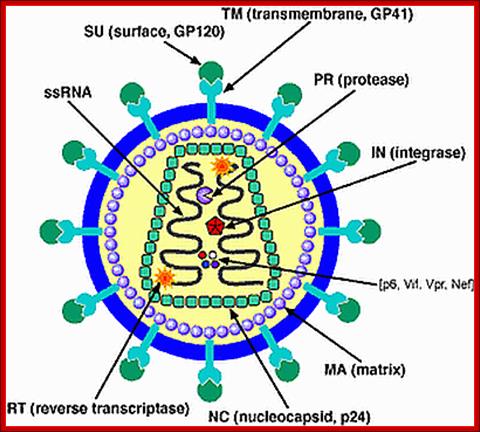
Retroviruses-Mechanism of Replication
Infection and Reproductive cycle:
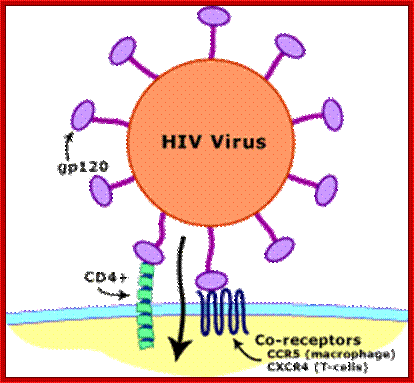
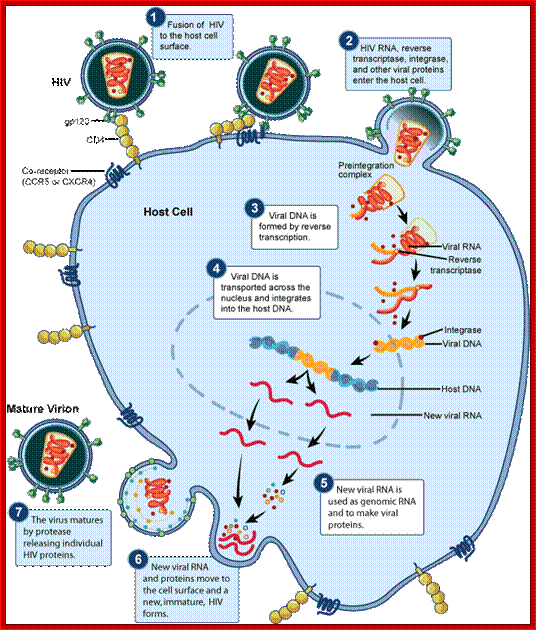
Transmission of HIV viruses is always through blood, either during intercourse between infected and uninfected gays or heterosexuals, sputum, using the same syringe by drug users. For the effective infection the inoculum’s size should be high i.e. the number of HIV particles entered into the system. Even one has sexual intercourse with HIV person, proper washing (either sex) can prevent (?), provided the sexual act is not violent in the sense there is rupture of epithelial cells. When the viral particles start circulating in the blood, they come in contact with a variety of cells, but the most compatible host cell is T-lymphocytes for they have the receptors for the binding of viral coat proteins. They can also bind to macrophages, B-lymphocytes and few other cells, but the most effective cells for infection are T helper lymphocytes for they have CD4 receptors with their associated proteins. Any cell that has CD4 receptors will be the target for HIV infection. Once they bind protein interaction leads to the fusion of viral envelop with the cellular plasma membranes; that leads to the internalization of the intact virus.
Viral glycoprotein gp120 is a trimeric protein contacts CD4 terminal domain of the T4 cell, this leads to activation of gp120, then it makes contact with chemokines receptor CCR5, this is a multipass membrane protein. These interactions make gp41 active and it cleaves gp120 and it collapses and the terminal domain of gp41 contact target cell and fuses with the T cell membrane; this leads to the entry of virus with capsid into the cell freed of membrane and the virus with capsid intact will be released into the cell and it gets activated. The required components for genome replication are already localized in the encapsulated virus and the replication is initiated and the required nucleotides move into the loosened capsid.
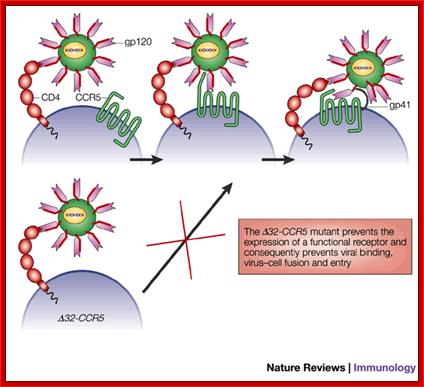
In some human population the CCR5 or CXCR4 gene contains 32 BP deletions called ~32CCR5. Humans who are homozygous for this mutation don’t get infected with the virus and resistant to HIV. If the person is heterozygous the infection is difficult.
http://.virology.net/Big_virology
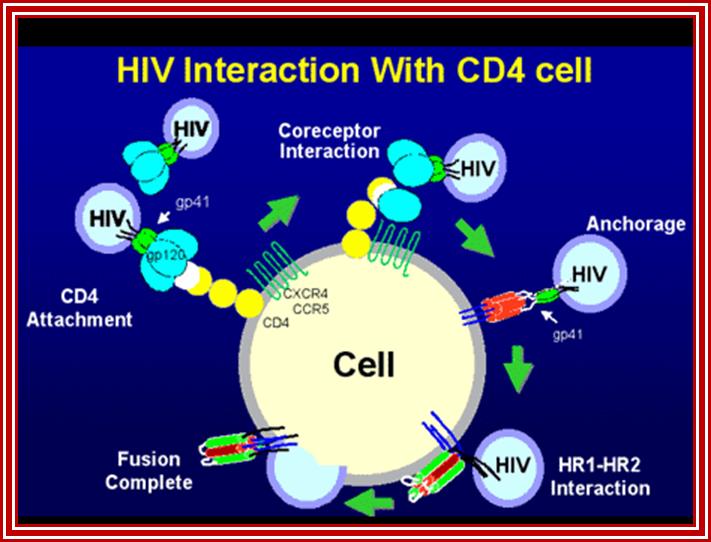
HIV interaction with CD4 cells. www.medscape.org
Tropism
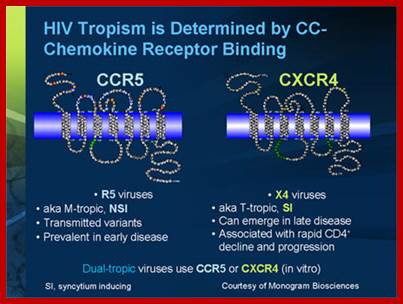
Viral tropism determines which host cells will be targeted. This is based on which cellular receptors or co-receptors they display on their cell membranes. In the case of HIV, tropism determines which cellular co-receptor, CXCR4 or CCR5, the virus will use to help mediate its entry into its host cell.
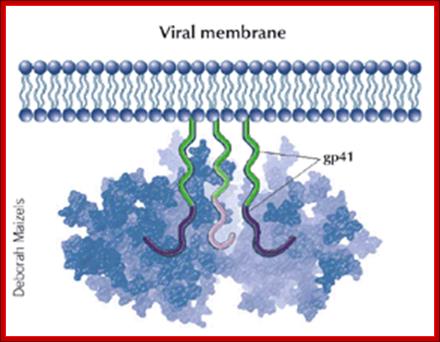
GP 41 protein- is a fusion protein responsible fro viral membrane fusion with target membrane3; http://www.medscape.org
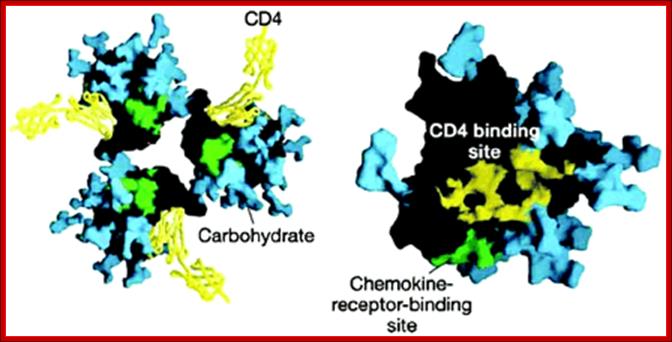
Peter D. Kwong; http://www.nature.com
Surface structure of HIV-1 gp120 complexed to CD4: (Left) The trimeric form of gp120 as assembled on the virus particle is shown, with soluble CD4 domains 1 and 2 (yellow) locked into the receptor binding pocket. Green shows the putative chemokine receptor binding site, and blue shows the carbohydrate moieties on this heavily glycosylated protein. A rotated (Right) and magnified view of a single gp120 surface is shown with the CD4-binding site colored yellow. [Reproduced from ref. 14 (Copyright 2002, Nature Publishing Group, (www.Nature.com)
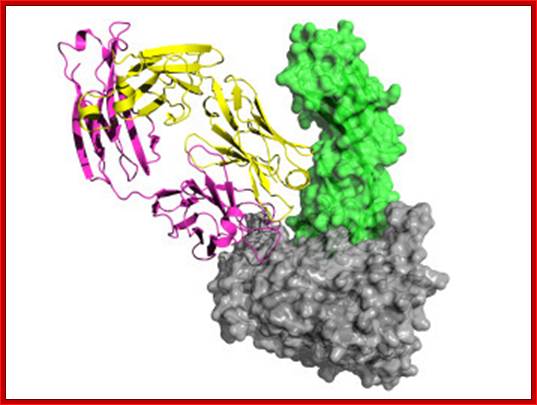
www.bbe.caltech.edu
This image of the complex, studied by the Caltech team, shows gp120 in gray, CD4 in green, the 21c antibody's light chain in yellow, and its heavy chain in magenta.
Vicriviroc-Revill P, et al

Gp41 structural feature with it fusion peptide located at amino terminal region. http://uhavax.hartford.edu

