Plasmid DNA Replication:
Col. E plasmid Replication
The term for such small circular DNA molecules which have different modes of transfer from one cell to the other and function were called as Plasmids by Joshua Lederberg in 1952. Plasmids are extra chromosomal DNAs, exist as ds circular molecules. The size and structural features and replication modes vary from one species of plasmid to the other. Some plasmids produce their own components for replication and others heavily dependent on host factors and enzymes. Many such episomal DNAs have been identified by their specific features such as, for their drug resistant features called R-plasmids, self-transmissibility F-plasmids, and antibiotic resistant plasmids and anti bacterial plasmids like colicin plasmids. They are involved in conjugation and transduction. Some plasmids resist to heavy metals, UV radiations, some produce restriction enzymes and some produce products that are involved in cellular metabolism. Agrobacterium plasmid called Ti plasmid (= 400kbp) is unique for its ability to transfer a segment of its own DNA called T-DNA into host plant cells, without the bacteria getting into the cell and induce tumors; it is an unique plasmid. Plasmids can be called as replicons for they replicate themselves and as they move from one bacterium to another, hence hey are called Mobilosomes. Some encode conjugative sex pilus essential for their own transfer. Such plasmids are constructed for specific DNA transfer, and such lab made plasmids often called vectors.
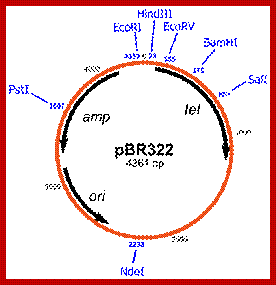
Plasmid as Cloning vector, where one introduce DNA from other sources into the plasmid which can be used the transfer that DNA to other systems. https://en.wikipedia.org
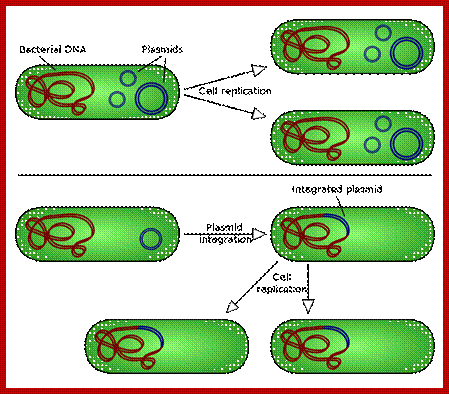
There are two types of plasmid integration into a host bacteria: Non-integrating plasmids replicate as with the top instance, whereas episomes, the lower example, can integrate into the host chromosome; https://en.wikipedia.org/
Characteristic Features of Some Plasmids:
|
Plasmid |
Size (kbp) |
Self transmissibility |
Copy nos. |
Features |
|
Col E1 |
6 |
No |
10-20 |
Colicin^+ |
R300 B |
9 |
No |
10-20 |
|
|
Psc-101 |
9 |
No |
4-6 |
TcR |
|
F^+ |
90 |
Yes |
1-2 |
F-pilius |
|
Col V |
90 |
Yes |
1-2 |
|
|
Ent-p30 |
90 |
Yes |
1-2 |
|
|
R1, R100 |
90 |
Yes |
1-2 |
Drug resistance |
|
R6K |
38 |
Yes |
10-20 |
|
|
RK2 |
56 |
Yes |
5-8 |
Broad host range |
|
Ti |
500 |
Yes |
1-2 |
Tumor inducing, transfer a part of its own DNA into host cell |
Rhi |
40-50 |
Yes |
1-2 |
Induces root nodules |
Host coded proteins in Replication:
|
Hosts |
RNAP |
DNA pol-I |
Dna-A |
Dna-B |
Dna-C |
Dna-G |
Dna-E |
Dna-T |
Col E1 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
|
PSC101 |
- |
- |
+ |
+ |
+ |
+ |
- |
|
|
R1, R100 |
- |
- |
+ |
+ |
+ |
+ |
- |
|
|
R6K |
+ |
- |
+ |
+ |
+ |
+ |
|
|
|
F^+ |
+ |
- |
+ |
+ |
+ |
+ |
|
|
|
RK2 |
- |
- |
+ |
+ |
+ |
+ |
|
|
|
RSF 1010 |
- |
- |
- |
- |
- |
+ |
|
|
|
|
|
|
|
|
|
|
|
|
Plasmid encoded proteins for initiation:
|
Plasmid |
Gene |
Protein |
Mol.wt (KD)/ subunits |
Function |
PSC 101 |
Rep-A |
Rep-A |
38/ 1 |
Helicase, 3’>5’ |
