximately between 4299 to 4328 bases.
phiX174 plasmid usage notes:
The genes identified in phage phiX174 are shown on the map. All genes are transcribed clockwise. Enumeration of phage DNA begins with the last nucleotide of the unique PstI site and continues clockwise around the viral (+) strand in the 5’=>3’ direction. The map shows enzymes that cut phiX174 DNA once. Enzymes produced by Thermo Scientific are shown in orange. The coordinates refer to the position of the first nucleotide in each recognition sequence.;http://www.thermoscientificbio.com/
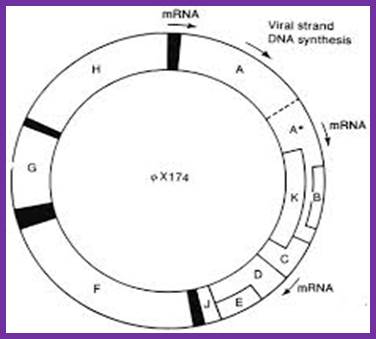
Overlapping reading frames of phiX174 phage http://www.helixsoft.nl/
The reading frame exhibits overlapping of gene segments. Small-A reading frame starts at 4494 ntd position and ends at 133ntd from the zero map position. The gene B starts at the right side of the A and ends in the terminal part of A. The K gene starts at the last part of A and slightly overlaps C. The end of the A gene overlaps the first codon of the gene C. The gene E overlaps D more than half of D.

![]()
Linearized PhiX genome with gene and intergenic regions and overlapping portion of certain genes.http://www.biochem.wisc.edu
Transcription appears to start at 3 sites, and the transcripts are polycistronic. A shift in the reading frame, for example generates E from D. This is a par excellent example to show, how a genome with its minimum size can generate more proteins. A length of 5386 ntds produces 11 proteins, i.e. equivalent to 586 ntds per proteins. In general most of the prokaryote genes have an average size of 1000bp.

phiX174 DNA sequence –X-ray film; Nobel laureate Sanger (original?)
The phi X 174 (or ΦX174) bacteriophage is a virus and was the first DNA-based genome to be sequenced. This work was completed by Fred Sanger and his team in 1977. Nobel Prize winner Arthur Kornberg used ΦX174 as a model to first prove that DNA synthesized in a test tube by purified enzymes could produce all the features of a natural virus, ushering in the age of synthetic biology. In 2003, it was reported by Craig Venter's group that the genome of ΦX174 was the first to be completely assembled in vitro from synthesized oligonucleotides. The ΦX174 virus particle has also been successfully assembled in vitro. https://www.revolvy.com/main.
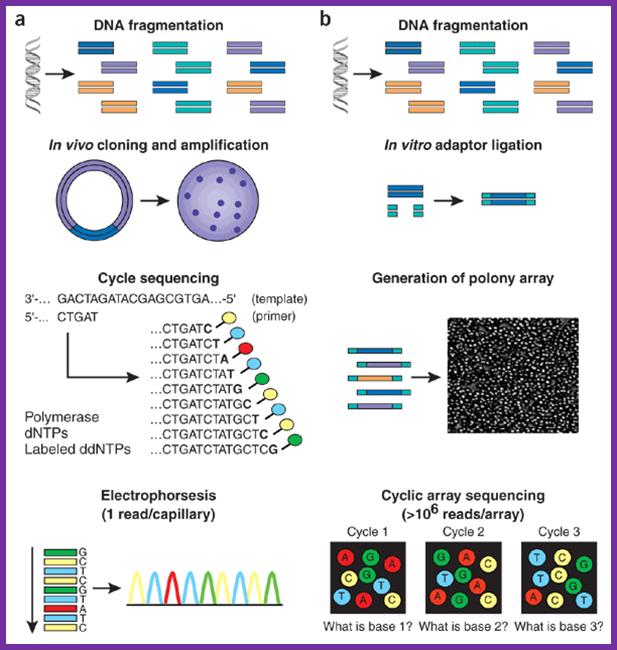
Array f(a) With high-throughput shotgun Sanger sequencing, genomic DNA is fragmented, then cloned to a plasmid vector and used to transform E. coli. For each sequencing reaction, a single bacterial colony is picked and plasmid DNA isolated. Each cycle sequencing reaction takes place within a microliter-scale volume, generating a ladder of ddNTP-terminated, dye-labeled products, which are subjected to high-resolution electrophoretic separation within one of 96 or 384 capillaries in one run of a sequencing instrument. As fluorescently labeled fragments of discrete sizes pass a detector, the four-channel emission spectrum is used to generate a sequencing trace. (b) In shotgun sequencing with cyclic-array methods, common adaptors are ligated to fragmented genomic DNA, which is then subjected to one of several protocols that results in an array of millions of spatially immobilized