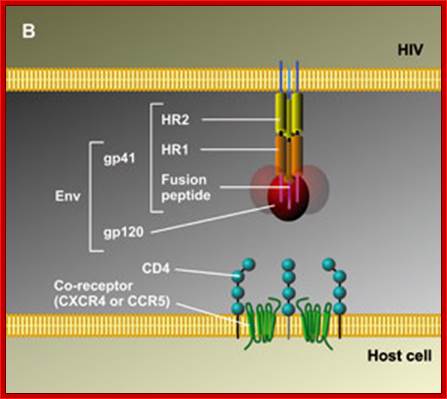
When HIV infects a CD4+ T cell (a), the viral glycoprotein gp120 first interacts with the CD4 receptor, then with the CCR5 or CXCR4 co-receptor, upon which the viral gp41 will bring the viral envelope in contact with the host cell membrane (b). The gp41 glycoprotein contains four major functional domains: starting from the N terminus towards the C terminus these are the fusion peptide, the heptad repeat 1 (HR1), the heptad repeat 2 (HR2) and the transmembrane domain that anchors gp41 into the viral lipid bilayer. Enfuvirtide is homologous to part of the HR2 region. When the N terminal fusion peptide of gp41 is inserted into the host cell membrane, the three HR2 domains of the gp41 trimer loop back in a triple hairpin and 'zip' themselves into three highly conserved hydrophobic grooves on the outer face of the HR1 trimeric bundle to form a six-helix bundle that pulls the outer membranes of the virus and the cell into close physical proximity, thus enabling the two membranes to fuse13. This process depends on an interaction of the heptad repeat HR2 with HR1. By being homologous to the HR2 domain, enfuvirtide blocks this interaction.

HIV Viral infection. Jürgen Drews; http://www.nature.com/
The figure gives a simplified view of the mechanism of entry of HIV-1 into host cells. The envelope glycoprotein of HIV-1 consists of two subunits, gp120 and gp41. After attachment of the virus to host cells that carry the CD4 receptor, gp120 interacts with the CD4 receptor, initiating a series of conformational changes in gp41 and gp120 that lead to the insertion of the hydrophobic amino terminus of gp41 into the host-cell membrane, and the formation of the fusion intermediate at the top of the figure. Subsequent conformational changes in gp41 bring the viral and cellular membranes close enough for membrane fusion to occur (a process known as gp41 zipping). By binding to gp41, enfuvirtide prevents the successful completion of gp41 zipping and subsequent viral entry. Ref. 43 © Macmillan Magazines Ltd (1998).
CCR5 with 32 bp deleted;

Deletion of 32bp leads to the deletion of 10-11 amino acids in the CCR5 protein, thus renders the CCR5 malfunctioning. Persons with homozygous condition for this trait are immune to HIV infection.

http://helicase.pbworks.com/
Evidence
from ![]() 32-CCR5 homozygous individuals has shown a
role for CCR5 in allograft acceptance, but has been fairly disappointing for
other inflammatory diseases. Of the 1,227 renal transplant patients screened,
1.7% of the patients were homozygous for the
32-CCR5 homozygous individuals has shown a
role for CCR5 in allograft acceptance, but has been fairly disappointing for
other inflammatory diseases. Of the 1,227 renal transplant patients screened,
1.7% of the patients were homozygous for the ![]() 32-CCR5 allele and only one of these
patients lost transplant function. Homozygous
32-CCR5 allele and only one of these
patients lost transplant function. Homozygous ![]() 32-CCR5 individuals have been described to
develop both rheumatoid arthritis and multiple sclerosis (MS). However, these
MS patients have not suffered more than a primary attack; in other words, they
have not succumbed to the normal relapsing syndrome. Certainly, the clinical
evidence from MS lesions in which CCR5 is seen to be highly expressed implies
that this receptor is involved in pathology.
32-CCR5 individuals have been described to
develop both rheumatoid arthritis and multiple sclerosis (MS). However, these
MS patients have not suffered more than a primary attack; in other words, they
have not succumbed to the normal relapsing syndrome. Certainly, the clinical
evidence from MS lesions in which CCR5 is seen to be highly expressed implies
that this receptor is involved in pathology.
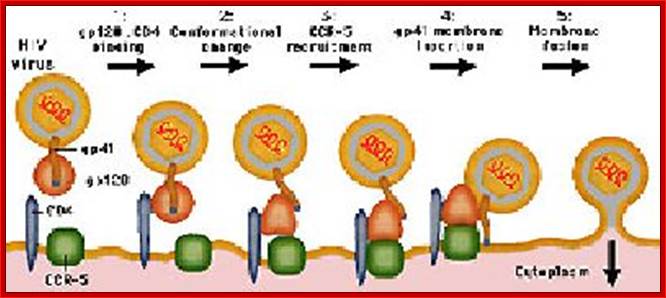
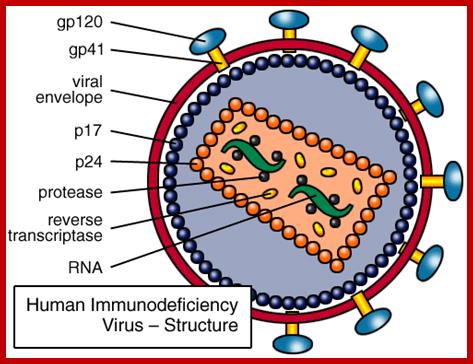
The diagram shows how the viral surface proteins p120 and p41 interact with host cellular CD4 receptors and surface CCR-5 leads to the integration of viral envelope membrane and cellular membrane and internalization of the viral protein. This process is not an endocytosis process. http://www.sctpn.net/1viralvoice
http://apbrwww5.apsu.edu/
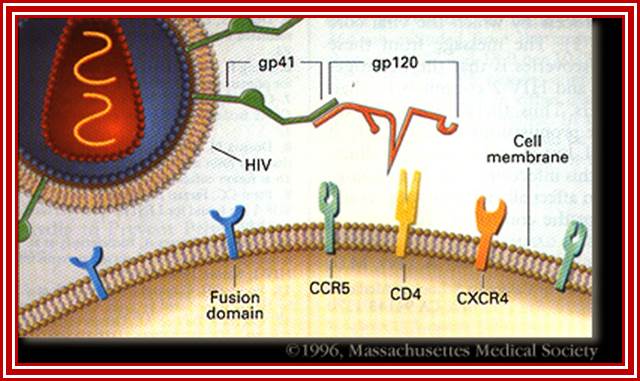
HIV-1 interacts with a cell-surface receptor, primarily CD4, and through conformational changes becomes more closely associated with the cell through interactions with other cell-surface molecules, such as the chemokine receptors CXCR4 and CCR5. The likely steps in HIV infection are as follows:

http://apbrwww5.apsu.edu/
The CD4-binding site on HIV-1 gp120 interacts with the CD4 molecule and binds to their cell surface.
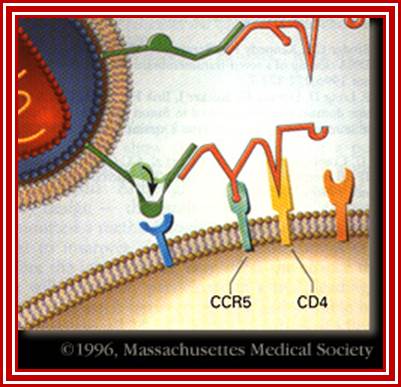
http://apbrwww5.apsu.edu/
Conformational changes in both the viral envelope and the CD4 receptor permit the binding of gp120 to another cell-surface receptor, such as CCR5.
http://apbrwww5.apsu.edu/
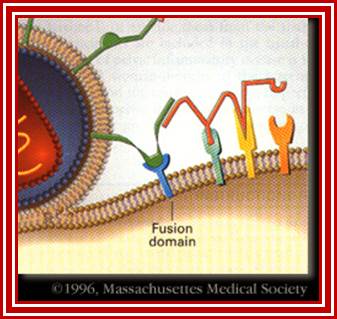
This second attachment brings the viral envelope closer to the cell surface, allowing interaction between gp41 on the viral envelope and a fusion domain on the cell surface. HIV then fuses with the cell.
During the M-tropic phase of HIV infection, the virus favors macrophages, which it invades by binding (through its gp120 protein) to the molecules CD4 and CCR5 on the macrophage surface. Eventually, however, HIV-1 can become dual-tropic. Such strains produce gp120 molecules capable of recognizing the CXCR4 protein on CD4-bearing T-cells. During this phase HIV-1 may infect both macrophages and T-cells. Still later, the bulk of the viral population may switch it's preference to the CXCR4 receptor and become T-tropic. T-tropic viruses readily destroy infected T-cells, contributing to the collapse of the immune system and the onset of AIDS. Alternatively, some viruses, such as certain strains of HIV-2 could attach to CXCR4 quickly leading to AIDS.
Entry of the virus:

Subsequent to the above mentioned events, the viral nucleoid with its capsule enters the cell. http://apbrwww5.apsu.edu/
Replication Mechanism:
The virus in the cytoplasm gets activated so that its capsid proteins are little bit loosened so that cytoplasmic components such as metal ions and nucleotides can enter into the capsid. Enclosed inside the capsid all the required factors are present, such as Reverse transcriptase, integrase and others. Using the already present enzymes and the template RNAs, there are two such templates, called the genomes are copied in a unique way to generate ds complementary DNA, which is slightly longer than the original size of the genomic RNA. The ends of the ds cDNA have repeats called long terminal repeats LTR. Using the LTRs, the enzyme integrase binds to circularize the ends, but no fusion of the ends. It is at this juncture the DNA is delivered into the nucleus. In the nucleus the ds circular modulated DNA binds to chromosomal DNA randomly, no sequence or site specificity; if any of the sites in the chromosomal DNA is free from histones and if chromosomes are replicating; and if the region of the DNA is transcribing; the integrase facilitates recombination and repair, thus the viral DNA gets integrated, which behaves as if it is the part of the host DNA.
The integrated genome can remain for any number of generations; if perchance the cells that get infected happened to be germ line cells; the viral genome is vertically inherited. This genome can be passed to many generations. During this passage of time, radiations or certain drugs can activate the viral genome. The LTR segments have all the components of promoter and enhancer elements. Using these elements the cellular enzymes transcribe the genome and the transcripts are spliced and the final unspliced, intact transcript is transported out of the nucleus. By the time the full length viral RNA (with 5’cap and 3’ poly (A) tail) enters cytoplasm from the nucleus, the required protein components are already made in cytoplasm. The capsid proteins using viral genomic RNA packaging sequences (psi) assemble the proviral particles and they are budded of as enveloped viruses containing glycoproteins on the membrane surface. The budded off viral particles circulate in the blood and further infect more number of cells, thus the viral particles spread and more number of T-cells gets incapacitated. This will have great impact on other cells involved immune system.
Post Entry Events:
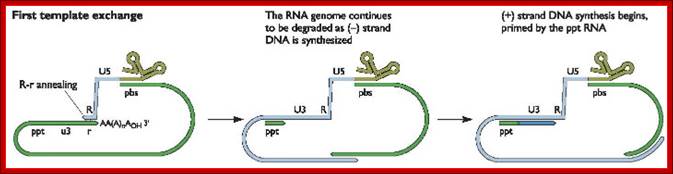
As the uncoated viral particle is released into the cytoplasm, required ions and ntds move into the relaxed viral core. The replication mechanism is unique; first it produces a small segment of negative sense cDNA for the first strand. Then the enzyme switches template ends and generates full length (-) sense cDNA as the first strand. The reverse transcriptase again switches the ends to generates (+) sense fragments then it completes the synthesis of (+) strand to produce ds DNA a little longer than the size of the original viral RNA.
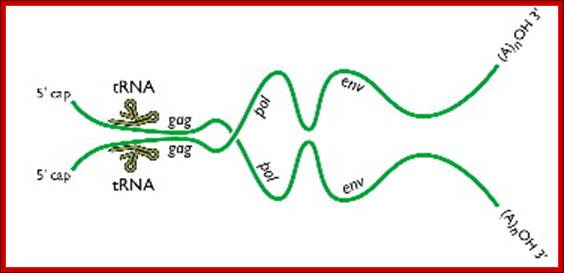
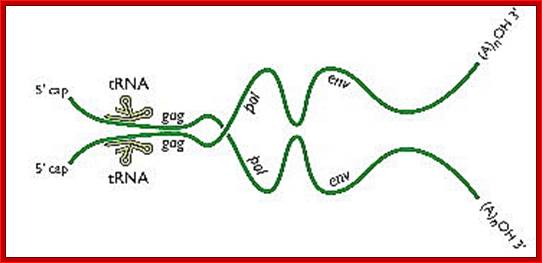
Two genomic RNAs (positive sense) ; http://www.twiv.tv/reverse-transcription
OMFG!
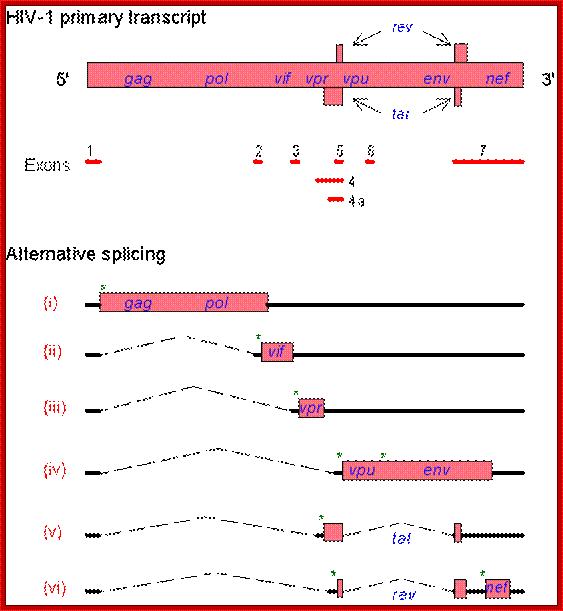
The HIV genome contains overlapping/alternate splicing sequences. Look at all
those overlapping genes!! HOW DOES HIV DO IT???? How does it KNOW how to
splice??? This
diagram is based on a fantastic map of the HIV-1, HIV-2, and SIV genomes,
available at
hiv-web.lanl.gov/;
http://www.mcld.co.uk/
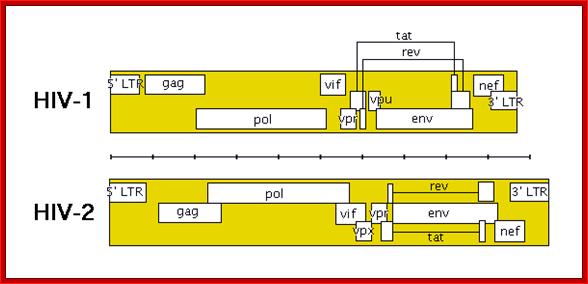
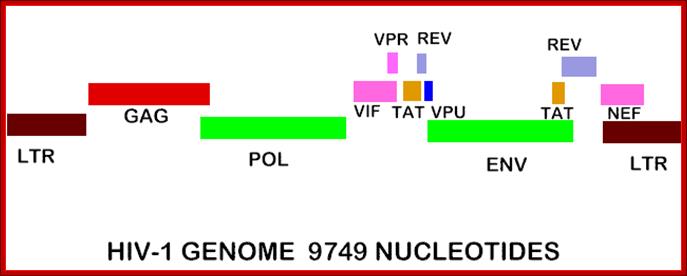
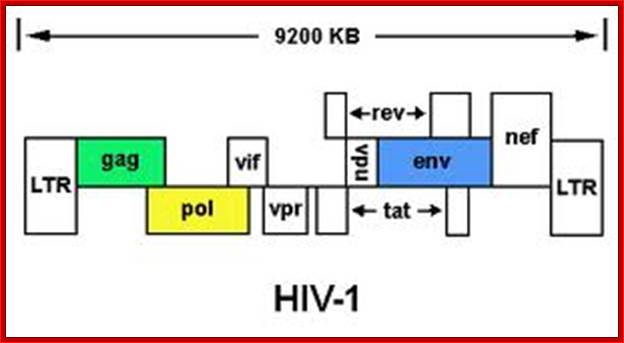
The genes in HIV's genome are as follows:
- gag (coding for the viral capsid proteins); pol (notably, coding for reverse transcriptase); (NB. gag and pol together can be expressed in one long strand called "gag-pol")
- env (coding for HIV's envelope-associated proteins)
And the regulatory genes:
- tat; rev; nef; vif; vpr; vpu (N.B. not present in HIV-2)
- vpx (N.B. not present in HIV-1); The HIV genome also has a "Long Terminal Repeat" (LTR) at each end of its genome - not quite a gene, but a sequence of RNA/DNA which is the same at either end and which serves some structural and regulatory purposes.
Pathmicro.med.sc.edu; http://www.bio.davidson.edu/
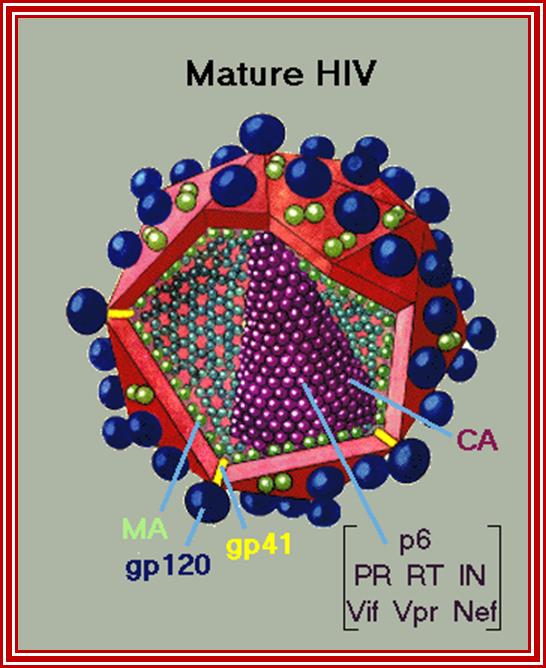
TAT: Trans-Activator of Transcription;REV: Regulator of Virion ;rotein expression;NEF: Negative Regulatory Factor;VIF: Virion Infectivity Factor;VPU: Viral Protein U;VPR: Viral Protein R; GAG-Capsid polyprotein,Pol-ENA dependent DNAPolymerase, ENV-envelop associatedproteins; LTR long terminal repeats. Image courtesy: Dr.Richard Hunt.
http://medstat.med.utah.edu
The above figure shows the complete RNA genome of HIV-1. The above HIV genome consists of nine open reading frame(ORFs). Three are prototypal of retroviridae-Gag.Pol.Env.
The POL gene codes for reverse transcriptase. HIV2 differs only in the context of VPX. You can also view the HIV-1 genome at NCBI Entrez Genome. Image courtesy of Dr. Richard Hunt.
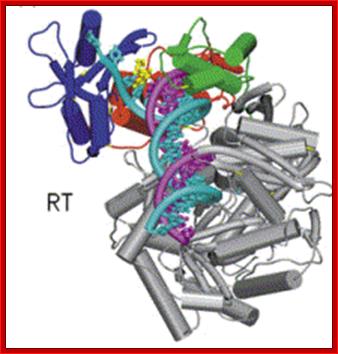
Reverse Transcriptase (RT). https://en.wikipedia.org

