HIV I and HIV II
HTLV I and II:
HTLVs cause T-cell leukemia and lymphoid malignancies.
· HTLV-I Infection, i.e. entry into T-cells is through its CD4 receptors. HTLV-II entry is through CD8 receptors; the virus does not produce any associated diseases.
· Infection of T-cells induces high count of WBC. Infected cells grow and proliferate in the absence of T-cell growth factors (TCGF) or interleukins-2, which are generally required for the growth of T-cells. If viral genome integrates it integrates randomly, and induces the synthesis of IL-2 and IL-2 receptors.
· In comparison to HIV genome, it has two extra genes called ‘tax’ and ‘rex’. The tax-gene product is a transcriptional activator can cause activation of several genes in host genome for cell division. Thus causes cell proliferation.
· It generates autocrine loop for the growth of the infected cells, also deregulates T cell-growth.
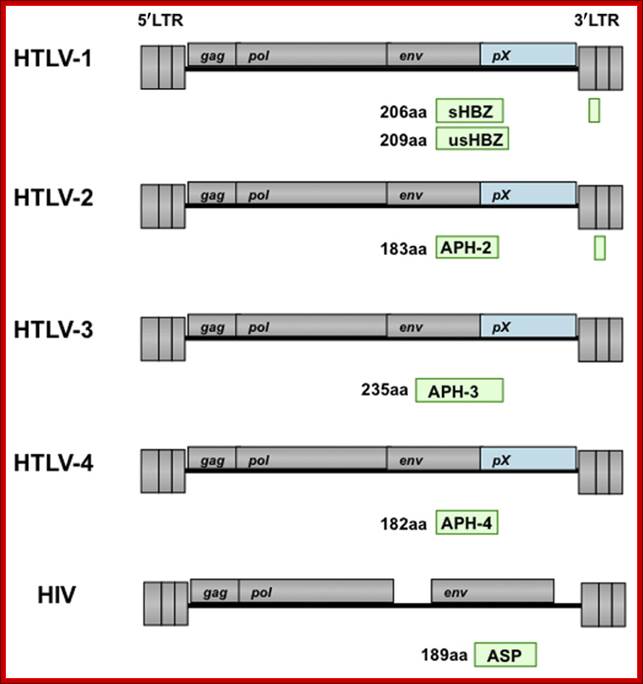
Human T-cell leukemia virus type 1 (HTLV-1) causes adult T-cell leukemia (ATL). The minus strand of HTLV-1 provirus encodes a bZIP protein donated as HTLV-1 bZIP factor (HBZ). Among the HTLV-1 regulatory and accessory genes, the tax and HBZ genes were thought to play critical roles in oncogenesis. However, HBZ is the only gene that remains intact and is consistently expressed in all ATL cases, while the tax gene is frequently inactivated by epigenetic modifications or deletion of the 5′ LTR. HBZ gene promotes the proliferation of ATL cells through its mRNA form. Moreover, HBZ induces T-cell lymphoma and systemic inflammation in vivo. HBZ fulfills its functions mainly through regulating HTLV-1 5′ LTR transcription and modulating a variety of cellular signaling pathways which are related with cell growth, immune response, and T-cell differentiation. Taken together, the multiple functions of HBZ render its predominant function in leukemogenesis of ATL. TiejunZhao1,2 and Masao atsuoka1*http://journal.frontiersin.org/
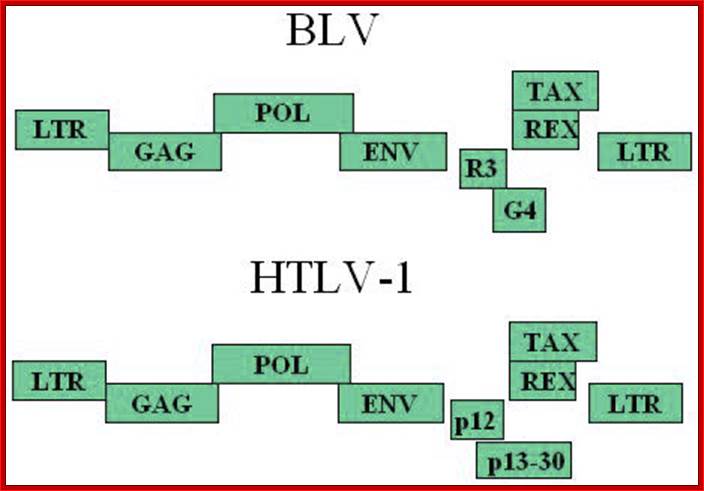
