Synthesis of Nucleotides:
To begin with nitrogenous bases are synthesized on phosphoribosyl groups, and then they are made into ribonucleotides, later they are converted to dNTPs. The biochemical steps are many and complicated; hence a simple representation of the pathway has been elucidated.
Pyrimidine synthesis:
Ribose + 3ATPs à pRpp + 3ADPs

Both the salvage and de novo synthesis pathways of purine and pyrimidine biosynthesis lead to production of nucleoside-5'-phosphates through the utilization of an activated sugar intermediate and a class of enzymes called phosphor-ribosyl transferases. The activated sugar used is 5-phosphoribosyl-1-pyrophosphate, PRPP. PRPP is generated by the action of PRPP synthetase and requires energy in the form of ATP as shown; Note that this reaction releases AMP. Therefore, 2 high energy phosphate equivalents are consumed during the reaction. http://themedicalbiochemistrypage.org/
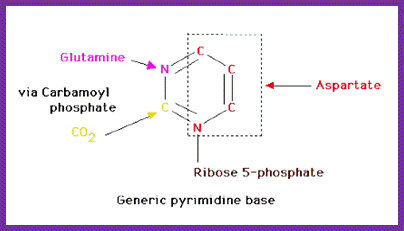
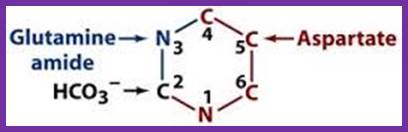
The starting compounds in the pathway are Aspartate and Carbamyl phosphate, which provide structural skeletal base to produce Orotic acid. The Orotic acid is then added to phospho ribosyl pyrophosphate (PRPP) to generate Orotate monophosphate. Elimination of carbon dioxide from Orotate generates rUMP.
Carbamoyl aspartate-> Dihydroorotate > Oratate>
Orotidylate >Uridylate monophosphate
Orotate monophosphate----à CO2 + rUMP
Then rUMP is converted to rUTP with kinase action.
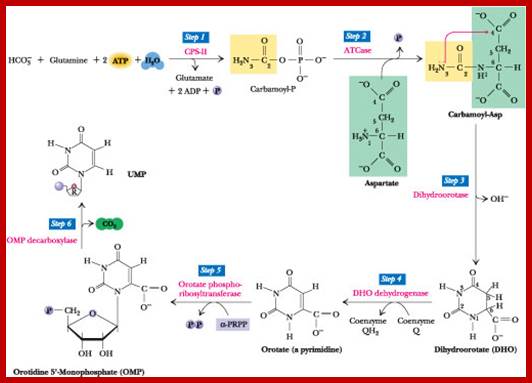
The de novo pyrimidine biosynthetic pathway. Step 1: Carbamoyl-P synthesis. Step 2: Condensation of carbamoyl phosphate and aspartate to yield carbamoyl-aspartate is catalyzed by aspartate transcarbamoylase (ATCase). Step 3: An intramolecular condensation catalyzed by dihydroorotase gives the six-membered heterocyclic ring characteristic of pyrimidines. The product is dihydroorotate (DHO). Step 4: The oxidation of DHO by dihydroorotate dehydrogenase gives orotate. (In bacteria, NAD+ is the electron acceptor from DHO.) Step 5: PRPP provides the ribose-5-P moiety that transforms orotate into orotidine-5'-monophosphate, a pyrimidine nucleotide. Note that orotate phosphoribosyltransferase joins N-1 of the pyrimidine to the ribosyl group in appropriate b-configuration. PPihydrolysis renders this reaction thermodynamically favorable. Step 6: Decarboxylation of OMP by OMP decarboxylase yields UMP. http://www.columbia.edu
In the synthesis of pyrimidine, the ring-system is completed first. Starting compounds are aspartate and carbamyl phosphate. The latter is produced in the cytosol from the amino residue of the amino acid glutamine, free bicarbonate and the terminal phosphate residue of ATP
rUMP + 2ATP = rUTP + 2rADP.