|
R1 |
Rep-A |
Rep-A |
33/ 1 |
Helicase, 3’>5’ |
|
R100 |
Rep-A |
Rep-A |
33/ 1 |
Helicase 3’>5’ |
|
R6K |
Pi-R |
Pi-R |
36/ 1 |
|
|
F^+ |
Rep-E |
Rep-E |
29 |
|
|
RK2 |
Trf-A |
Trf-A |
32-43 |
|
|
RSF 1010? |
Rep-C |
Rep-C |
31 |
|
|
PT 181 |
Rep-C |
Rep-C |
38 |
|
Replication in Col E1.Plasmid DNA:
Initiation on leading strand:
- The plasmid elicits resistance to colicin (bactericidal) products by ColE1.
Cole E1 –Colicin protein; http://www.chembio.uoguelph.ca/
- It doesn’t require de novo protein synthesis. Even in the presence chloramphenicol, replication of the plasmid continues.
- A characteristic feature of the plasmid is its unidirectional replication (?).
- For its replication it relies entirely on host products. It also produces few proteins such as colicin a resistant proteins and ROM proteins (Rnase-I modulating proteins).
- The origin of Coli-1 is located in a 600bp region flanked by a promoter for initiation of transcription. The region at which the DNA melts consists of AT rich sequences.
- About 555bp (it is a promoter region) left of the AT site, transcription is initiated at by host’s RNAP (a Holozyme, a pentamer). Transcription extends into the origin region. As transcription continuous into the region, the DNA melts into a bubble (it can also be called transcription bubble).
- The RNA produced doesn’t code for any protein and the transcript is called RNA-II. The RNA thus produced spontaneously organizes into secondary structures i.e. several stem- loops, which pairs with the DNA template on which it is produced (or pairs with mother template).
- This hybridization is a crucial step.
- An enzyme called RNase-H (the term derived from the fact this enzyme chews up only the RNA that is hybridized to DNA) at this point recognizes certain hybrid sequences and nibbles and produces a terminal with 3’OH. Now this RNA acts as the primer for leading strand. Immediately DNA pol-I recognizes this region and incorporates dNTPs and extends the chain to about 300-400ntd long. Then DNA Pol-III Holozyme displaces DNA Pol I and extends the DNA chain to synthesize continuous strand.
- The position or site at which Holozyme displace DNA Pol I to extend is called “pas” site (primosome assembly site).
- Leading strand initiation is sensitive to Rifamycin that means RNA Pol, unlike the Primase, produces the RNA primer.

ftp://ftp.tugraz.at/pub/Molekulare_Biotechnologie
Initiation on lagging strand:
· In the replication bubble, just about 15 bp downstream of the ori site, on the lagging strand, small secondary structure forms (stem loop structure), which acts as the recognition site for the assembly of another primosome complex.
· Primosome assembly is hierarchical and starts with Pri- A.
· Pri-A, Pri-B and Pri-C first assemble on the n-pas site, soon Dna-B assisted by Dna-C joins the Pri-ABC group in ATP dependent manner. At this point Dna-G, the primase joins.
· This primase is a monomer and resistant to Rifamycin unlike normal RNA-pol complex. The primase lays 11 ntds long RNA primer, whose ends are used by the DNA pol III Holozyme.
· Wherever the opened DNAs’ single strand is available, it is covered with SsBs.