Schematic representation of the Bovine leukemia virus BLV and Humna T-cell leukemia virus HTLV-1 genomic structures; Three structural genes code for the viral capsid (gag), the reverse transcriptase / integrase (pol) and the envelope(env). A series of other open reading frames (ORFs) at the 3'-end of the genome encode regulatory proteins: Tax, Rex, R3 and G4 (BLV) or p12 and p13/p30 (HTLV). These ORFs are translated from complex single or multiple spliced mRNAs (see 15,26,90 for further details). Tax and Rex are essential proteins required for transcriptional and post-transcriptional activation of viral expression. The R3 and G4 proteins are dispensable for infectivity but are involved in the maintenance of high viral loads; .http://bioscience.org/
Schematic structure of (a) the Bovine Leukemia Virus (BLV) genome and (b) the viral particle. http://www.mdpi.com/

HTLV-http://www.ffe.com.tw/
![]()
![]()
![]()
HIV; https://commons.wikimedia.org/
HTLV infection:
Retrovirus entry relies on interactions between the viral envelope glycoproteins (Env) and one or more, cell receptor(s). In the case of HTLV-1, a first receptor, the ubiquitous glucose transporter GLUT1, was discovered in 2003. It was later demonstrated that Neuropilin 1 (NRP-1) is also a cell receptor for HTLV-1.

Retrovirus entry relies on interactions between the viral envelope glycoproteins (Env) and one, or more, cell receptor(s). In the case of HTLV-1, a first receptor, the ubiquitous glucose transporter GLUT1, was discovered in 2003 (1). We later demonstrated that Neuropilin 1 (NRP-1) is also a cell receptor for HTLV-1 (2). NRP-1 is a cell surface receptor initially characterized as a receptor for Semaphorin 3a, a regulator of axon motility. NRP-1 is also a receptor for various VEGF isoforms and plays therefore an important role in angiogenesis. NRP1 is over-expressed upon T-cell activation and is recruited within the immunological synapse, which resembled the structure allowing the cell-cell transmission of HTLV-1; http://cochin.inserm.fr/
NRP-1 is a cell surface receptor initially characterized as a receptor for Semaphorin 3a, a regulator of axon motility. NRP-1 is also a receptor for various VEGF isoforms and plays therefore an important role in angiogenesis. NRP1 is over-expressed upon T-cell activation and is recruited within the immunological synapse, which is a reassembled structure allowing the cell-cell transmission of HTLV-1.
HTLV gene expression:
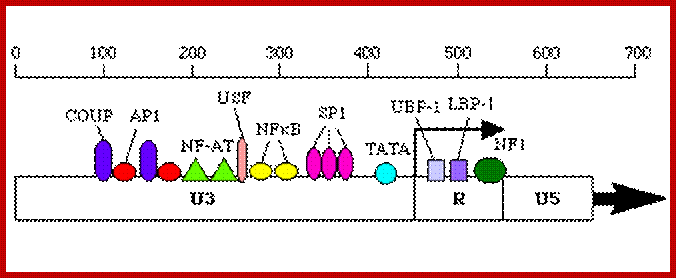
Promoter elements and their Transcriptional complex bind to their proximal regions of the promoter elements, shown in the diagram.
