Viroids Virusoids And Prions
There are a group of genetic RNAs, whose size is very small ranging from 240 to 375 ntds long, naked (i.e. not covered or enclosed by any capsid proteins, infectious and pathogenic agents, are called VIROIDS.
There are another type of genetic RNAs, incapable of replicating by themselves and reside in other viruses and depend upon them for their replication. They can cause diseases; they are called VIRUSOIDS or SATELLITE RNAs.
There is another class of infectious agents, causing serious diseases, which are neither DNA or nor RNA, but conformationally changed proteins, which can propagate by inducing conformational changes in other proteins of the same kind; thus their number increases with time; this is a class by itself but unique and they are called PRIONS.
Viroids, virusoids and prions are unusual infectious agents characterized by having a very small genome and in the case of prions, possibly no genome at all: by Reed Wickner
Viroids:
Viroids are now considered as the most potent pathogenic agents, which cause diseases that can devastate certain crop plants and cause serious economic problems, at least some parts of the world. More than 25 to 30 different viroids, with numerous variants in each type, have been identified. Viroids are common plant pathogens which cause serious economic problems. Twenty five or more different viroid sequences have been determined and numerous variants identified:
|
Avsunviroidae: |
e.g. avocado sunblotch viroid, peach latent mosaic viroid |
|
|
Pospiviroidae: |
Subgroup 1: |
potato spindle tuber viroid, coconut cadang cadang viroid, tomato plant macho viroid |
|
Subgroup 2: |
citrus bent leaf viroid, pear blister canker viroid |
|
· No proteins are encoded
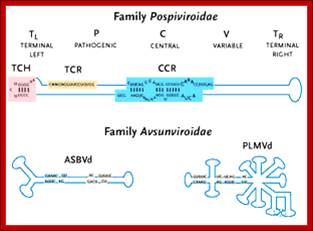
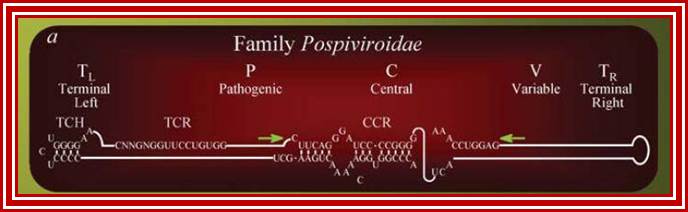
The structure of a Pospiviroid is indicated schematically below:

Structure of B group Viroids shown; http://www.mcb.uct.ac.za/
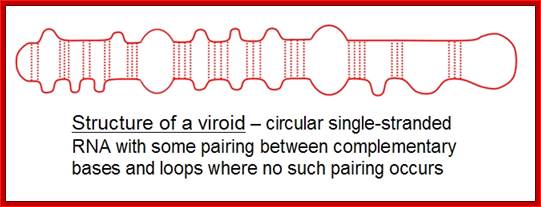
It is a circular single-stranded RNA with certain regions are complementary base-paired; www.cronodon.com
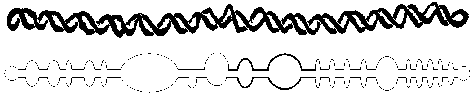
Potato spindle viroid RNA genome and its folded with base pairs and bulbs; www.public.iz –uni-duesseldorf.de
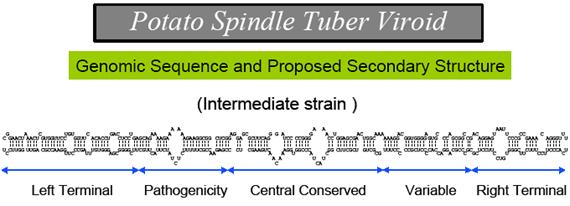
This is potato spindle viroid; http:// www.apsnet.org
Pospiviroidae are a large group, only 2 members of the Avsunviroidae are currently known. The RNA genomes of viroids are 246-375 nucleotides in length and share many similarities. The RNA folded into secondary structure with central conserved domain, double stranded pathogenic domain and open bulges called RH terminal domain.
- They are all single stranded covalent circles
- There is extensive intramolecular base pairing
- A DNA-directed RNA polymerase makes both plus and minus strands
- Replication does not depend on the presence of a helper virus
- Almost look like ds rods of RNA with short open unpaired regions.
- They are resistant to DNase but sensitive to RNase activity.
· Replication is independent of helper virus.
· The Viroidal RNA does not encode any information for a protein.
· DNA directed host RNA polymerase II synthesizes and replicates both plus and negative RNA strands.
· The ds RNA (partial), rigid, rod like, is not associated with any proteins, strange?
· Replication shows rolling circle mode.
The classification described above is based on analysis of the central conserved region (CCR). Members of the Avsunviroid group are clearly different to those described above . They lack a CCR and possess a ribozyme activity (a ribozyme is a catalytic RNA molecule, in this case RNA cleavage is the ribozyme activity). Additionally it is speculated that Avsunviroids may replicate in chloroplasts whereas Pospiviroids replicate in the nucleus and nucleolus.
Three enzymatic activities are required for viroid replication, an RNA polymerase, an RNase and an RNA ligase. Avsunviroids probably replicate via a symmetric rolling circle mechanism, whereas Pospiviroids probably use an asymmetric mechanism. By this I mean the +ve infecting circular RNA strand of a viroid serves as a template to make a large linear multimeric -ve strand. RNA pol II is probably the enzyme which does this. Pospiviroids with an asymmetric replication pathway they make +ve RNA from this long linear molecule. A host RNase activity cleaves the +ve strand into unit viroid lengths. This molecule is then ligated to form a circular viroid. In Avsunviroid replication the long -ve RNA is self cleaved by the associated ribozyme activity. The RNA circularizes to form a -ve circle. A second rolling circle event makes a long linear +ve strand which is again cleaved by the ribozyme activity.