http://www.cabm.rutgers.edu
Reverse transcriptase showing both the p66 and p51 subunits. Both subunits have the palm, thumb, fingers, and connection subdomains. The RNase H domain is located on the p66 subunit only. Every 50th amino acid residue is labeled.
This ribbon representation of the RT top figure, active domain illustrates its hand-like structure, showing fingers (blue), palm (pink) and thumb (green). The active site (red atoms), where DNA is elongated, is in the palm region. Also shown is an NNRTI drug (yellow) in the pocket where it binds. The enzyme has two enzymatic functions. Firstly it acts as a polymerase where it transcribes the single-stranded RNA genome into single-stranded DNA and subsequently builds a complementary strand of DNA. This provides a DNA double helix which can be integrated in the host cell's chromosome. Secondly it has ribonuclease H (RNase H) activity as it degrades the RNA strand of RNA-DNA intermediate that forms during viral DNA synthesis. The HIV-1 RT is an asymmetric 1000-amino acid heterodimer composed of p66 (560 amino acids) and p51 subunits (440 amino acids). The p66 subunit has two domains which are polymerase and ribonuclease H. The polymerase domain contains four subdomains which have been termed “fingers”, “palm”, “thumb” and “connection” and it is often compared to a right hand (figure 1). The role of the p66 subunit is to carry out the activity of RT whereas it contains the active sites of the enzyme. The p51 is believed to play mainly a structural role.
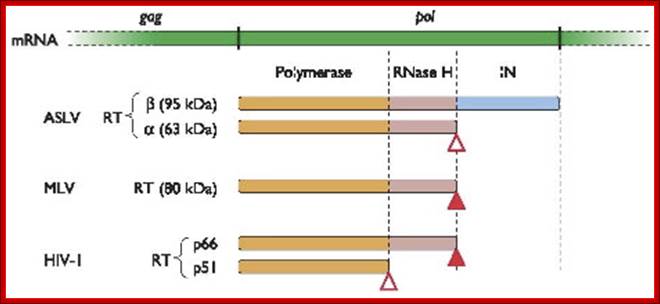
Reverse transcriptase proteins from different retroviruses and their domains. In HIV the 66 kd protein contains both Polymerase and RNase domains, but 51kd protein contains just polymerase domain. ASLV RT contain two subunits and MLV contains only one subunit. http://www.twiv.tv/
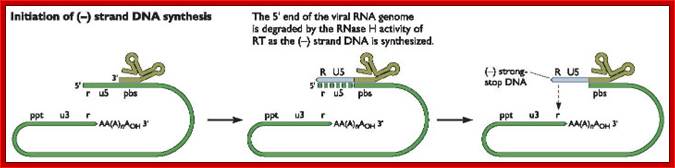
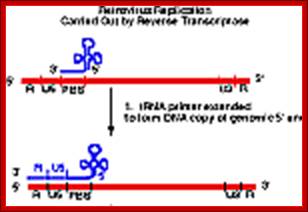
The Reverse transcriptase uses the 3’OH group of lys-tRNA bound to PB site, as primer; the tRNA is base paired to 18 ntds long primer binding site, incorporates dNTPs and extends the primer till the 5’end of viral RNA template.
- The new strand produced is complementary to the genomic RNA and it has negative sense. It’s newly formed (-) DNA segment includes U5 and R segments.
- On reaching the template end, enzymes Rnase-H domain becomes active and starts removing the RNA template from the 5’ end and removes nucleotide by nucleotide (7-8 ntds blocks), when it reaches (Primer Binding Site) PBS i.e. the tRNA 3’end, it stops. Perfect execution, how?
- Here an enzyme exhibits both polymerase and RNase-H activity for they have different domains.
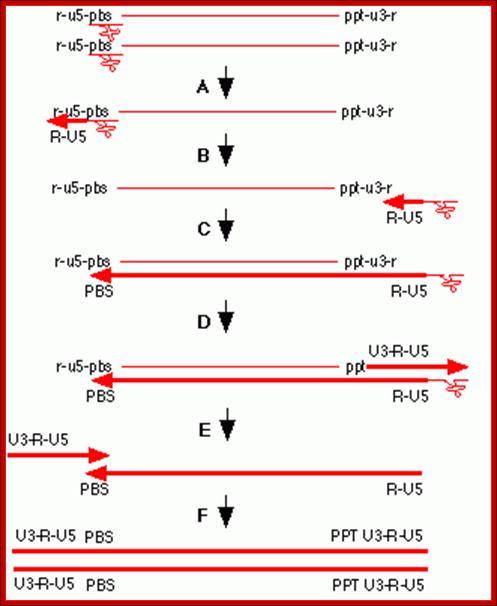
- The C-DNA segment having complementary sequences to 5’R’ and U5 can switch the ends, which can be achieved by 5’R’ sequence of the C-DNA base pairing with 3’ R region. The R regions have perfect direct repeats, so the 5’ R part of C-DNA can perfectly base pair with 3’ R segment, thus the genome gets circularized.
![]()
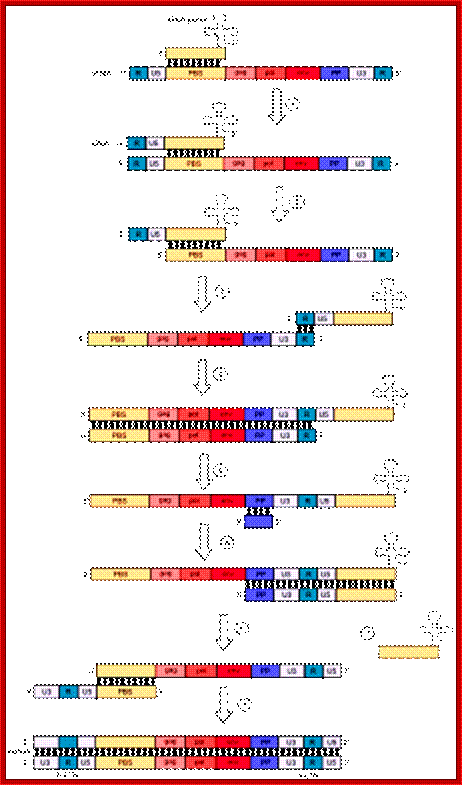
Replication HIV viral RNA genome into ds CDNA with long terminal repeats
- In circularized form, the primer-tRNA is still covalently linked to the 5’ end of the C-DNA, there by the C-DNA segment-containing R-U5-tRNA is base paired to 3’R sequence at the 3’ end of R’ repeat sequences of the viral RNA.
- Now the 3’ end of the R part of C-DNA acts as the primer and the Reverse transcriptase, which is still bound to the template incorporates dNTPs and the synthesis progresses till the end of the genomic RNA strand which culminates at the end of PB, thus a full length (-) C-DNA as the first strand is produced.
http://www.twiv.tv
- How the enzyme is still bound to the template is not clear. Whether the enzyme still bound to the template, how it switches the templates ends is speculative, but very interesting.
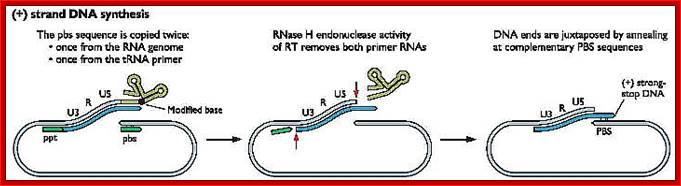
- At this position and juncture, the Rnase-H switches direction and starts removing the parental RNA strand from its 5’ end till it reaches what is called poly purine tract found at the junction between Env and U3. Surprisingly the RNA strand from the 3’ end containing U3, R3 and poly- (A) tail, is also removed till the polypurine tract. Thus polypurine tract, of eight nucleotides long, acts as the primer for the second strand synthesis.
Now the enzyme switches its activity from RNase-H to polymerase and starts assembling dNTPs using polypurine tract (AAAAAAAA) OH3’end and extends till the end of primer binding region of tRNA i.e. 18 ntds long.
- How only 18 ntds of tRNA are copied not more than, it is yet to be properly explained.
Then the polypurine tract (8-10ntd long A s and Gs) and tRNA, which is still covalently linked to the 5’ end of the first cDNA strand is removed by the Reverse transcriptase, with its associated RNase-H activity.
At this stage one more end switching takes place, again by circularization. The enzyme is still intact with its template.
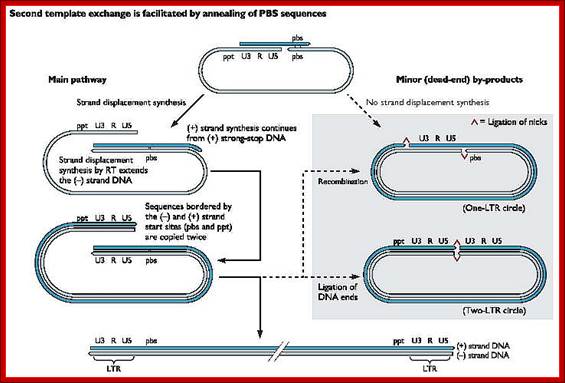
End switching is possible because the second strand produced has 5’ U3—R—U5—PB>3’ and the PB part of the second strand can easily base pair with PB region of the first C-DNA strand. When this is accomplished, the reverse transcriptase extends 3’ end of the (+) strand and progressively synthesizes the second (+) DNA strand to its end.
- Then it switches to the other side and extends the 3’ end of the first c-DNA (-). This complete the production ds cDNA from the ss genomic RNA, but with longer dsDNA than the original size of the genomic RNA.
- Both terminal regions of ds cDNA contain U3-R-U5 sequences as repeats, so they are called long terminal repeats (LTR). The dynamics of the enzyme in performing various functions at precise point and precise timings is just remarkable, a feat no other enzymes have endowed with such remarkable features.
http://www.twiv.tv/reverse-transcription
Completion of replication; this happens within the proviral particles and in cytoplasm; remember there are only two RNA genomes per viral particle. Infection to be effective; the quantum of the virus infected should substantially in good numbers.
http://diveintohiv.com/hiv-life-cycle;This is to exemplify the events
Another view of Replication; remember though the genomic RNAs have positive sense they are not translated for they are not released into cytoplasm. So any copying of RNA genome is deemed both replication and transcription.
Integration:
The entire process of replication completes within the viral core, which is similar to that of Reo virus. The ds viral C-DNA, unusually, has 4-6 base pairs of inverted repeats at its ends. The sequences are 5’AATG—CATT 3’.
5’AATG----------------------------------------------------------CATT ’(+)
3’TTAC----------------------------------------------------------GTAA5’(- )
The integrase protein found within the core, now binds to the end sequences; two subunits to each end. Protein-protein interaction brings the two ends together; the genome looks almost circular; this is referred to as Intasome- a tetrameric protein
Once it is believed that the ds C-DNA gets circularized with integrase then moves into the nucleus. There it integrates into the linear chromosomal DNA, which is free from nucleosomes.
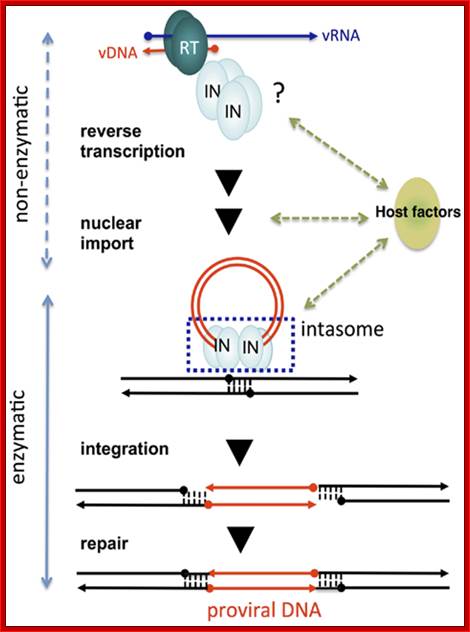
Non-enzymatic and enzymatic roles of HIV-1 IN that help establish proviral DNA. After entry into cells, retroviral genomic RNA (vRNA) is reverse transcribed into DNA (vDNA) by RT. Then, vDNA is transported into the nucleus (nuclear import) and finally integrated into host chromosomal DNA (black lines). IN forms a functional tetramer with vDNA ends (intasome) to assist with integration. Subsequent repair processes by host DNA repair machinery establishes the proviral DNA, in which retrovirus DNA is stably integrated into host chromosomal DNA. IN tetramer structures for its enzymatic (integration) and non-enzymatic functions (reverse transcription and nuclear import) and possible involvement of host factors are schematically depicted. The vRNA, vDNA, and host DNA are shown as blue, red, and black lines, respectively. An arrowhead shows the direction of each genome from the 5′- to the 3′-end. The ends of ds viral cDNA has repeats of U3, R and U5 in the same order at both ends which provided the sequence elements for the binding of integrase, which in turn cuts and remove two nucleotides from each end. With the bound ends to integrase forms a circular model ready for the entry into the nucleus. journal.frontiersin.org.
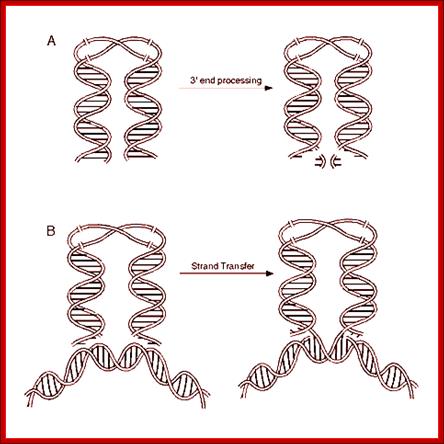
cDNA 3’end processing leads to the binding to host DNA, provided it is free from Histones, and integration via DNA repair
- Circularization of the ends without covalent bond formation is achieved by the binding of integrase molecules to their respective ends. This leads to circular form without circularization and maintains linear form as well.
- Integrase consists of N terminal and C terminal domains assist to generate aggregates of integrases. The central domain is catalytic core
- In this form the c-DNA is delivered to the nucleus through the nuclear pore complex. How? Perhaps they use importin regulated proteins.
- Movement, of the DNA into the nucleus, is protein mediated and ATP dependent, but what proteins are involved in the transport is not clear. Whatever proteins involved should show specificity for c-DNA and they should possess signal sequences such as NLS for the facilitated transport into the nucleus.
Integration of the c-DNA is greatly facilitated, if the cells are in cell division mode and the cell is transcriptionally active, for it requires open DNA, in the sense DNA should be free from conglomerate of proteins that normally associated with chromatin structures.
- It is only during chromosomal DNA replication (most often), and transcription process, the host DNA available for the integration of viral cDNA.
Integration of cDNA is not site specific or biased for any set of sequences, but it is random, which means it can integrate into any region of the gene, can be into promoter, or with in a coding region or to any other region including spacer regions found between the genes.
· Integrases, bound to the ends deliver viral cDNA on to relatively free regions of host chromosomal DNA.
The integrase enzyme has several functions, endonuclease activity, which can use 6 or more base pair segment. The integrase, on one hand, cleaves the host DNA to produce at least 6 ntd long sticky tails.
· On the other hand, it cleaves the c-DNA ends with 6bp long inverted repeats to produce two ntds long sticky ends.
Proteins located at the ends, as well as located on chromosomal DNA, facilitate the ends of c-DNA and host DNA to align and base pair. Gaps left on host DNA are filled and nicks are ligated. The viral cDNA looses two or more nucleotides.
· The host DNA at flanking ends of cDNA gains extra length of duplicated segments.
Once the viral genome as ds cDNA integrates in to host genome, it stays stable for many generations until and unless it is activated.