The rUTP can be used to generate rCTP by amination at 5’C of rUTP.
rUTP + NH2 = rCTP.
The rUTPs and rCTPs are used to produce Deoxy pyrimidine nucleotides.
Ribonucleotide CTP, by ribonucleotide reductase at 2’ sugar position is converted to dCTP.
rCTP à dCTP + (O)
However, synthesis of Deoxy ribothymidine starts with dUTP, and it is converted to dUMP by specific dephophorylation. Then by the action of Thymidillate synthase a mythyl (CH3) group is added to the 5C’ position of the base to form dTMP. The dTMP is converted to dTTP.
dUTP à dUMP + 2Pi
dUMP + CH3 à dTMP
dTMP + 2ATP = dTTP + 2 ADP
Purine synthesis:
The purine pathway is shaded; the de novo purine pathway starts with PRPP biosynthesis, while salvage pathways use PRPP to transform purine bases to monophosphate nucleotides. Arrows indicate a multi-step pathway; Jiménez et al. BMC Biotechnology 2008 8:67 doi:10.1186/1472-6750-8-67
To start with 5’ribose phosphate is converted to 5’phopspho ribosyl pyrophosphate (5’P ribose alpha PP) in separate steps.
5’ ribose + 3ATP = 5’P ribose3’ PP.
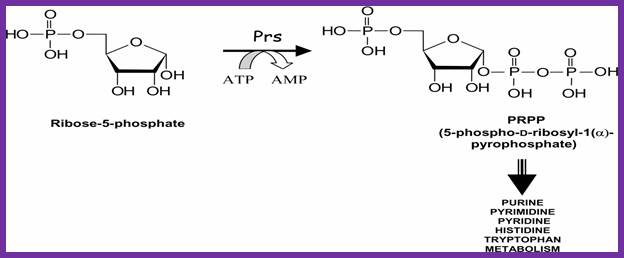
Synthesis of PRPP
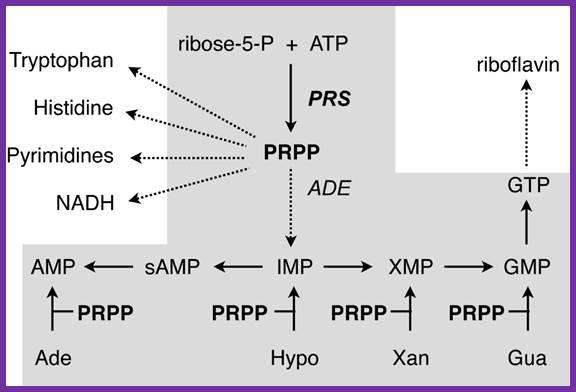
Metabolic contribution of PRPP to the purine biosynthesis and other anabolic pathways. Laura-Nadine Schuhmacher; http://www.keywordsking.com
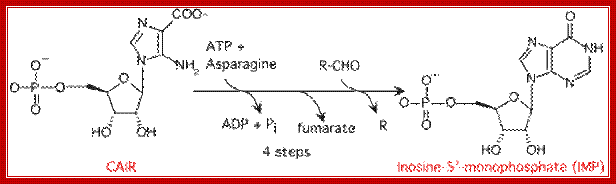
In a series of reactions, an Inosine ring is built up step by step on C1’ of the sugar to produce Ribosyl Inosine-Monophosphate (rIMP).
First committed step is addition of glutamine to 5’pR3’pp, then in a series of reactions Inosine monophosphate (rIMP) is generated.