Viroidal RNA Replication; once inside the cell it is taken into the cell nucleus, where the plus strand copied to produce negative strand. The negative strand acts as a the template for producing positive strands which is done by host enzymes (it must be a reverse transcriptase). The RNA transported out and it can infect host cells; http://www.microbiologytext.com
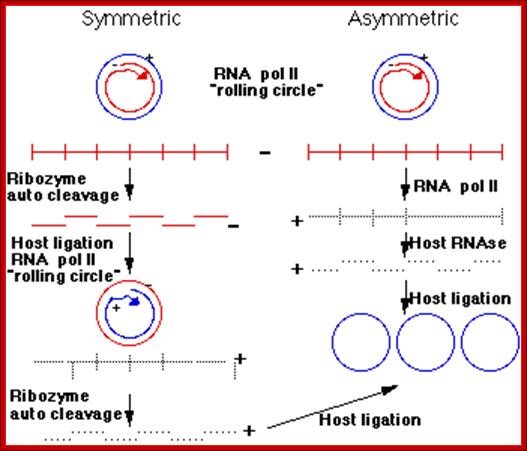
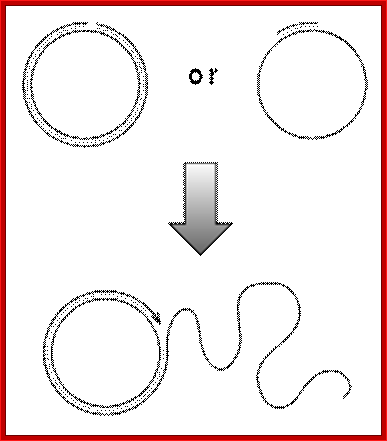
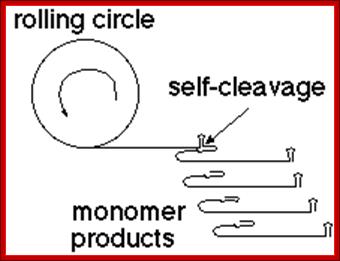
Rolling circle replication; http://en.wikipedia.org/
Probably there is more than one mechanism responsible for viroid pathogenesis. Recent evidence suggests that one pathway is due to viroid RNA activating a ( in plants) RNA activated protein kinase, or PKR (analogous to the PKR enzyme activated by viral RNAs in mammalian cells). Protein synthesis is reduced and this causes pathogenic effects. In the case of potato spindle tuber viroid, there is a good correlation between a strains’ pathogenicity and its ability to activate PKR in vitro.

Depending upon certain distinguishing features they have been classified into two groups; group-A and group-B, with few subgroups in each.
|
Group A viroids |
Group B viroids |
|
Ex. Avocado sun blotch viroids, Peach latent mosaic viroids |
B-1: Potato spindle tuber viroids, Coconut-cadang cadang viroids, Tomato plant Macho viroids. B-2: citrus bent leaf viroids, Pear blister canker viroids |
|
|
|
|
Group A viroids |
Group B viroids
|
|
This group is lacking in centrally Conserved region called (CCR).
|
Group- B does contain central Consensus domain (CCR).
|
|
They possess Ribozyme activity. Replicate in chloroplasts.
|
They lack Ribozyme activity, They Replicate in the nucleus and Nucleolus.
|
|
For viroid replication, they require RNA polymerase, an Rnase and an RNA ligase.
|
Replication of the viroids require RNA polymerase, an Rnase and RNA ligase.
|
|
Replication of the viroids require RNA polymerase, an Rnase and RNA ligase.
|
Replication of the viroids require RNA polymerase, an Rnase and a RNA ligase.
|
|
Replication of the viroids require RNA polymerase, an Rnase and an RNA ligase.
|
Replication of the viroids require RNA polymerase, an Rnase and RNA ligase.
|
|
Concatameric, multimeric (-) RNA.
|
They replicate by asymmetric Mode, it means they make a Long, multimeric and linear – RNA strand by rolling circle Mechanism.
|
|
They replicate by asymmetric Mode, it means they make a Long, multimeric and linear – RNA strand by rolling circle Mechanism.
|
This long RNA is again copied Into linear + strand. RNA polymer- Raze -II performs this process. Host Rnase cleaves +RNA into Viroidal units and ligated to gen- erate + circular molecules.
|

Tomato macho viroids: Infected-left and healthy plants-right
General Viroidal RNA structural features:
CC CCGG(LH)- variable-central-conserved--GGUGGCC-(RH)
A short list of Viroids:
|
Viroid name |
Acronym |
Size of the genome (ntds) |
|
Potato spindle tuber viroid |
PSTV |
359 ntds. |
|
Avocado sun blotch viroids |
(ASBV) |
247 |
|
Citrus exocortic viroid |
CEV |
371 |
|
Chrysanthemum stunt viroid |
CSV |
354 |
|
Coconut cadang viroid |
CCCV |
246 |
|
Hop stunt viroid |
HSV |
297 |
|
Cucumber pale fruit viroid |
CPFV |
303 |
|
Peach latent mosaic viroid
|
(PLMV) |
|
|
Tomato plant Macho viroid
|
(TPMV) |
|
|
Pear blister canker viroid
|
(PBCV) |
|
|
|
|
|
- Most of the viroids identified, so far, are plant viroids, which cause considerable damage to commercial crop plants.
- They have single stranded RNAs, circular, and 80- 90% of the RNA is base paired to form a rod like filament, but with some unpaired region as open loop
- Viruses do not code for any protein, they are not associated with any proteins. (?
- They may be located in cytoplasm or in cytoplasmic plastids, because these are the sites of replication.
- Host RNA polymerase-II, host RNase and host RNA ligases are used for the replication of Viroidal RNA.
- But some Viroidal RNA possesses self-cleaving property, and behaves as Ribozymes.
· Generally, replication begins at a specific site, called replication initiation site, similar to that ssDNA replication. But the negative strand formed continues to elongate, in rolling circle mode, thus it generate a long, multimeric, linear but concatameric RNA.