The diagram from WIKIPEDIA
- If the genome happens to be integrated in germ line cells, it is inherited vertically to the next generation, and remains undetected and with out disease symptoms.
Several viral genomes can be integrated into the same chromosome at several positions, or into different chromosomes, if the viral inoculum is very high.
Production HIV viral particles:
The viral cDNA in integrated form can be stable, inactive and it will be a part of the host genome, forever. Until another viral infection or any other factors activates it, the long genome with its LTR ends remains normal. In this state the viral genome remains inactive for many generations. If the integration takes place in germ line cells, this can be inherited. When activated, whatever may be the factor or means, the entire genome is transcribed into a full length viral RNA, then it is poly adenylated and capped as in other cellular mRNAs. The pre mRNA is spliced at defined positions and transported into the cytoplasm, where each of the transcripts (spliced) representing each segment of the coded genes are translated to generate viral protein products.

A general representation of the transcription of the integrated viral cDNA; the transcript later undergoes processing such as 5’capping and 3’ polyadenylation and splicing to generate different products. www.web-books.com

http://pinwallpaper.xyz/www. zdrav40.ru
The cDNA genome of the HIV codes for the above proteins.
Transcript mRNAs are spliced and transported into cytoplasm for translation; except for rev response element binding protein, the other proteins stay put in cytoplasm. The final transcript is full length mRNA is bound by rev proteins and transported out of the nucleus.
The viral ds c-DNA structurally has features similar to eukaryotic genes or gene clusters. It has LTR (made up of 5’–U3-R-U5----//----U3-R-U53’) sequences at either end.
- Genes, Gags—Pol (ref-nef)--Env are in between LTR blocks; they are in the same sequence.