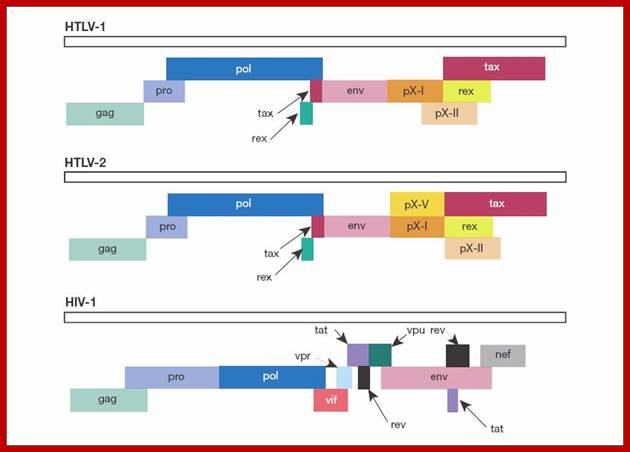
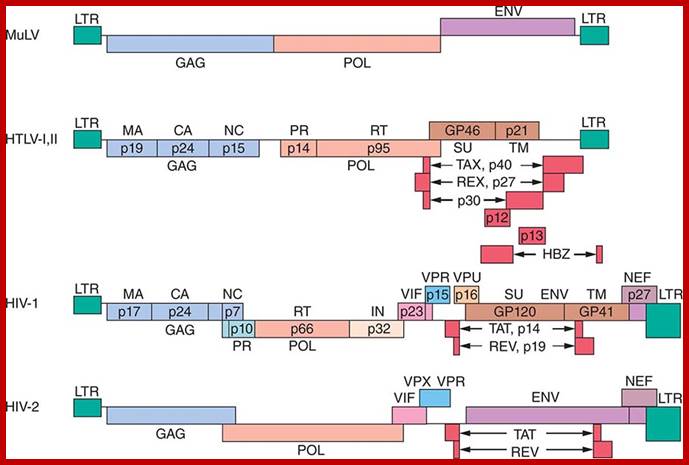
Genomic organization of human T-cell lymphotropic virus (HTLV) type 1, HTLV type 2, and HIV. Adapted from Mandell GL et al, eds.Principles and Practi; http://www.obgyn.net/
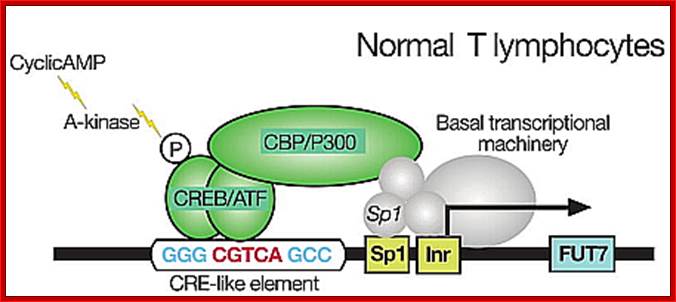
Normal T lymphocyte cells (uninfected) transcriptional activation; http://lookfordiagnosis.com/
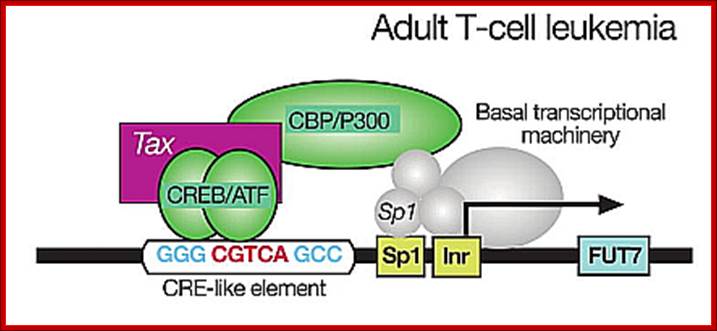
Adult T lymphocyte infected with HTLV where TAX gene products activate host cell transcription; thus cells enlarge their population. http://lookfordiagnosis.com/
Genome once enters into the target cell, it produces transcripts which undergo splicing and mRNAs are transported into the cytoplasm for translation. The ‘tax’ gene plays very important role is activating host cell genes as well as its own
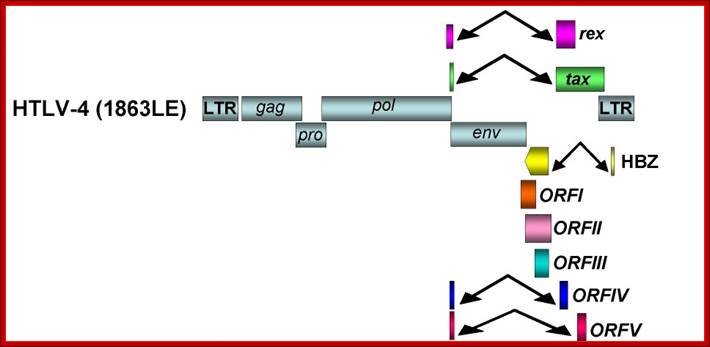
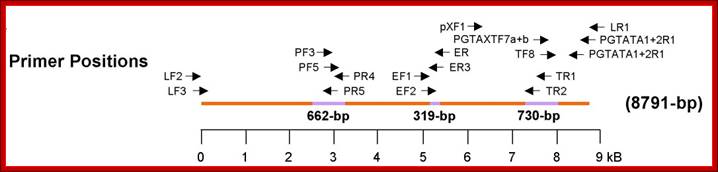
Organization of the HTLV-4 genome (a) and schematic representation of the PCR-based genome walking strategy (b). (a) shown are non-coding long terminal repeats (LTR), coding regions for all major proteins (gag, group specific antigen; pro, protease; pol, polymerase; env, envelope; rex, regulator of expression; tax, transactivator), HTLV basic leucine zipper (HBZ), and 3' genomic open reading frames (ORF) of unknown function. Putative splice donor (sd) and splice acceptor (sa) sites are indicated. (b) Small proviral sequences (purple bars) were first amplified from each major gene region and the long terminal repeat using generic primers as described in methods. The complete proviral sequence was then obtained by using PCR primers located within each major gene region by genome walking as indicated with arrows and orange bars. Switzer et al; http://www.retrovirology.com/