PCR colonies or 'polonies'15. Each colony consists of many copies of a single shotgun library fragment. As all polonies are tethered to a planar array, a single microliter-scale reagent volume (e.g., for primer hybridization and then for enzymatic extension reactions) can be applied to manipulate all array features in parallel. Similarly, imaging-based detection of fluorescent labels incorporated with each extension can be used to acquire sequencing data on all features in parallel. Successive iterations of enzymatic interrogation and imaging are used to build up a contiguous sequencing read for each array feature. http://www.nature.com/
Replication Cycle events:
|
Stage |
Duration |
Events |
|
SsDNA(+)>dsRf DNA |
0 to 5 minute |
Adsorption, penetration, complementary strand synthesis |
|
Rf > RF DNA |
5-20 minutes |
Transcription of (-) -) strand, |
|
RF > ss DNA(+) |
20-40 minutes |
Generates(+) strand by rolling circle mode |
PhiX genome with individual genes and with some overlapping genes such as A, A*, B, K, C, D and E
Life cycle:
Infection is through gp-G spike binding to host cell’s lipopolysaccharide, especially N-acetyl glucose amine component of the outer membrane.
The penetration site is a region at which the outer and inner membranes are closed together, like an attachment sites in higher forms.
A distinctive phospholipase found in this region has a role in viral infectivity. DNA penetrates along with gp-H, so the protein is called pilot protein. As soon as the ssDNA enters into cytoplasm, it gets coated with host ssBs and immediately the DNA is rendered super coiled.
With in 20-30 minutes of infection the viral genome produces sufficient number of A proteins and also A* products. The A* has a non-specific SS DNA endonuclease activity..
During host’ DNA replication, single stranded DNAs generated, are subjected A* endonuclease digestion. By this act, virtually, it shuts of host DNA synthesis.
The super coiled SSDNA is replicated to produce dsRF (replication form) of DNA, which in turn produces ssDNA (+), which in turn generates more RF forms and generate nearly 500 or more viral particles, and they are released by cell lyses.
Synthesis of (-) strand DNA:
The phage DNA has positive sense. The SS super coiled DNA generates stem loop structures at Or sites for minus strand synthesis. The origin region is about 100 ntds long, found between gene F and G. This region has two stem loop structures.
ORI (-):
(+)5’---TGTA--TTATA-loop-1-T—G-–A-loop-2--TGTCCTT—3’-3
Loop2- 5’ TT GTC CTT—3’ (--) strand
<---- GA.ppp5’ primer
The stem loop structure act as the recognition point for the assembly of primosome complex which initiates the synthesis of minus strand.
At the base of the second loop there are sequences, which are required for the primase to lay primer. Primosome complex formation initiates with Pri-A, it is supported by pri-B, pri-C and Dna-T, which assemble on the loop 2 structure. Dna-B is a helicase assisted by Dna-C in ATP dependent manner joins the Pri-A complex.
Then Dna-G joins the helicase, DNA-G is host RNAP called primase. This RNAP is different from the normal RNAP, which is involved in gene transcription and Rifamycin sensitive, while the RNA primase is insensitive to Rifamycin.