https://sites.tufts.edu/
--------------U3---------------------------------------I>+1-R-I--U5-
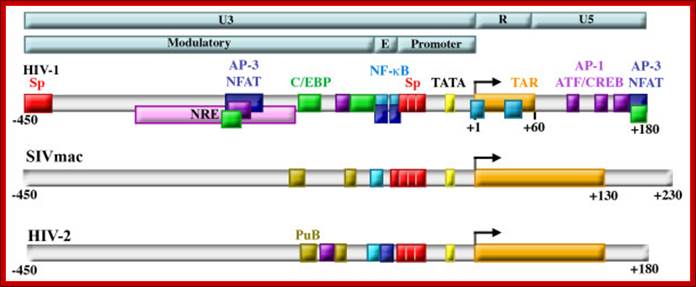
Structure of retroviral LTRs. Retroviral LTRs are divided into the U3, R, and U5 regions, and the U3 region is further divided into the Modulatory, Enhancer (E) and Promoter regions (top bars). HIV-1, HIV-2, and SIV all contain highly conserved promoters containing TATA boxes (yellow) and Sp factor binding sites (red) and enhancers (labeled E in light blue bar) containing NF-κB binding sites (blue). The R region of each contains a trans-acting responsive element (TAR) (orange) that forms an RNA stem loop structure upon transcription that binds to the viral protein Tat. A negative regulatory element (NRE, pink) was identified that was subsequently shown to serve as both activator and repressor by binding NFAT proteins (dark blue), AP-1 proteins (purple), and C/EBP factors (green). The modulatory regions of SIVmac and HIV-2 also contain purine box arrays (PuB, gold) and sites that bind members of the Ets family (teal). Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Kilareski EM, Shah S, Nonnemacher MR, Wigdahl B - Retrovirology (2009)
5’LTR----- +1>TAR-GAG-POl-REGu-ENV I RRE I NEF- -LTR3’
Structural and Functional Features of the Integrated Viral Genome:
The LTR s at 5’ and 3’ of the prophage genome have absolutely the same sequence, it can be said they are the direct repeat elements.
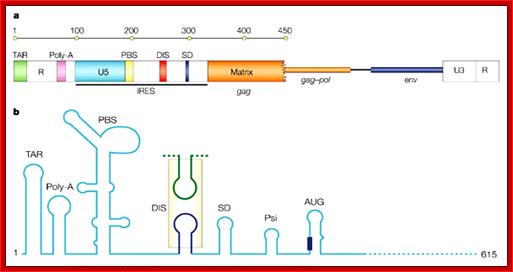
- At 5’ end of the Genome the LTR consists of U3 (200-1200 bp long), R (10- 80 bp) and U5 (80-100 bp). Next to U5 are the following, 18 bp primer binding site, and then a splice donor site (AG splice junction sequence). Next to it is the sigma sequence for viral packaging which extends 400 450 bp into the gag gene.
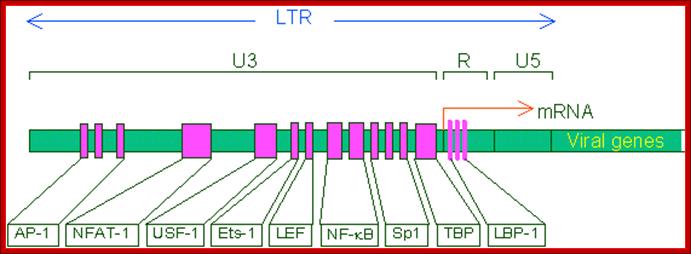
This line drawing represents the promoter blocks of the U3 and R and the regulatory factors that bind to them. The position of transcriptional initiation is marked.
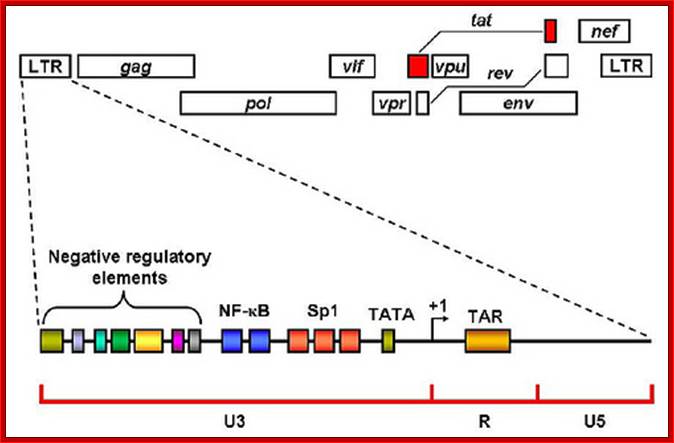
In addition to the structural genes (gag, pol and env), the HIV-1 genome contains 4 accessory (vif, vpr, vpu, nef) and 2 regulatory (tat, rev) genes, the products of which are responsible for establishing sophisticated interactions between the virus and human host. HIV-1 Tat is a multifunctional protein that contributes to several pathological symptoms of HIV-1 infection as well as playing a critical role in virus replication. Tat is a robust transactivating protein that induces a variety of effects by altering the expression levels of cell and virus genes. The functions of Tat are therefore primarily related to its role in modulation of gene expression. In this review the functions of HIV-1 Tat that have been well documented, as well as a number of novel functions that have been proposed for this protein, are discussed. Since some of the functions of Tat vary in different cell types in a concentration dependent manner and because Tat sometimes exerts the same activity through different pathways, study of this protein has at times yielded conflicting and controversial results. Due to its pivotal role in viral replication and in disease pathogenesis, Tat and the cellular pathways targeted by Tat are potential targets for new anti-HIV drugs.www.microbiologybytes.com; Bizhan Romani et al. https://microbiologybytes.wordpress.com
The U3 region has promoter and enhancer elements. Starting with 4-5 inverted repeats at the 5’ end, there are two 70- 100 bp long enhancer elements.
- Next to it, at (-) 80 from the START site, CCAATATA sequence is present for the binding of SP1 transcriptional factor.
Start site is the nucleotide at which transcription initiates or starts; it means it is the position at which the first nucleotide of RNA is incorporated. Start nucleotide is the first nucleotide at 5’ end in the mRNA and it is also the nucleotide to which cap is added.
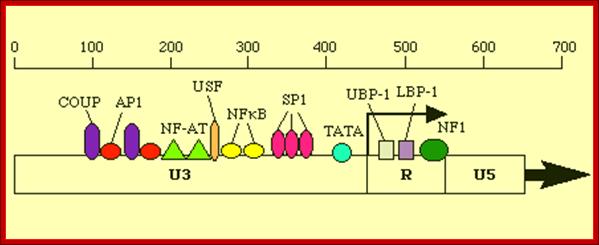
Another view of the structural feature of enhancer-promoter elements involved in initiating transcription; the sites for the binding of various transcriptional factors are labeled; http://www.microbiologybytes.com/
Next to the sp1 binding (CCAATTATA) segment is TATA region (--) 25 to 30 ntds from the start site, at which host RNA polymerase-II enzyme complex assembles and initiates RNA synthesis. AP1, NF-AT elements act as enhancer elements. Actually the start sequence starts within R segment.
- The leader sequence, a noncoding 5’ end of the gene extends beyond U5 and PB region. In HIV the R region contains all the promoter components for the binding of the following factors, such as NF-AT—NRF---NFkB---SP1---TBP and RNAP II complex at TATA-------- START- R---U5--
Following are the few promoter elements of integrated viral genomic elements:
Promoter elements of:
|
Virus |
5’ end promoter elements
|
|
ALV |
5’ ---EF-II---EF-I------CCAATTATA------R
|
|
MMTV |
5’ ---GRE---GRE—GRE—CCAATT------TATA----R
|
|
HTLV |
5’ ---tax+---tax+---tax+-------TATA-----R
|
|
HIV |
5’ –AP1-NFAT—NRF---NFkB-Sp1--CCAATTATA---TATA----R
|
|
MLV |
5’----Lva-NP1--LVb-NP1--LVc--GRE--[CAAT]—TATA---R
|
In stimulated T cells chromosomal loosening and freeing the LTR region free of nucleosomes; which leads to the assembly of transcriptional complex.
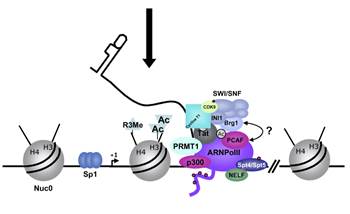
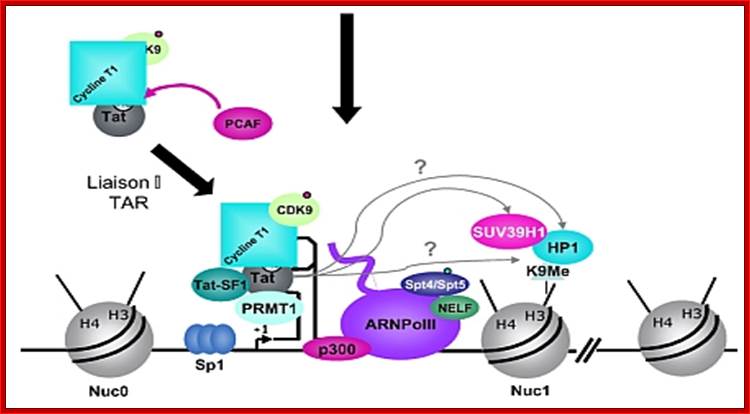
Chromosomal remodeling leads to the assembly of transcriptional complex. http://www.igh.cnrs.fr/ Mol.biol
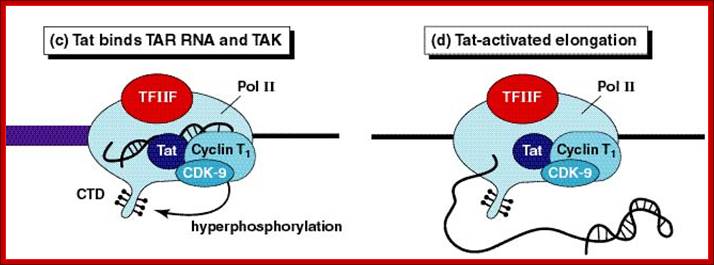
Role of TAT protein binding to TAR region of the RNA facilitate the transcriptional elongation.
When activated, transcription of the entire genome starts at one end to go full length and produces a poly cistronic mRNA, which is a common feature and it is an unusual feature for eukaryotic systems with certain exceptions,
The gag-pol transcript, in most the time translates up to the end of the gag and terminates, so only gag and its components are produced. These precursor proteins are further cleaved by their own proteases.
- Occasionally while translating shifting its reading frame at the end of gag segment, it continues and produces longer translated product. This product generates gag, pol, protease and integrase.
- While the Env transcript produces a single protein, which is further cleaved by its own protease to generate gp121 and TM-41 components.
Very interesting aspect of the viral genome transcription in relation to the production viral particles is the synthesis of full length viral RNA.
- In higher systems, unprocessed mRNAs are not transported out of the nucleus. This problem has been over come by one of the properly spliced transcript bound to Rev proteins moves out of the nucleus and translated. The rev protein moves back into the nucleus using NLS and Ran-GDP. The rev protein binds to rev response element found at the 3’ end of the full length mRNA. The binding of this protein facilitates the transport of unspliced full length viral RNA, which is absolutely essential for the production of viral particles.
- By the time full length viral RNA enters into the cytoplasm the required proteins for viral production are made in adequate numbers.
- Some of the proteins required for viral membrane such as gp121, TM41 and matrix proteins are translated on Rough Endoplasmic Reticulum (RER) and processed at different levels in Golgi complex and ultimately they are delivered to plasma membranes or they form vesicular membranes in which the said proteins are organized.
Once the full length viral RNAs are transported, the RNA is initially recognized by viral proteins by binding to sigma or what is called packaging sequences, which is located between PB and gag extending 410- 420 ntds into gag coding region.
- All most all viruses, whether they are DNA viruses, (ssDNA or dsDNA), RNA viruses (monopartite or multipartite) or Phage nucleic acids, all have specific packaging sequences, at specific sites in their genomes.
- Once the initial binding starts, aggregation of other proteins ensues to form a protein capsid around 2 genomes. Viral capsid also contains Reverse transcriptase and ultimately, they are anchored on to the inner surface of the host cell membranes at certain sites and then the particle is budded of by exocytosis.
This is where the virus acquires envelop membrane and membrane proteins. Viral assembly of B and D type occurs in cytoplasm, later they are budded off.
- But the assembly of C-type viruses takes place at the inner surface of the host membranes. During this process, the virus can also acquire cellular proteins like HLA 1, HLA-2, MHC 1 and MHC 2 and others.
- Most interesting part of viral production is maturation. Once the viral particles are budded off, most of the precursor proteins assembled is subjected to proteolytic cleavage in site-specific manner. This leads to certain conformational changes and reorientation of the components inside into matured form of virus.
In HIV, the binding of Transcriptional activating factors SP1 activates the transcription. The binding of nuclear factors like NFs, especially NFkB (NF-kappa B) to enhancers, increases the efficiency of genome transcription.
- At the other end of the integrated viral genome it has the same U3-R-U5 segments. At the junction point of Env and U3 is polypurine tract rich in A s and Gs. Also at this end of the end of R segment, a poly-A signaling sequence is present at 30 to 35 ntds upstream of the polyadenylating site, which is also the end of R region.
- Transcripts, as they are produced, capped at the 5’ end and polyadenylated at the 3’ end.
The pre mRNAs are also processed; initially splicing produces more of gag-pol mRNA and the same is poly adenylated. In this single splicing event Env is spliced out.
- The transcript moves out; on translation, it invariably produces gag protein, which on proteolysis generate matrix, capsid and nucleo capsid proteins.
Sometimes, the terminator codon UGA (or UAG) found in between gag and pol is read though to produce fusion protein, which is also processed into gag-members, proteases, integrases and reverse transcriptase, which within itself contains Rnase-H activity.
- The read through is performed by using glutamyl transferase suppresser tRNA.
As these two different proteins are in different reading frames and context, translation of gag is more frequent by >50% and the read through product is just 5%.
Another two splicing events produces Rev Transcript, which is promptly transported, translated, and the REV protein enters into the nucleus, which is absolutely required for the transport of unspliced full length mRNA.
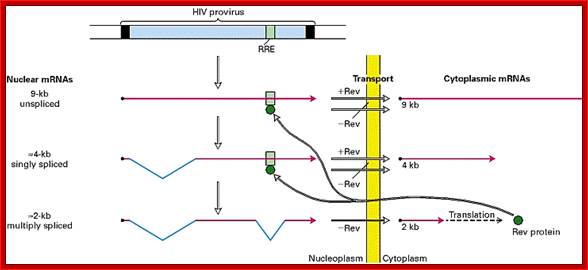
The full transcript is processed differently at a particular frequency to generate specific transcripts, which are transported out and translated. Some of the translated products enter the nucleus, such as Rev, which is required for the transport of the full length processed (capped and polyadenylated) viral transcript out of the nucleus for encapsulation and budding. http://www.web-books.com/
http://www.internationaltherapeutics.com/
Viral RNA packaging and budding:
As more and more viral required proteins are produced, the full length transcript is transported out of the nucleus with the help of Rev Proteins. Along with many transcripts for regulatory factors are also produced and the same are transported out of the nucleus, where in cytoplasm they are translated.
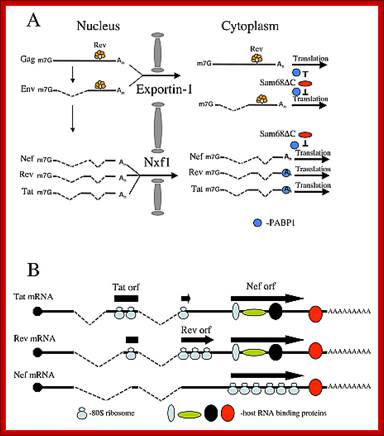
Transport of many regulation factors such as Tat, Nef, and others.