· Infection leads to the expansion of infected T-cell. The transformed T-cells become bigger in size and show multinucleation. Interestingly, the viral genome does not get integrated into the host chromatin DNA in most of the cases.
· Host cell (T-cell) receptor genes for HTLVs are located in human chromosome 17.
· HTLV-I and II’s tax-gene codes for a protein that activates the expression of viral genes and it also activates some cellular genes.
· The Rex- gene codes for the expression of single spliced or unspliced full length mRNA (participates in transport of unspliced or single spliced mRNAs out of nucleus into cytoplasm) for generating viral proteins which are required for the formation of viral particles.
· HTLV-I infects CD8 cells but patients show no sign of associated diseases.
· T cell infected with HTLV transmits to other healthy cells by cell to cell synapses. That is the means of expansion of HTLV population.
However both strains of HTLVs can transform and immortalize infected cells at low frequency.
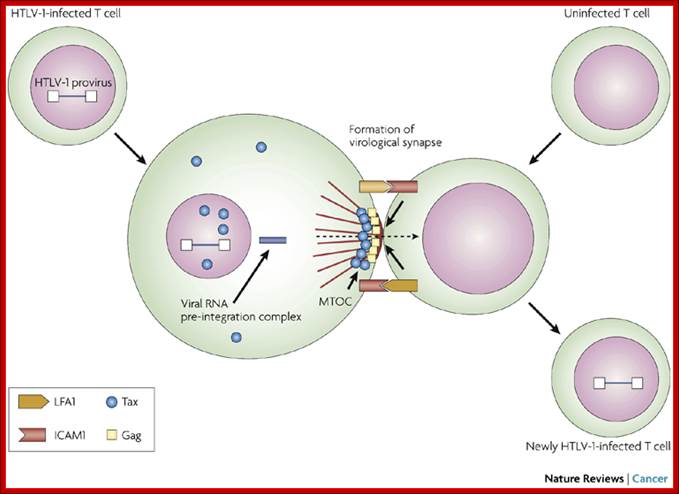
Unlike many other viruses, cell-free human T-cell leukaemia virus type 1 (HTLV-1) virions are largely non-infectious. This figure illustrates the cell–cell contact required to create a virological synapse through which the viral genome is transmitted from one cell to another. The roles played by lymphocyte function-associated antigen 1 (LFA1), and intercellular adhesion molecule 1 (ICAM1) in forming cell–cell contact are shown. Tax contributes to the formation of a microtubule organizing centre (MTOC). Cell–cell transmission of HTLV-1 occurs through a virological synapse.