This primosome complex with its helicase-primase moves in 5’--->3’ direction on (+) strand. While it is moving, when it comes in contact with 5’GTC sequence, it halts and produces a complementary RNA strand of 10 to 11 ntds long. It slides backwards for laying the primer, just like E.coli primase producing primer on the lagging strand. The primosome moves in 5’ to 3’ and lays primers at every 1000 to 1500ntds intervals.
The host DNA pol-III binds to primers and extends 3’ OH group of the primers till it reaches another primer and stops. DNA pol-I removes the primers and fills the gaps. A DNA ligase seals the nicks.
The synthesis of (-) strand is akin to the synthesis of discontinuous strand in E.coli.
The double stranded DNA produced is now called RF form (replicative form). The replicative form also gets super coiled. Super coiling is the most important pre-requirement for replication and transcriptional activities. The replicative form of DNA is now subjected to transcription of some genes. It is now known transcription is initiated at three sites and generates polycistronic RNAs.
The prominent promoter for initiation of transcription is at the left of A gene.
Though the transcript looks like a polycistronic one, its translation mostly generates gp-A, a 60 KD protein. This protein has both sequence specific endonuclease and ligase properties. The truncated A gene product is A*. Its translation initiates near half the way from the 5’ end of A transcript. The A* has a property to recognize host DNA and digest it, with out affecting the translational machinery.
At this stage the RF form of DNA produces protein A in sufficient numbers and A* in less quantity. The mode of replication is similar to E.coli’ lagging strand synthesis, producing Okazaki like fragments.
(+) SS-DNA + pri-A, B, C+Dna-T+Dna-B+Dna-G at nPAS primosome Primosome ABC complex
SsDNA + primosome complex+4dNTPs+DNAP-III+DNAP-I + DNA ligase---->
-------> ds replicative form of DNA (RF-DNA).
The super coiled replicative forms are called RF1 and the nicked replicative forms are called RFs.
A list of proteins required for (-) strand synthesis:
|
Gene |
product size (kd) |
Function |
|
Pri-A |
76. Monomer |
It has a helicase like activity |
|
Pri-B |
11.5.dimer |
Assist pri-A |
|
Pri-C |
23. Monomer |
Assists pri-A |
|
Dna-T |
22. Trimer |
Assist primosome assembly |
|
Dna-B |
50. Hexamer |
Helicase5’>3’ |
|
Dna-C |
29.monomer |
Assists Dna-b |
|
Dna-G |
60.monomer |
Primase |
|
Rep-A |
65.monomer |
An helicase 3’> 5’ |
|
|
|
|
|
SsB |
19. Tetramer |
Binds ss region all the time and they are replaced when other factors bind |
|
Top and Ligases |
|
Removal of super coils, sealing of nicks |
|
DNAP-III |
|
Host DNA polymerase |
|
DNAP-I |
|
Host enzyme responsible for removing primers and filling the gap |
Synthesis of (+) strand DNA:
The origin for (+) strand synthesis is located at the 5’ end of the gene A. The site is about 29 ntds long and has certain sequences for recognition; an A-T rich region, a consensus sequence, and gp-A binding site.
5’--4305-G I A-4306—3’
5’--4299.CAACTT G I ATATTAATAACACTATAGACCAC.4328---3’
(A-T rich) (gp-binding site)
The gp-A is globular protein; it has an active site with an amino acid sequence –tyr-val-ala-lys-tyr-val-asn-lys-. The gp-A binds to a site in the + strand of RF DNA. It also binds to a recognition region where it identifies the cleavage site. Binding melts the RF at AT rich region and creates a replication bubble with a replication fork.
This event leads the enzyme (gp-A) to cut DNA between G at 4305 and A at 4306, producing G3’OH and 5’P-A. The enzyme also binds to 5’p of A nucleotide covalently through its own OH group of tyrosine amino acid residue leaving 3’G-OH free.
With a nick in the (+) strand and gp-A bound to 5’-A, yet another protein Rep-A, which is a helicase binds to the (-) strand at this position. This complex acts as the primosome for the assembly host’s DNAP-III.