· The synthesis of (+) strand may vary.
· In A-type, the linear (–) RNA is cleaved by its Ribozyme activity, and circularized by RNA ligase. The circular (—) RNA is copied again in rolling circle mode, and then they are cleaved into full-length Viroidal segments and circularized.
· In B type the linear (–) RNA produced is copied into linear (+) RNA, which is then cleaved and circularized.
· The site of replication differs; A type replicates in cytoplasm and chloroplasts and B- type in nucleus.
· Interestingly replication is inhibited by Actinomycin-D and alpha Amanitin.
Bare-nacked Viroids; http://www.the-scientist.com/
Viroids, which cause a disease in one species, don’t affect the other species. They are generally transmitted through insects, which live on plant sap. The mechanism of disease manifestation is still not very clear. At least, what has been found in .Potato spindle tuber viral, infected plants, can be taken as an example to explain the process? In PSTV infected plant sap a specific protein is found which is heavily phosphorylated, than the proteins found in uninfected plant. This is more or less similar to that of interferon infected cells where a protein kinase activated by a double stranded RNA, inhibits protein synthesis by phosphorylation of e-EF-2a. Perhaps the ds Viroidal RNA may activate a cellular protein kinase, which might have the role in causing disease. Inhibition or reduction of cellular protein synthesis has wide implications and it is perhaps the cause for the disease.
Sequence elements outside the hammer-headed ribozyme catalytic core enable intracellular activity: Anastasia Khorovva et al.
http://maptest.rutgers.edu/
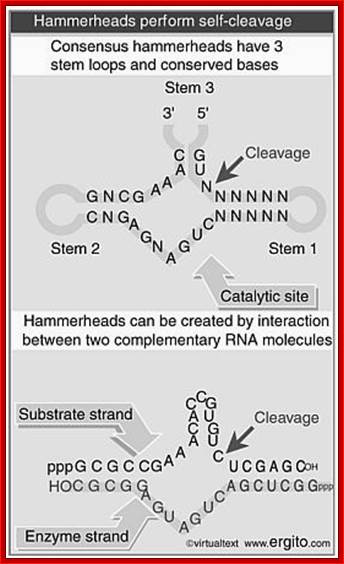
Hammer-headed ribozyme
The mode of self-splicing activity in type-A viroids, and self-splicing group-I introns of rRNA and mitochondrial RNA show same or similar mechanisms. Based on these studies some people believe that these Viroidal RNAs are considered as escaped introns, or molecular fossils having self replicating, self catalyzing RNAs activity once existed in pre cellular world that formed about 3.2 to 3.5 billion years ago.
The hammerhead ribozyme (HHRz) is a small, naturally occurring ribozyme that site-specifically cleaves RNA and has long been considered a potentially useful tool for gene silencing. The minimal conserved HHRz motif derived from natural sequences consists of three helices that intersect at a highly conserved catalytic core of 11 nucleotides. The presence of this motif is sufficient to support cleavage at high Mg2+ concentrations, but not at the low Mg2+ concentrations characteristic of intracellular environments. Here we demonstrate that natural HHRzs require the presence of additional no conserved sequence elements outside of the conserved catalytic core to enable intracellular activity. These elements may stabilize the HHRz in a catalytically active conformation via tertiary interactions. HHRzs stabilized by these interactions cleave efficiently at physiological Mg2+ concentrations and are functional in vivo. The proposed role of these tertiary interacting motifs is supported by mutational, functional, structural and molecular modeling analysis of natural HHRzs.

Tomato Chlorotic Dwarf Viroid; http://www.agf.gov.bc.ca/
Another example - viroids are infectious RNA molecules (e.g. the 359 nt RNA of Satellite Tobacco Ringspot Virus) that replicate by rolling circle transcription from an RNA template. The polymeric product self-cleaves to yield individual monomer units of the genome.
|
Self-cleavage of viroid RNAs |
|
|
Processing of the viroid genome depends on the presence of 13 required nucleotides, and the formation of a specific secondary structure surrounding the cleavage site.
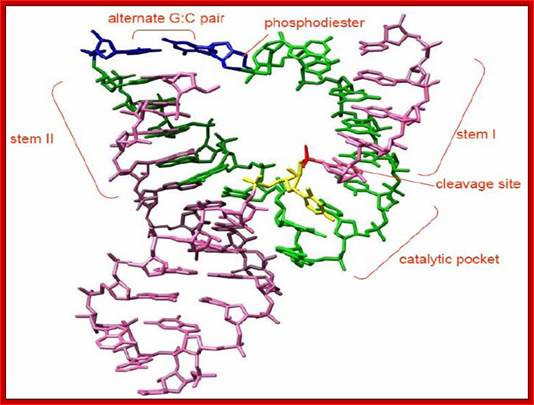
Hammer Headed Ribozyme:

The minimal consensus structure for the hammerhead ribozyme is shown above. The sequences shown as outlines are required for efficient cleavage. Residues indicated by N can be any base and N´ indicates the complement of the base with which it is paired. The Hat the cleavage site means any base but G. The lines at the ends of stems I, II , and III indicate that any number of bases may connect the two halves of each stem, or they need not be connected at all. In the 3-D structure to the left and the secondary structureabove, the strand cleaved is shown in red and the rest of the molecule is shown in blue. http://www.tulane.edu/
Detail of the cleavage site: The
above figure shows a secondary structure, a "hammerhead", that
chelates magnesium, and forms a self-cleaving RNAZyme structure. This can be
put to use in many ways.