Role of
cellular factors in HIV capsid assembly and virus budding.
Following synthesis, Gag polypeptides (p55) and
the viral accessory protein Vif associate with a host protein, HP68. HP68, an
ATP binding protein, appears to interact with the NC region of Gag and promotes
progression of Gag-containing assembly intermediates into immature capsids at
the host cell plasma membrane (Zimmerman, et al., Nature 415, 88-92, 2002). A
cellular factor Tsg101, which functions in vacuolar protein sorting (Vps), is
required for HIV-1 budding. Tsg101 binds to an essential tetrapeptide (PTAP)
motif within the p6 "Late" domain of the Gag protein and also to
ubiquitin. Depletion of cellular Tsg101 by small interfering RNA (RNAi) arrests
HIV-1 budding at a late stage, and budding is rescued by reintroduction of
Tsg101. Vps28 binds to Tsg101 and also appears to be essential for budding.
Finally, dominant negative mutant Vps4 proteins that inhibit vacuolar protein
sorting also arrest HIV-1 budding (Garrus JE, et al., Cell 107, 55-65, 2001). A
similar role for Tsg101 in HIV and Ebola virus budding is observed by
Martin-Serrano J, et al., Nature Medicine 7, 1313-1319, 2001. Figure provided
by courtesy of Drs. Jaisri Lingappa and Wes Sundquist, it was
redrawn by John Weddle. https://www.aidsreagent.org
In cytoplasm Gag mRNA is translated and the proteins are threaded through ER and processed and packaged in Golgi complex and the same are deposited in the plasma membranes. Even the matrix proteins are found at the inner surface of the plasma membrane.
In the upstream of the Gag protein there are several sequences such as IRES for ribosomal entry and Psi just upstream of the Gag.. This Psi sequence is used for the assembly of viral proteins into proviral particle. Then the proviral particle is gets associated with membrane bound Matrix proteins then with membrane associated P120 and p41 and the virus is budded off.
5’LTR of the HIV RNA is used for the assembly
A pair of HIV genomes; www.microbe.tv/twiv/reverse-transcription/
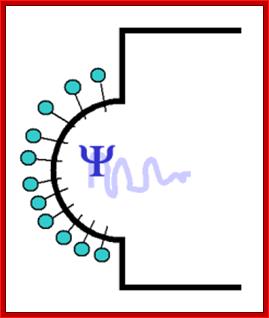
Assembly of viral capsid proteins cell membrane around;step1
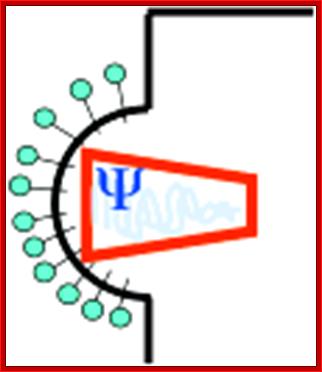
Assembly of viral capsids around genomic RNAs, step2
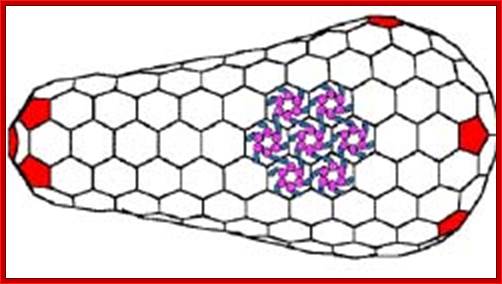
Purple CAN terminal domain and Blue CAC terminal domain; capsid proteins are hexagonally arranged subunits. They are the targets for developing long lasting vaccines.
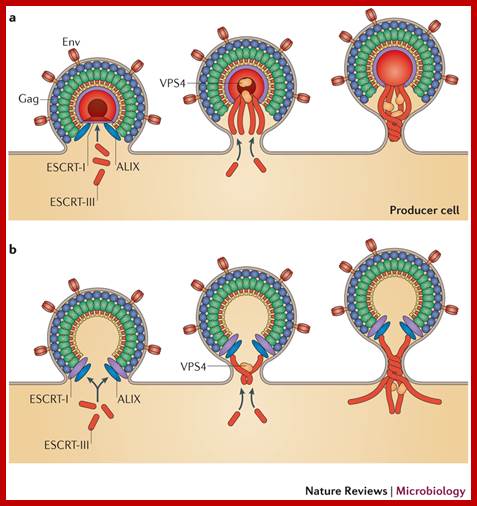
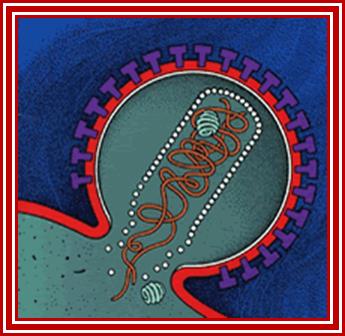
Viral budding in the process.
HIV-1 assembly, release and maturation; Eric O. Freed; http://www.nature.com
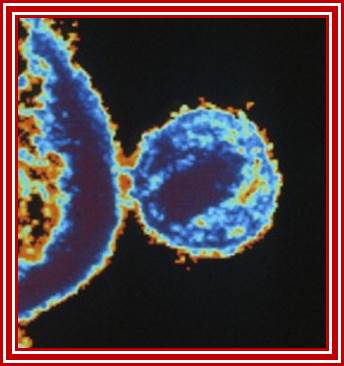
Viral budding
- Full-length viral transcript can be translated to generate precursor proteins, some are partially digested, and all put together assemble into viral particle and later they are budded off. However most of the mRNA are generated as spliced products and translated.
- Full length mRNA is produced as the last product and the same is used for the production capsid bound particle which later is used for budding with encapsulation of Matrix and envelop proteins.
Disease diagnosis:
Clinically diagnoses are done by using ELISA or take home kits, which give 90 to 96% of certainty of infection. Though expensive, clinicians claim that the PCR techniques good results and quick also.
Drugs:
- Vaccines against gp120, fails because of higher mutation rates in the gene.
- Several drugs such as 3’AZido 3’ deoxy Thymine (AZT), which blocks DNA synthesis by binding to reverse transcriptase enzyme.
- Similarly di deoxy ionosine and di deoxy thymidine are also used.
- Inhibitors of proteases are currently being used and experimented.
- In most of the cases combination of drugs are used.
- Immunization by using various combinations of GP120 epitopes has been employed.
- Antisense RNA against the viral RNA can be employed.
- One can try CD4 toxins or gp-41 toxins such as monoclonal antibodies against such proteins can be prepared and can be used.
- One can employ tat inhibitors or inhibitor for integrase.
- Cyclosporin-A can be used to suppress T-cell activation.
- There are many target elements against which one can employ strategic chemicals.
- There are claims vaccines against HIV are in the process of trail?
It is puzzling how and why the cure for HIV has not made available, in spite of the best possible brains, best equipments and billions of dollars was and is made available for research labs, universities and hospitals.
- Abacavir AKA Ziagen, a well-known anti-HIV drug may perhaps help against HIV but on the other hand ironically increases the risk of heart attack, reveals.
Fixed-dose combinations
Fixed dose combinations are multiple antiretroviral drugs combined into a single pill.
|
Brand Name |
Drug Names (INN) |
Date of FDA Approval |
Company |
|
September 26, 1997 |
|||
|
November 15, 2000 |
|||
|
September 15, 2000 |
|||
|
August 2, 2004 |
|||
|
August 2, 2004 |
|||
|
July 12, 2006 |
Fixed-dose combinations- from wikipedia;
Fixed dose combinations are multiple antiretroviral drugs combined into a single pill.
|
Brand Name |
Drug Names (INN) |
Date of FDA Approval |
Company |
|
September 26, 1997 |
|||
|
November 15, 2000 |
|||
|
September 15, 2000 |
|||
|
August 2, 2004 |
|||
|
August 2, 2004 |
|||
|
July 12, 2006 |
One good outcome of many years of research on retroviruses is that retro viral genomes can be selectively used for developing them as viral vectors for transferring foreign genes into mammalian cells for gene therapy.
Retroviral vectors:
A retroviral vector is used to infect compatible animal cells with the desired gene or genes. Mostly the vector size should small, containing LTR regions on both sides. The desired Recombinant DNA is introduced next to psi region. The upstream LTR provides upstream promoter elements and the down stream LTR provides Poly-A signal sequences. If the clone gets integrated, it can express the gene of interest. If some one wants to propagate this construct one has to use GAG, pol and envelope genes or they have to provide as helper viral packages.
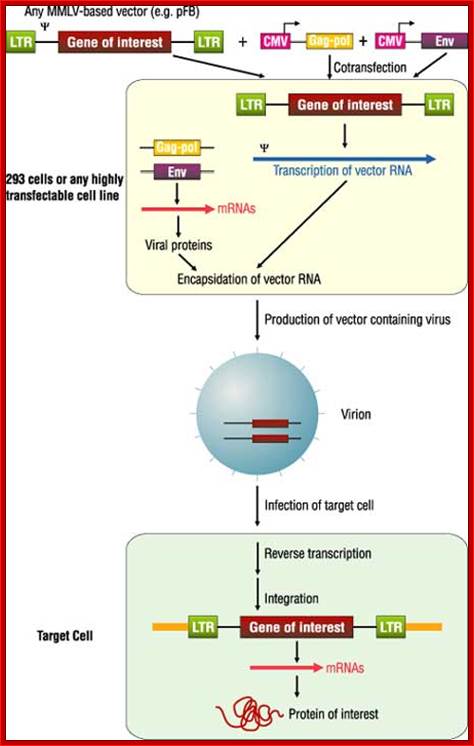
Fig. Recombinant DNA using HIV genome as a vector; http://www.biolab.cn/
Nonreplicating retroviral vectors contain all of the cis elements required for transcription of mRNA molecules encoding a gene of interest and packaging of these transcripts into infectious virus particles .The vectors typically comprise an E. coli plasmid backbone with the gene of interest inserted into a pair of 600-bp viral long terminal repeats (LTRs). The LTR is divided into three regions. The U3 region, containing the retroviral promoter/enhancer, is flanked in the 3¢ direction by the R region, which contains the viral poly-adenylation signal (pA). The U5 region follows and, along with R, contains sequences that are critical for reverse transcription. Expression of the viral RNA is initiated within the U3 region of the 5¢ LTR and is terminated in the R region of the 3¢ LTR. Between the 5¢ LTR and the coding sequence for the gene of interest resides an extended version of the viral packaging signal (Y+), which is required in cis for the viral RNA to be packaged into virion particles