The Rep-A binds to (-) strand, gp-A binds to 5’end of A of + strand; at this point DNAP-III (Holozyme) binds to the nick region in the RF with G3’OH as a free end. At the same time the Holozyme also gets associated with Rep-A and gp-A. This complex drives through the RF DNA in 3’>5’ direction on (--) strand, because the rep-A, a helicase, a motor protein moves in 3’ to 5’ direction. As other components are associated with the helicase, they also move with the motor protein.
The DNA pol-III uses the 3’G-OH as the primer end and extends the chain in complementary fashion in an uninterrupted manner. The (-) strand synthesis is like continuous strand synthesis in E.coli. During this progression of the complex ds replicative form of DNA unwinds ahead of the fork. The unwinding is facilitated by the action of Topoisomerase-II (Gyrase) ahead of the fork. The (+) strand, whose 5’ end is bound to gp-A, peels off in form of a loop.
When the replication complex progresses the entire length and encounters the newly synthesized (+) origin sequences, the gp-A, which is still bound to the 5’end of the (+) strand, performs three exquisite reactions. (1) First it cuts the newly made (+) strand exactly at the same site, same nucleotides i.e. between 5’G and 3’A, where the first cut was made to produce G with 3’OH group. (2) the 5’p-A covalent bond to the first is transferred to the cut free 3’G-OH and ligates it. Thus the old (+) strand is converted into circular ss DNA and (3) while the 5’p-A bond is transferred to 3’G-OH of the same strand, (it becomes circular), the 5’P of the newly made strand is covalently added to its another tyrosine residue, which is located in the same catalytic site of the enzyme. The Rep-A and the Holozyme associated with the (-) strand continue to produce complementary copy (i.e. + strand).
With one more round of replication one more circular ssDNA is released and the replication progresses relentlessly. This way it can produce any number of copies.
This process is augmented, by the replication of + ssDNA produced to form RF form of DNA. The RF form of DNA immediately undergoes super coiling. This is the stable form of DNA.
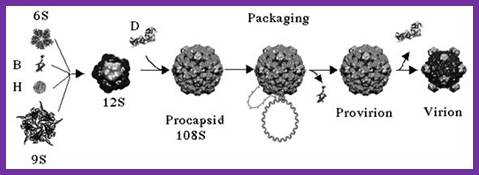
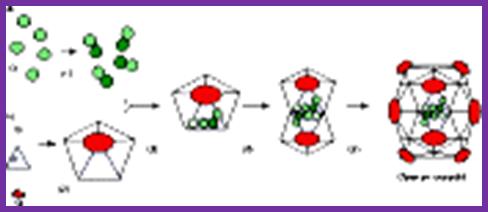
The assembly of a macromolecular structure proceeds along an ordered
morphogenetic pathway, and is accomplished by the switching of proteins between
discrete conformations as they are added to the nascent assembly1, 2, 3. Scaffolding proteins often play a
catalytic role in the assembly process1, 2, 4, rather like molecular chaperones5. Although macromolecular assembly
processes are fundamental to all biological systems, they have been
characterized most thoroughly in viral systems, such as the icosahedral Escherichia coli bacteriophage ![]() X174 (refs 6, 7). The
X174 (refs 6, 7). The ![]() X174 virion contains the proteins F, G,
H and J7, 8. During assembly, two scaffolding
proteins B and D are required for the formation of a 108S, 360-Å-diameter
procapsid from pentameric precursors containing the F, G and H proteins6, 9. The procapsid contains 240 copies of
protein D, forming an external scaffold, and 60 copies each of the internal