Therapeutic;
Anti-Hepatitis
C ribozyme May 11, 2000 "Administration of LY466700 to chronic Hepatitis C
patients has now been initiated in a clinical trial designed to study safety
and to assess the effect of the compound on HCV viral RNA levels following a 28
day dose-response regimen. The drug will be administered by a daily
subcutaneous injection to approximately 20 patients."
http://www.hepnet.com/hepc/news051100.html
ANGIOZYMETM "is the first chemically synthesized ribozyme to be
studied in human clinical trials. ANGIOZYME(TM) specifically inhibits formation
of the VEGF-r (Vascular Endothelial Growth Factor receptor), a key component in
the angiogenesis pathway." http://www.slip.net/~mcdavis/database/angio183.htm
Anti-HIV ribozyme "The Hammerhead anti-gag ribozyme catalytically cleaves
HIV-1 RNA within the gag open reading frame, blocking protein synthesis of the
gag-encoded p24 capsid protein (1). The Hammerhead anti-gag ribozyme is
introduced into cells through through transformation of target cells with a
ribozyme RNA expression vector" http://www.niaid.nih.gov/daids/dtpdb/000681.htm
Diagnostics
Allozymes
- Allosteric ribozymes "are a class of ribozymes that are activated to
cleave a reporter RNA in the presence of a target analyte. The resulting signal
from the cleavage of the reporter RNA can be readily measured. Allosteric
ribozymes have multiple diagnostic applications, including detecting and
quantifying a wide range of nucleic acids, proteins and small molecules."http://www.rpi.com/diagnost.jsp.
HalfzymesTM - broadly applicable
and is well suited for direct nucleic acid screening of blood products for
viral contamination, determination of viral drug resistance, and for the
detection of single nucleotide polymorphisms (SNPs) relevant to human health. In
the absence of nucleic acid target, the technology lacks sequences required to
form the catalytic core and to properly dock a tethered substrate RNA that
serves as a reporter. A target nucleic acid supplies these sequences. http://www.rpi.com/diagnost.jsp
The idea of an RNA molecule binding to a ligand (similar to the Allozymes) is not going to seem strange for very long! Winkler et al. described, in the Oct 31, 2002 issue of Nature, that mRNA can be involved in allosteric regulation. The example given was vitamin B1 biosynthesis in E. coli, where the mRNA encoding enzymes for biosynthesis can bind to thiamine. When this binding occurs, the mRNA changes conformation and the ribosome can no longer bind to the ribosome binding site. Halfzymes - maybe they work like this. You start with a target, and design a ribozyme to match it so that you can form an activating secondary structure.
Origins and spread; A number of features of viroids suggest they may have originated in the hypothetical prebiotic RNA world. They can possess a ribozyme activity. They are GC rich which would attenuate the low fidelities of replication activities. They are circular and so do not require start and stop functions for replication. They move within a plant, probably in association with host proteins via the phloem vascular channels and plasmodesmeta cell contact points. They seem to form a quasi-species population and can recombine. Spread, at least in commercial crops, seems to be very dependent upon the activities of Man. Viroids of a particular type are widespread in some areas and absent in others.
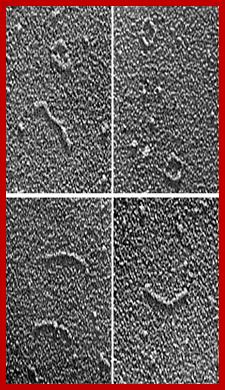
Viroids under EM; http://schaechter.asmblog.org/
1. Environments are composed of molecules occupying locations in a metric space with continuous or discreet time.
2. There are an infinite variety of molecules possible.
3. Reactions are transformations of molecules in close proximity to other molecules in close proximity, some of the resulting molecules being of types different from the reactants.
4. There are an infinite number of possible reactions having positive probabilistic rates of reaction.
5. Nucleotide elements of polymers spontaneously change at a low rate compared with the rate of nucleotide replication or complementation. These changes are called mutations.
6. There exist nucleotide molecules which will replicate or complement a nucleotide polymer (template) under conditions which occur periodically.
7. There are periodic conditions under which the replicated or complemented polymer will separate from its template.
8. A nucleotide polymer species is called enduring if the probability of it or a replica surviving a replication cycle (polymerizing and separating period) is greater than one half. An unbounded number of distinct nucleotide polymer species are enduring.
9. There are periodic conditions under which these nucleotide molecules will spontaneously bond together, forming or lengthening polymers.
10. Some combinations or permutations of nucleotide molecules interact with other molecules differently than do other combinations of nucleotide molecules.
These conditions imply that there is some source of the nucleotide molecules (either new or recycled); and that those molecules are periodically available.
The periods may be continuous and simultaneous. Condition 9, bootstraps the process so there will be polymers to replicate.
Condition 10 guarantees that the genetic information matters; the arrangement of nucleotides in a polymer effects how it interacts with other molecules.
Because more than half of them survive, enduring molecules which replicate themselves will grow to outnumber the polymer species which don't endure. Certain species of nucleotide polymers will replicate more easily or have higher survival rates; such attributes will cause their numbers to increase relative to the others.
Notice that the nucleotide game has replication, competition, and evolution; all without membranes or proteins.
How will this game play out? Evolution of the Nucleotide Game continues.
l
The first symptoms of PSTVd infection in tomato (Figure 2) are growth reduction and chlorosis in the top of the plant. Subsequently, this growth reduction may develop into stunting, and the chlorosis may become more severe, turning into reddening and/or purpling. In this stage, leaves may become brittle. Generally, this stunting is permanent; occasionally, however, plants may either die or partially recover. stunting begins, flower and fruit initiation stop.
PSTV on Tomatoplants; https://www.apsnet.org
PSTV; 1971 yılında ABD Tarım Biriminden T. O.
Diener,
Viroids are plant pathogens that consist of a short stretch (a few hundred
nucleobases) of highly complementary, circular, single-stranded RNA without the
protein coat that is typical for viruses. The smallest so far is a 220 nucleobases
scRNA (small cytoplasmic RNA) associated with the rice yellow mottle
sobemovirus (RYMV)[1]. In comparison, the genome of the smallest known viruses
capable of causing an infection by themselves is around 2 kilobases in size.