Vector developed for the required gene: http://www.biolab.cn/
Although the relative speed and simplicity of the transient high-titer virus production methods used here are attractive for most applications, the gag-pol and env open reading frames (ORFs) are all followed by an internal ribosome entry site (IRES) linked to a downstream drug-resistance gene so that these vectors may also be used to establish stable producer lines. The antibiotic-resistance genes used in the gag-pol and env vectors are different from each other, and from those used in most of the more popular retroviral vectors; hence, any env vector may be used with the gag-pol vector and with a wide range of antibiotic-resistant retroviral vectors to produce triple-stable viral producer lines. The position of the antibiotic-resistant gene as the second ORF in a dicistronic expression cassette, as opposed to its expression from a second cassette on the same plasmid, ensures that expression of the viral packaging proteins is co-maintained with the antibiotic-resistant genes by including antibiotics in the media.
Presently scientists in various labs are working on how to produce vaccine against HIV. The genome undergoes frequent mutation, thus it is difficult to generate a vaccine against the virus. Some vaccines developed happen prevent spreading, but does not provide complete immunization. Recently using data base search they have found certain parts of capsid proteins are least mutated. By recombinant method, it is possible isolate such segments and generate vaccines, so it will be stable and long lasting.
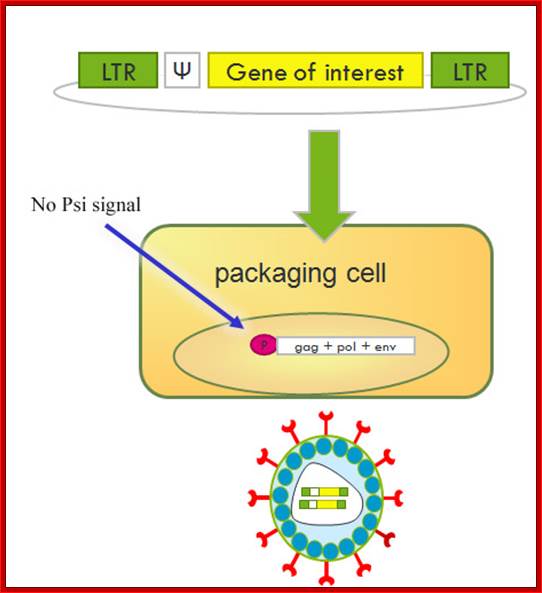
Retro viral products; http://www.giga.ulg.ac.be/
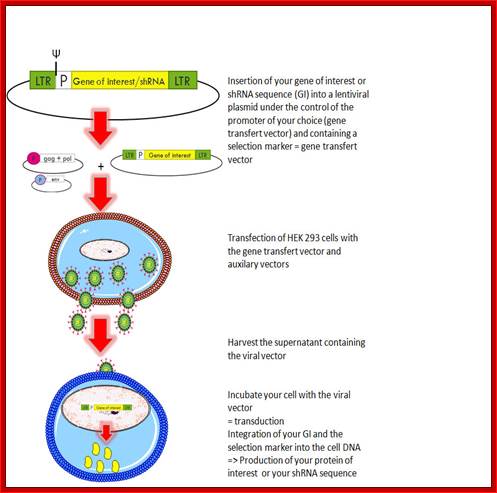
Retroviral vector is used for treatment with a designed gene. http://www.giga.ulg.ac.be/

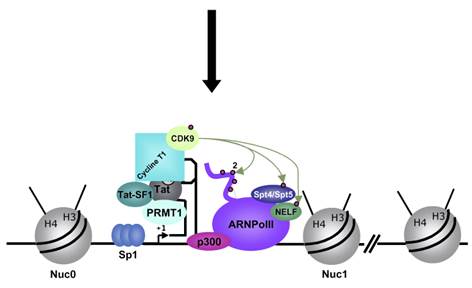
second
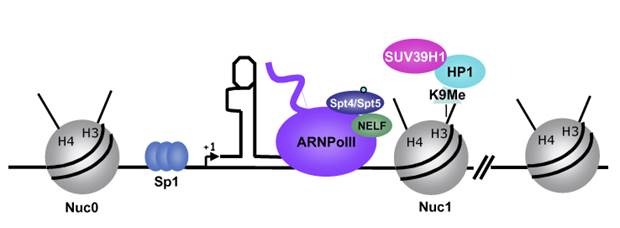
first
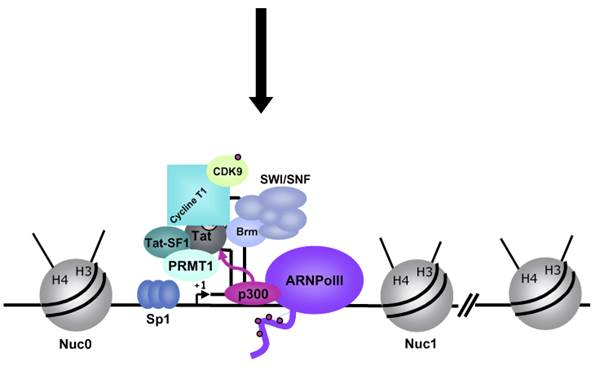
third
fourth

Fifth
Modèle de l’activation du promoteur du VIH-1 dans le contexte chromatinien. http://www.igh.cnrs.fr