Virological synapse for the expansion of infected cell population; Masao Matsuoka & Kuan-Teh Jeang http://www.nature.com/
Fluorescent labeling to show synapse between infected and fresh cells www.mailce.com/lenfosit-nedir.html
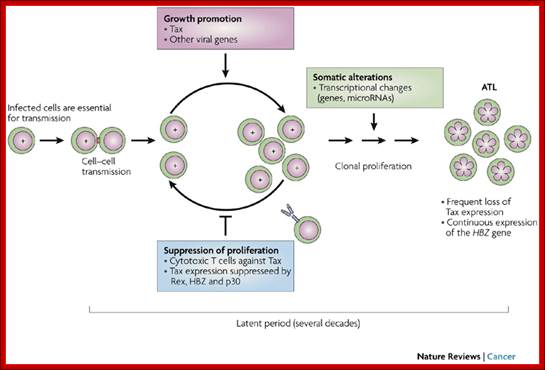
Cell–cell transmission of human T-cell leukaemia virus type 1 (HTLV-1), early expression of Tax and subsequent cytotoxic T lymphocyte (CTL) selection against Tax are shown. HTLV-1 basic leucine zipper factor (HBZ) is continuously and durably expressed in infected cells. A T cell with '+' in the nucleus represents an infected cell with integrated provirus. An uninfected T cell (empty nucleus) is poised to become infected through a virological synapse (small red rectangle) created by cell–cell contact with an infected cell. The clonal expansion of infected cells is promoted through the actions of Tax and HTLV-1 accessory proteins, and selected against by host CTLs. After a prolonged asymptomatic period of 20–40 years, aneuploid adult T-cell leukaemia (ATL) cells ('flower cells', with flower-shaped nuclei) emerge in approximately 5% of infected individuals. Most HTLV-1-infected individuals remain life-long asymptomatic carriers.
Starting from infected cells and transmission to fresh cells by synapse and the same expansion activation to produce adult T cell (ATL) population. Masao Matsuoka & Kuan-Teh Jeang; http://www.nature.com/

Anatomy of a retroviral virological synapse; Quentin Sattentau; http://www.nature.com/