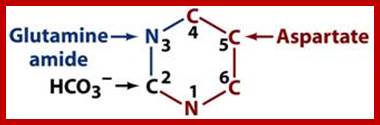
The biosynthetic origins of purine ring atoms;N1 arises from the amine group of Asp, C2 and C8 originate from formate N3 and N9 are contributed by the amide group of Gln C4, C5 and N7 are derived from Gly C6 comes from HCO3− (CO2); -Pyrimidine ring formation-First, synthesis of the pyrimidine ring. Next, attachment of ribose-phosphate to the ring. Further modification of the pyrimidine ring; https://en.wikipedia.org; http://www.uh.edu

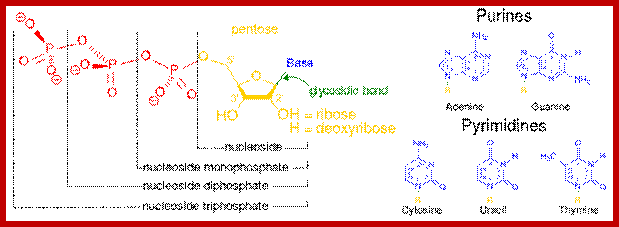
https://en.wikipedia.org

The amine group at 2nd C position is provided by Glutamine; required energy is provided by ATP. Polynucleotide chin elongating; https://www2.chemistry.msu.edu
![]()
http://www.columbia.edu/
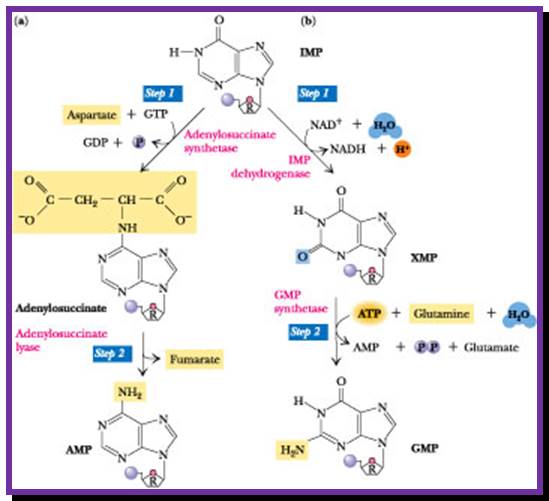
Then rIMP is converted to rGMP via Xanthine monophosphate and rAMP by another series of reactions. Ribonucleotide reductase converts ribose purines to deoxy purines. Further kinase reactions result in dGTP and dATP
rIMP àrXAMP à rGMP (amination to rXAMP at 2C’ position)
rGMP + 2ATP à rGTP or rATPs.
rXAMP à rAMP (by amination at 6C’ position).
rAMP + 2rATPs-à rATPs,
rGMP + 2rATPs -à rGTPs
rATPs and rGTPs by reductase action are converted into dATPs and dGTPs.
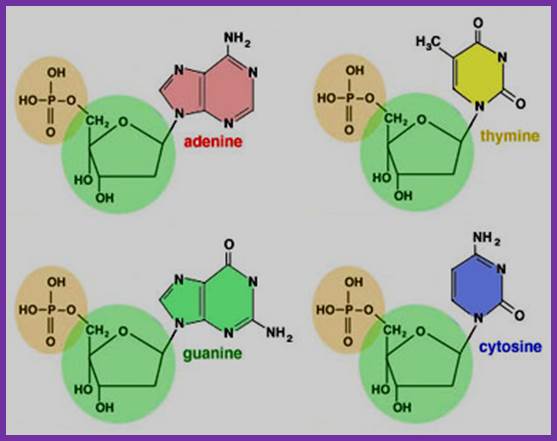
Nucleotides

http://www2.fiu.edu/
Importance of Nucleotides:
· They are the monomeric building blocks of DNA and RNA whose synthesis depends upon the availability of the triphosphate nucleotides. Synthesis of nucleotides is also regulated at various levels.
· ATP is the most important among the energy rich molecules and that is the source for most not all energy transducing processes in the cell (in an organism).
· Protein synthesis requires both ATPs and GTPs.
· ATPs and UTPs play an important role in carbohydrate synthesis as carriers- UDPGs & ADPGs
· GTPs, CTPs UTPs and ATPs are involved in glycosylation of proteins and lipids.
· GTPs are involved in Actin polymerization to F-actins.
· GTPs are involved in signal transduction process.