· Initiation, though takes place on both leading and lagging strands in the replication fork, the fork movement is only in one direction, and so replication is unidirectional.
Regulation of Initiation:
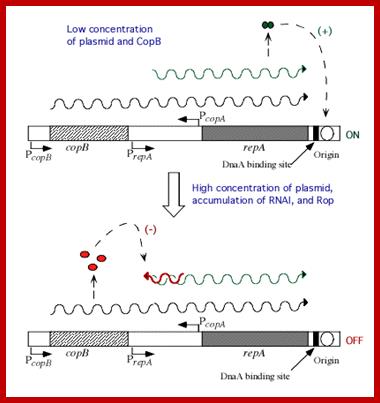
- Replication of the Col.E1 plasmid, in bacterial cells, is rapid. The copy number is more than 20.
- Species RNA-I, that is transcribed on the opposite strand from about 445 ntds upstream of the ori site, which is about 108 nucleotides long, and it is complementary to the RNA II. RNA-I can hybridize with RNA-II at its 5’ end.
- If RNA- I hybridize with RNA II before the latter can form secondary structure and anneal with its template, initiation is blocked. This is because primer mediated by RNase-H doesn’t form, hence RNA-I is a transacting regulatory factor of Col E 1 replication.
- Interesting thing about this regulation is that the 3’ end of the RNA-I forms three loops and binds to the three loops of RNA-II at its 5’ end. The rest of the RNA-I is linear does not form secondary structures and prevents further loop formation in RNA II.
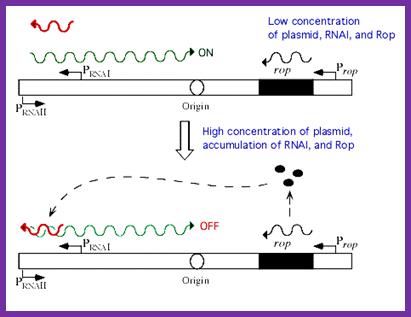
Regulation of plasmid colE1 copy numbers by antisense mechanism; http://www.sci.sdsu.edu/
Regulation of plasmid R1 copy numbers by antisense RNA: http://www.sci.sdsu.edu/
Inhibitory control over the RNA-II can be exercised only when the length of RNA II is just 15-200ntd long and before it hybridizes with the leading strand.
- Col E1 plasmid also produces a protein named as RoM, 7.5 KD (RNA One Modulator) or it is also called Rop (Repressor of primer). The ROM protein by binding to RNA-I, enhances the binding of RNA-I to RNA-II, thus prevent primer formation. Mutation in ROM gene, increase the copy number by six fold.
- Another feature of the ColE1 plasmid is its incompatible nature, where other kinds of plasmids cannot grow side by side with the plasmid in the same cell.
Required Components Col.E1 Plasmid Replication:
|
Protein |
Gene |
Mol.wt (KD) |
Subunits |
Functions
|
|
Dna-A |
Dna-A |
50 |
1 |
Binds to box A |
|
Pri-A |
|
76 |
1 |
Primosome initiation |
|
Pri-B |
|
11.5 |
2, dimer |
Primosome formation |
|
Pri-C |
|
23 |
1 |
Primosome formation |
|
Dna-B |
Dna-B |
50 |
6, hexamer |
Helicase, 5’ŕ3’ direction |
|
Dna-C |
Dna-C |
29 |
1 to with Dna-B |
Helps Dna-B in loading on to lagging strand in the fork region |
|
SsBs |
SsBs |
19 |
4, tetramer |
Bind to p-s-p backbone |
|
RoM |
|
7.5 |
Monomer |
Facilitates RNA-I and RNA-II hybridization |
|
RNA-pol |
|
> |
Pentamers |
Produces RNA primers on leading strand |
|
DNA-pol-I |
|
103 |
1 |
Removes primers and fills the gap |
|
DNA pol-III |
Many |
>512 |
>12 |
New Dna synthesis on both leading and lagging strands |
|
DNA-ligase |
|
|
1 |
Ligates ends |
-o-