The human pathogen hepatitis-D is similar to viroids. Viroids were discovered
and given this name by Theodor O. Diener, a plant pathologist at the
Agricultural Research Service in Maryland, in 1971.
Viroid RNA does not code for any known protein; some even lack the AUG
initiation codon. The replication mechanism involves interaction with RNA
polymerase II, an enzyme normally associated with synthesis of messenger RNA,
and "rolling circle" synthesis of new RNA. Some viroids are
ribozymes, having RNA enzyme properties which allow self-cleavage and ligation
of unit-size genomes from larger replication intermediates. It has been
proposed that viroids are "escaped introns".
Viroids are usually transmitted by seed or pollen. Infected plants can show distorted growth. The first viroid to be identified was the Potato spindle tuber viroid (PSTVd). Some 33 species have been identified
Taxonomy:
Family: Pospovroae,
Genus Pospiviroid; type species: Potato spindle tuber viroid,
Genus
Hostuviroid; type species: Hop stunt viroid
Genus Cocadviroid; type species: Coconut cadang-cadang viroid
Genus Apscaviroid; type species: Apple scar skin viroid
Genus Coleviroid; type species: Coleus blumei viroid 1;Family Avsunviroidae
Genus Avsunviroid; type species: Avocado sunblotch viroid
Genus Pelamoviroid; type species: Peach latent mosaic viroid
There
has long been confusion over how viroids are able to induce symptoms in plants
without encoding any protein products within their sequences. Evidence now
suggests that RNA silencing is involved in the process. First, changes to the
viroid genome can dramatically alter its virulence. This reflects the fact that
any siRNAs produced would have less complementary base pairing with target
messenger RNA. Secondly, siRNAs corresponding to sequences from viroid genomes
have been isolated from infected plants. Finally, transgenic expression of the
noninfectious hpRNA of potato spindle tuber viroid develops all the corresponding
viroid like symptoms.
This evidence indicates that when viroids replicate via a double stranded
intermediate RNA, they are targeted by a dicer enzyme and cleaved into siRNAs
that are then loaded onto the RNA-induced silencing complex. The viroid siRNAs
actually contain sequences capable of complementary base pairing with the
plant's own messenger RNAs and induction of degradation or inhibition of
translation is what causes the classic viroid symptoms.
Replication
HDV's unique replication distinguishes it from other animal viruses. Although, HDV infection depends on the presence of Hepatitis B Virus (HBV), HDV replication does not require the presence of HBV. HBV is needed for the production of HDV virions. After entering the cell nucleus and uncoating, the HDV genome uses the cell’s RNA polymerase II to transcribe two types of RNA: (1) circular, plus sense RNA, called the antigenome; and (2) shorter, polyadenylated, linear RNA. The circular RNA is used as a template for new genomic RNA. The linear RNA is transported to the cytoplasm where it undergoes translation to new HDAg.
Three RNAs of HDV
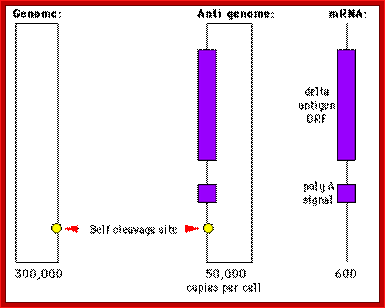
Source: http://www.mcb.uct.ac.za/; http://www.tulane.edu/
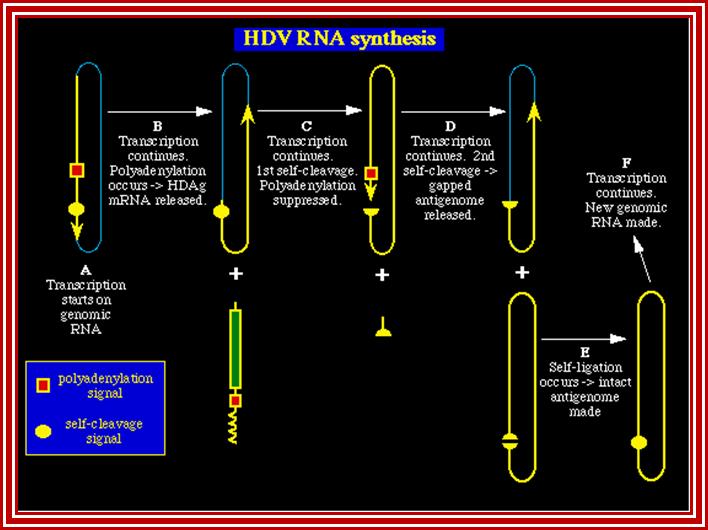
HDV is unusual because it is the only human negative-stranded RNA virus that links together the transcription of both the antigenome and the mRNA. Using a mechanism known in many plant viruses as the double rolling circle model, the genome transcribes both RNAs with a single initiation. In Step A {Figure 2}, transcription starts and continues through the polyadenylated and self-cleavage sites. Polyadenylation processes the RNA and releases the HDAg mRNA that travels to the cytoplasm (Step B). The antigenomic RNA strand undergoes self-cleavage and continues transcribing for another 1.7kb (the length of the genomic RNA). Self-cleavage somehow prevents normal downstream fragment degradation. Usually after mRNA processing, the unused RNA is degraded. Not in this case! During this continued transcription, polyadenylation is suppressed (Step C). After transcribing 1.7kb, the second self-cleavage occurs. This releases the unit-length antigenome RNA (StepD). The antigenome (+) undergoes self-ligation to form the circular template for the genomic RNA (-) (Step E). The virus uses RNA polymerase II again to transcribe copies of the genome.
Assembly of the HDV virion requires the envelope proteins (HBsAg) of the helper hepadnavirus, HBV. Without the envelope the virus cannot replicate properly. This is why coinfection or super infection with HBV is necessary.
 "A pseudoknot-like structure required for efficient
self-cleavage of hepatitis delta virus RNA. Perrotta,
Anne T.; Been, Michael D. " Nature v350,
n6317 (April 4, 1991):434.