scaffolding protein B, the capsid protein F, and the spike protein G9, 10. Maturation involves packaging of DNA
and J proteins and loss of protein B, producing a 132S intermediate6, 7. Subsequent removal of the external
scaffold yields the mature virion. Both the F and G proteins have the
eight-stranded antiparallel
X174 virion contains the proteins F, G,
H and J7, 8. During assembly, two scaffolding
proteins B and D are required for the formation of a 108S, 360-Å-diameter
procapsid from pentameric precursors containing the F, G and H proteins6, 9. The procapsid contains 240 copies of
protein D, forming an external scaffold, and 60 copies each of the internal
scaffolding protein B, the capsid protein F, and the spike protein G9, 10. Maturation involves packaging of DNA
and J proteins and loss of protein B, producing a 132S intermediate6, 7. Subsequent removal of the external
scaffold yields the mature virion. Both the F and G proteins have the
eight-stranded antiparallel ![]() -sandwich motif8, 11 common
to many plant and animal viruses12, 13. Here we describe the structure of a
procapsid-like particle at 3.5-Å resolution, showing how the scaffolding
proteins coordinate assembly of the virus by interactions with the F and G
proteins, and showing that the F protein undergoes conformational changes
during capsid maturation. http://www.nature.com/
-sandwich motif8, 11 common
to many plant and animal viruses12, 13. Here we describe the structure of a
procapsid-like particle at 3.5-Å resolution, showing how the scaffolding
proteins coordinate assembly of the virus by interactions with the F and G
proteins, and showing that the F protein undergoes conformational changes
during capsid maturation. http://www.nature.com/
Packaging of genome into capsid:
As this RF-form of DNA builds up in numbers, at which time both ds RF DNAs and ssDNA are found in the same cytoplasm. The RF form of DNA is transcribed to generate few polycistronic mRNAs. It is important to note that the strand used for transcription is (-) strand and not the (+) strand.
Generation of RF form is very important for it is this form that is stable, and it is the stable form that is capable of transcription. Transcription and translation leads to the production of viral particles. Important viral proteins required for viral assembly and products viral particles are gp-F, G, B, H and D; they all assemble in a sequence to produce a pro-head not conformationally fully formed structure. Another protein that plays a key role is gp-C, which binds to the cut 5’ end of the +DNA and by rolling mode; the (+) strand is peeled off from the (-) strand. With gp-C bound to the 5’end, while the entire length unwinding, another protein called gp-J associates with (+) strand all along the length of the DNA. This leads to the packaging of the single stranded DNA into the not yet fully formed pro-head.
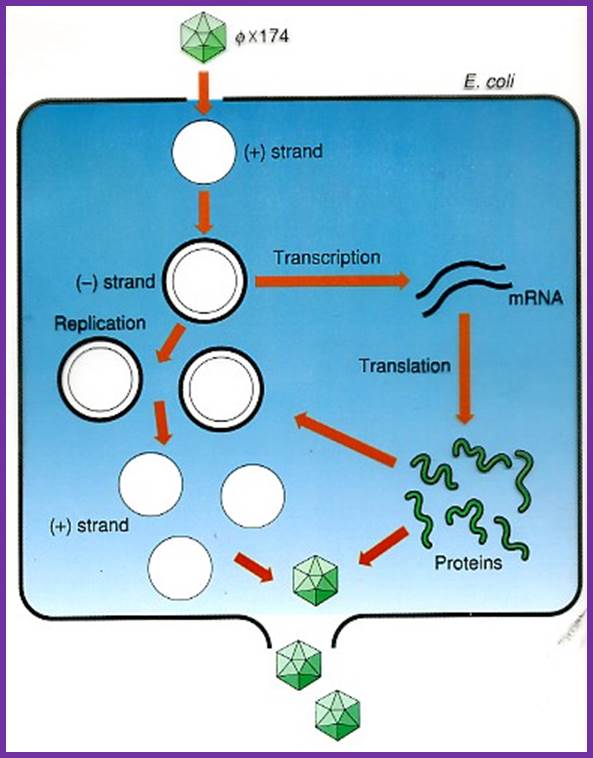
Life cycle of the phage; http://www.dls.ym.edu.tw/
Once the DNA packages into the pro-head certain conformational changes are induced and final form of phage particle are produced. Cell lyses leads to the release of the viruses.