· ATPs are involved in Tubulin polymerization into Microtubules.
· Many metabolic pathways are regulated by ATPs and GTPs.
· ATPs and GTPs are also precursors for cAMPs and cGMPs respectively, which act as second messengers.
· ATP and GTP molecules also play an important role in cell cycle and developmental pathways.
· Adenine nucleotides are the structural components of coenzymes such as NAD, NADP, FMN, and FAD, CO-enzyme A.
· S-Adenosyl methionine is the precursor for ethylene.
· SAM is also used in methylation of DNA.
Purine Nucleotide Salvage Pathway: Adenine phosphorosyl transferase and Hypoxanthine-guanine phosphoribosyl transferase: (Wikipedia, the free encyclopedia):
APRTase is an enzyme involved in the purine nucleotide salvage pathway. It functions as a catalyst in the reaction between adenine and phosphoribosyl pyrophosphate (PRPP) to form AMP. Deficiency of APRT in human beings may lead to kidney stones formed of adenine and salts.
2, 8-Dihydroxy-adenine urolithiasis is also known as "adenine phosphoribosyl transferase deficiency
APRT is functionally related to hypoxanthine-guanine phosphoribosyl transferase (HPRT).
Hypoxanthine-guanine phosphoribosyl transferase (HGPRT) is an enzyme encoded in humans by the HPRT1 gene.
HGPRT is a transferase that catalyzes conversion of hypoxanthine to inosine monophosphate and guanine to guanosine monophosphate. This reaction transfers the 5-phosphoribosyl group from 5-phosphoribosyl 1-pyrophosphate to the purine. HGPRT plays a central role in the generation of purine nucleotides through the purine salvage pathway.
HGPRT catalyzes the following reactions:
|
Substrate |
Product |
Notes |
|
- |
||
|
Often renamed as HGPRT. Performs this function only in some species. |
||
|
Only certain HPRTs. |
HGPRTase functions primarily to salvage purines from degraded DNA to reintroduce into purine synthetic pathways. In this role, catalyzes in the reaction between guanine and phosphoribosyl pyrophosphate (PRPP) to form GMP.
Role in disease:
Mutations in the gene lead to Hyperuricemia:
· Some men have partial (up to 20% less activity of the enzyme) HGPRT deficiency that causes high levels of uric acid in the blood, which leads to the development of gouty arthritis and the formation of uric acid stones in the urinary tract. This condition has been named the Kelley-Seegmiller syndrome.[3]
· Lesch-Nyhan syndrome is due to HPRT mutations resulting in extremely ineffective enzyme activity.[4]
· Some mutations have been linked to gout, the risk of which is increased in Hyperuricemia.
Application of Hybridomas:
The B cells contain enzyme, which enables them to survive when fused to myeloma cells when grown on HAT medium to produce monoclonal antibodies. The antibodies are produced from cells called hybridoma cells. A hybridoma, which can be considered as a hybrid cell, is produced by the injection of a specific antigen into a mouse, procuring the antibody-producing cell from the mouse's spleen and the subsequent fusion of this cell with a cancerous immune cell called a myeloma cell. The hybrid cell, which is thus produced, can be cloned to produce many identical daughter clones. These daughter clones then secrete the immune cell product.
The method of selecting hybridomas is by use of HAT medium, which contain hypoxanthine, aminopterin, and thymidine. The aminopterin inhibits enzyme dihydrofolate reductase (DHFR), which is necessary in the de novo synthesis of nucleic acids. Thus, the cell is left with no other option but to use the alternate salvage pathway, which utilizes HGPRT. In the HAT medium, HGPRT- cell lines will die, as they cannot synthesize nucleic acids through salvage pathway. Only HGPRT+ cells will survive in presence of aminopterin, which are the hybridoma cells and plasma cells. The plasma cells eventually die as they are mortal cell lines, thus only hybridoma cells are left surviving. The hybrid cell (hybridoma cell) can be cloned to produce many identical daughter clones. These daughter clones subsequently secrete the monoclonal antibody product.