"A pseudoknot-like structure required for efficient
self-cleavage of hepatitis delta virus RNA. Perrotta,
Anne T.; Been, Michael D. " Nature v350,
n6317 (April 4, 1991):434.
On the role of RNA silencing in the pathogenicity and evolution
of viroids and viral satellites:
On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites; http://www.pnas.org/
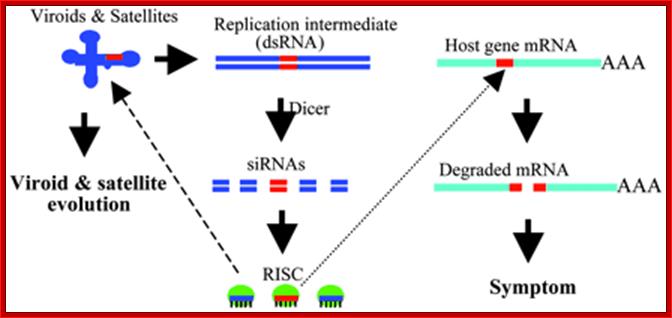
A model for the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellite: Replication of the sub viral RNAs generates dsRNA intermediates, which are processed by Dicer into 21- to 25-nt siRNAs, and these siRNAs are then incorporated into siRNA-ribonuclease complexes (RISC). If significant sequence identity exists between a region in the sub viral RNA genome and a region in host gene mRNA (shown in red), RISC will target the host gene for degradation leading to symptom development. RISC can also target the sub viral genome for degradation, forcing the sub viral RNA to evolve and to adopt and maintain RNA silencing-resistant secondary structure.
Virusoid and Satellites;
Virusoids are genetic RNAs but have no independent existence. The size of these RNAs is several hundred nucleotides long, circular, live within capsid core of another virus, called helper virus. They totally depend on the helper virus for their replication.
Five Virusoid RNA genomes are 220-338 nucleotides long, circular, single stranded and possess a ribozyme activity. They can replicate in the cytoplasm using an RNA-dependent RNA polymerase. This enzymatic activity is common in plants but not found in animal cells. They depend on a helper virus for replication. This helper virus also encapsidates them, e.g: Subterranean clover mottle virus satellite RNA: Helper - Sobemovirus
A Virusoid genome does not code for any proteins, but instead serves only to replicate itself. Virusoids can replicate in the cytoplasm and possess a ribozyme activity. RNA replication is similar to that of viroids, but each requires that the cell be infected with a specific "helper" virus. Five virusoids are known, and the helper viruses for these are all members of the Sobemovirus family. An example of a "helper" virus is the subterranean clover mottle virus, which has an associated virusoid. Virus enzymes may aid replication of the virusoid RNA. The virusoid is incorporated into the virus particle and transmitted as a "satellite," a separate nucleic acid not part of the viral chromosome. Replication of the helper virus is independent of the virusoid.
Virusoids belong to a larger group of infectious agents called satellite RNAs, found in bacteria, plants, fungi, invertebrates and vertebrates. Satellite genomes encode proteins, satellite viruses encode capsid proteins but are still dependent upon a helper virus for replication. Examples include:
- Barley yellow dwarf virus satellite RNA: Helper - Luteovirus Tobacco ring spot virus satellite RNA: Helper - Nepovirus These agents may modify the symptoms of infection by their helper virus. They do no interfere with the replication of their helper virus and are therefore differentiated from defective interfering particles that are associated with many viral infections.
Virusoids are circular single-stranded RNAs, dependent on plant viruses for replication and encapsidation. The genome of virusoids consists of several hundred nucleotides and only encodes structural proteins.
Virusoids are similar to viroids in size, structure and means of replication (rolling-circle replication). Virusoids, while being studied in virology, are not considered as viruses but as sub viral particles. Since they depend on helper viruses, they are classified as satellites. In the virological taxonomy they appear as Satellites/Satellite nucleic acids/Subgroup 3: Circular satellite RNAs.
The term virusoid is also sometimes used more generally to refer to all satellites
Names of few virusoids:
Barley yellow dwarf virus: Helper virus-luteo virus.
Tobacco ring spot virus: Helper virus-nepovirus.
Subterranean clover mottle virus: Helper- sobemo virus.

- Most of them are found in plant systems, but the presence of such satellite RNA in animal systems is not an unusual feature, ex. Hepatitis delta virus (HDV).
- Virusoids replicate in cytoplasm using RNA dependent RNA polymerase. This kind of enzyme activities is common in plants than in animal systems.
They can be spread by vegetative propagation, within seeds or by direct inoculation either by insects or man.
There
are similar infectious agents which infect animals, e.g. newt satellite 2
transcript. One such agent infecting humans is the hepatitis
delta virus (HDV).
HDV was first identified in the 1970s in Australia as a nuclear antigen, the
delta antigen. Subsequently, it was found to be the cause of a particularly
virulent form of hepatitis known as type D hepatitis.
Common
in indigenous natives of S.America, method of transmission not understood. They
can be transmitted prenatally. In the West, transmission is associated with
drug abuse and transfusion of blood products. It seems prudent to assume that
it can also be transmitted sexually. No specific treatment for the infection.
The delta antigen is associated with a defective pathogen which is obligatorily
associated with Hepatitis B helper virus:

This virus has a circular single stranded RNA genome of about 1700 nucleotides. It has a ribozyme (RNA cleavage) activity. This is the smallest known genome for an animal virus.
RNA and delta antigen (195 AA) are packaged in a Hepatitis B particle. No DNA intermediate has been detected during the replication phase and it is thought that replication occurs by RNA directed RNA synthesis using a DNA dependent RNA polymerase. Certain parts of the genome and the pattern of replication is of course similar to a viroid. One difference is that mRNA and a protein - the delta antigen - are made.
The encapsidated RNA is (-) ve sense strand so (+)strand RNA synthesis to make the mRNA is required. These RNAs are nuclear associated and in any case circular RNAs are not good templates for protein synthesis. About 10% of the antigenomic strand is cleaved and polyadenylated and serves as a 800nt mRNA. RNAse activities tend to be exonucleolytic rather than endonucleolytic. The fact that the genomic strands are circular probably contributes to the agent’s stability.
The different RNAs made by this virus are indicated in the diagram shown below: The genomic strand is 70% Watson Crick base paired and rod like in gross structure.
The delta antigen is a 22kd nuclear phosphoprotein essential for replication and particle formation. It is basic and associates specifically with the RNA genome thereby stabilizing it. Two forms are made differing by 19 amino acids at the C-terminus. The large form is a dominant inhibitor of genome regulation and directs genome packaging into HBV virus particles. This packaging is due to farnesylation of a Cys residue 4 AA from the C-terminus. Recently a host protein interacting with the delta antigen has been identified. In fact sequence similarities suggest it is a cellular homologue of the delta antigen. This suggests that it may be able to modulate viral replication. It may also suggest that HDV originated from a viroid like element which then "captured" a cellular transcript. Probably the genomic strand transferred onto the mRNA for the cell homologue of the delta antigen. This CAN BE copied into the antigenomic strand and stabilize as part of the genome.
These agents all have a small genome. They replicate, spread, and if appropriate, form particles, in much the same way as all of the other viruses you will hear about during this course. One could almost say that there is a continuum between naked replicating transmissible RNAs, depended packaged transmissible RNAs, simple viruses and complicated viruses like the herpesviridae and poxviridae. Organelles and obligate cellular bacterial parasites like rickettsia would seem to represent a different lineage completely (because of the independent capacity for protein synthesis).
Replication
HDV's unique replication distinguishes it from other animal viruses. Although, HDV infection depends on the presence of Hepatitis B Virus (HBV), HDV replication does not require the presence of HBV. HBV is needed for the production of HDV virions. After entering the cell nucleus and uncoating, the HDV genome uses the cell’s RNA polymerase II to transcribe two types of RNA: (1) circular, plus sense RNA, called the antigenome; and (2) shorter, polyadenylated, linear RNA. The circular RNA is used as a template for new genomic RNA. The linear RNA is transported to the cytoplasm where it undergoes translation to new HDAg.
HDV is unusual because it is the only human negative-stranded RNA virus that links together the transcription of both the antigenome and the mRNA. Using a mechanism known in many plant viruses as the double rolling circle model, the genome transcribes both RNAs with a single initiation. In Step A. transcription starts and continues through the polyadenylated and self-cleavage sites. Polyadenylation processes the RNA and releases the HDAg mRNA that travels to the cytoplasm (Step B). The antigenomic RNA strand undergoes self-cleavage and continues transcribing for another 1.7kb(the length of the genomic RNA). Self-cleavage somehow prevents normal downstream fragment degradation. Usually after mRNA processing, the unused RNA is degraded. Not in this case! During this continued transcription, polyadenylation is suppressed (Step C). After transcribing 1.7kb, the second self-cleavage occurs. This releases the unit-length antigenome RNA (StepD). The antigenome (+) undergoes self-ligation to form the circular template for the genomic RNA (-) (Step E). The virus uses RNA polymerase II again to transcribe copies of the genome.
Assembly of the HDV virion requires the envelope proteins (HBsAg) of the helper hepadnavirus, HBV. Without the envelope the virus cannot replicate properly. This is why coinfection or super infection with HBV is necessary.
Prions:
Stanley Prusiner first proposed the term “prion” in the early 1980s. He found evidence of neurological diseases caused by agents that appeared resistant to the processes that normally destroy nucleic acids. Once highly controversial, the idea won Prusiner the Nobel Prize in Physiology or Medicine in 1997. A prion is an infectious protein which is hypothesized to infect tissues and "reproduce" first by changing its own conformation, and secondly by inducing other benign proteins to do the same. The conformation that is induced is much larger than the normal protein confirmation should be (called an amyloid fold) and following aggregation of these prions, they can cause damage to tissues by expanding within them and causing "holes" to form.
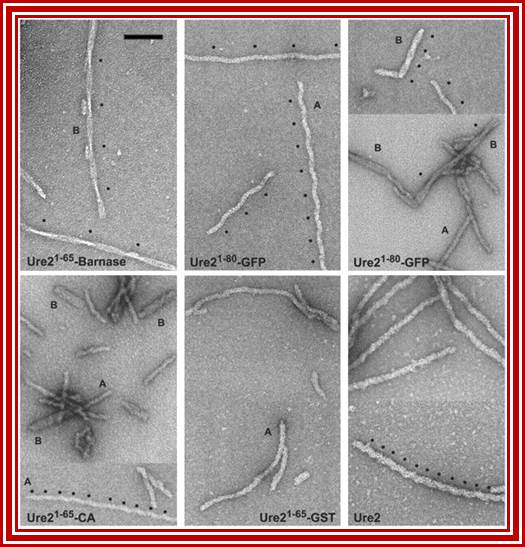
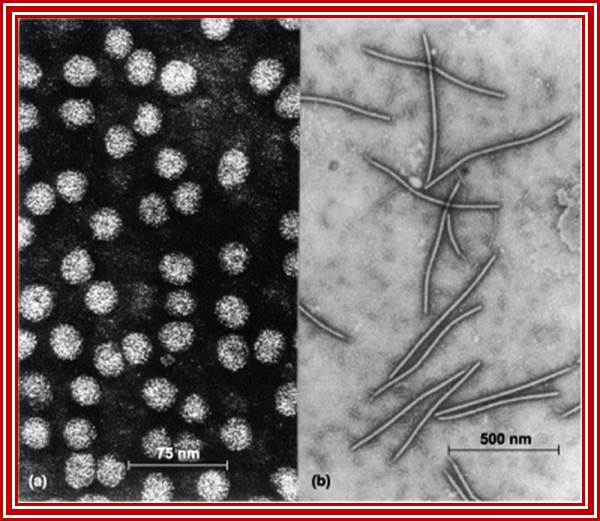
Amyloid filaments of proteins containing the Ure2p prion domain visualized by negative-staining EM; http://www.pnas.org/
A Case Study of Self-Replication and Evolution of Prions:
Replication Mechanisms:
Prions are the auto catalytically replicating proteins responsible for BSE, CJD, Scrapie and GSS. These diseases have genetic, sporadic and infective features, suggesting an interesting patho-physiology, which any molecular mechanism must explain. The prion protein is very stable resisting high temperatures, formaldehyde and UV light. The infective agent PrP-Sc differs from the host-produced protein PrPc (present on the plasma membrane of neurons) only in conformational form . PrP-Sc is structurally stable, rich in Beta-sheets, and having a lower free energy of formation. It is thought that a high activation energy limits the rate of conversion of PrPc into PrPSc, so that PrPc is much more common even though PrPSc is thermodynamically more stable. The conversion is thought to be due to the induction by PrPSc of the conversion of PrPc alpha-helices to PrPSc beta sheets (See figure below. Left PrPc, Right PrPSc). (Bieschke et al 2004), (Prusiner 1998), (Wille et al 2002), (Cohen and Prusiner 1998, see figure 0aa )
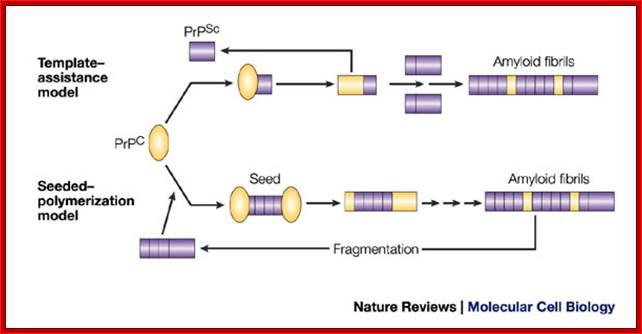
In the template-assistance model, the normal cellular form of PrP (PrPC) physically interacts with the infectious PrPSc form and induces PrPC to be transformed into the PrPSc conformer. PrPSc somehow acts as a template, allowing an otherwise energetically unfavourable protein-folding event to occur. The resulting PrPSc–PrPSc homodimer then dissociates to allow the formation of a new heterodimer and the generation of further PrPSc molecules. Over time, PrPSc molecules assemble into the higher-molecular-weight fibrils.
In the seeded-polymerization model, conversion of PrPC to PrPSc occurs as a consequence of the interaction of monomers of PrPC with oligomers of PrPSc. These oligomers act as seeds and recruit PrPC monomers. Eventually, the seeds go on to form higher-order amyloid fibrils, which themselves do not have seeding potential. However, fragmentation of the fibrils generates new infectious seeds to which PrPC molecules bind and are converted to PrPSc. In a variation of this model (not shown), the PrPC and the PrPSc forms are in a reversible thermodynamic equilibrium and the PrPSc form becomes stabilized only by interaction with other PrPSc molecules.
How do mammalian prions form? Mick F. Tuite and Brian S. Cox; http://www.nature.com/

Diseases caused by prions are characterized as spongiform encephalopathy. Holes develop throughout the brain. Cells begin to die from the abnormal prions’ ability to link together and form long protein rods. The tissue damage causes progressive degeneration in neurological control that can affect things like balance, muscular control, mood, and sleep (depending upon the area of the brain most affected). All prion-caused diseases are fatal.
Prions have both genetic and infectious etiologies. All mammal cells carry a gene that produces the normal form of the protein, PrPC. Sporadic or inherited point mutations in this gene can cause disease. But prions can also cause diseases by passing between individuals. The abnormal form can induce conformational change in the normal prions already present in the neurological tissues, producing more abnormal forms. These forms can then cause further conformational changes in a positive feedback loop.
The function of PrPC is not completely understood. It is concentrated in the cellular membranes of neurons and may play a role in synaptic communication. Mice missing the prion gene appeared to develop no ill effects, but also failed to develop symptoms of the disease when exposed to prions.
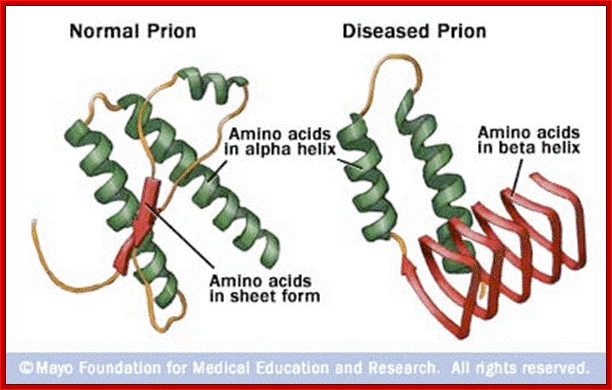
The infective protein is insoluble in water, contains a different folding pattern dominated by beta helices, and is resistant to formaldehyde, proteases, ethyl alcohol, and some forms of radiation. This resistance to standard methods of sterilization is the likely cause of most iatrogenic CJD cases. The insoluble infective protein groups together inside the cells in tissue of the central nervous system, forming amyloid fibrils. These groupings of the infectious prion lead to the vacuoles present in nervous tissue and sponge-like appearance of neural tissue among advanced cases (Aguzzi and Polymenidou, 2004; Voet and Voet, 2004).http://nawrot.psych.ndsu.nodak.edu/
The general public became interested in prions during the 1990s when Great Britain discovered an epidemic of bovine spongiform encephalopathy, or Mad Cow Disease. The discovery that people might be affected by prions ingested from beef made headlines around the world. The United States announced the first case inside its borders in late 2003. Midwestern deer and elk populations are affected by chronic wasting disease, a prion disease of concern to hunters and game animal farmers