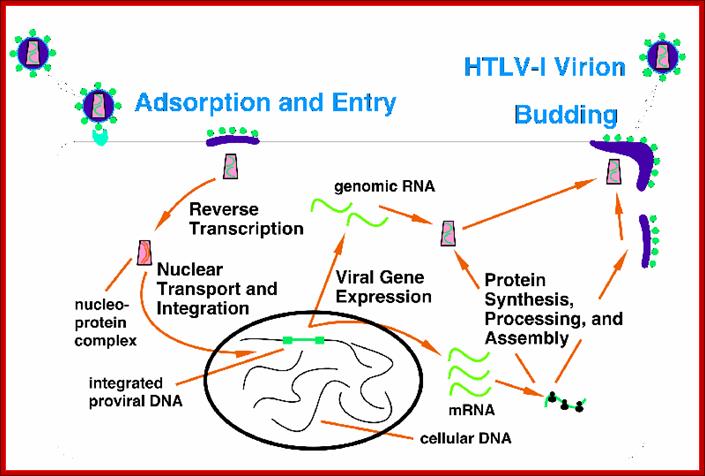
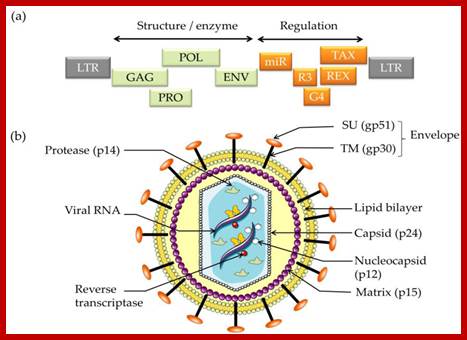
HTLV life cycle; http://www.bioscience.org/
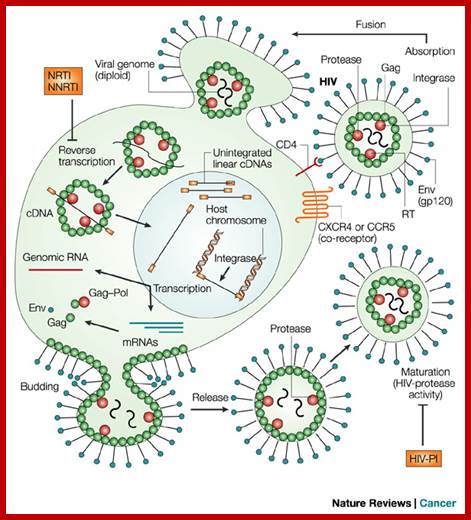
http://summer2012microbiologyuncg.blogspot.in/
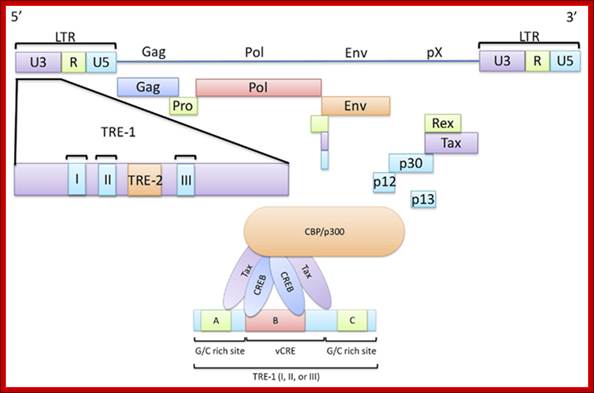
HTLV Tax; http://c431376.r76.cf2.rackcdn.com/31352/fmicb-03-00406-HTML/image_m/fmicb-03-00406-g001.jpg
HIV I and II:

HIV1; http://www.abovetopsecret.com/
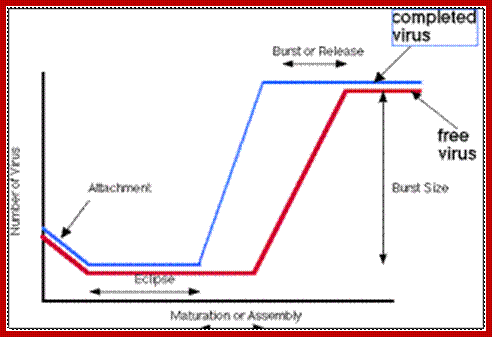
Infection and release- Time scale;
Virology-microbiology-b.blogspot.in 2009

HIV2 http://www.newsbackatcha.com/
 CD4 is a cell-surface
glycoprotein found on the mature helper T cells and immature thymocytes, as
well as on monocytes and macrophages. (Some cytotoxic T cells have CD4 protein
as well.) Normally, about 65% of T cells in the blood are CD4+ (have CD4
protein protruding from their membrane). http://www.mcld.co.uk/
CD4 is a cell-surface
glycoprotein found on the mature helper T cells and immature thymocytes, as
well as on monocytes and macrophages. (Some cytotoxic T cells have CD4 protein
as well.) Normally, about 65% of T cells in the blood are CD4+ (have CD4
protein protruding from their membrane). http://www.mcld.co.uk/
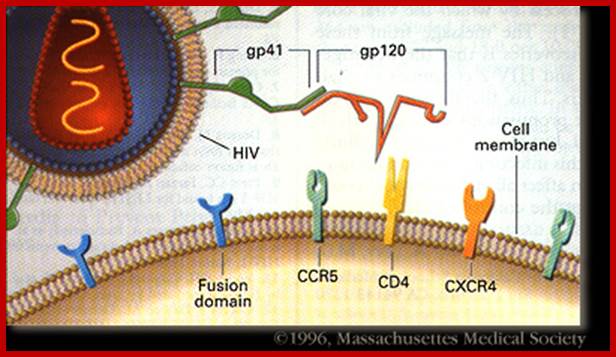
https://web.stanford.edu/

Interaction of R5 and X4 HIV-1 gp120 with different cell surface molecules may result in stimulation of distinct signaling pathways. R5 HIV-1 gp120 binds to the preassembled CD4–CCR5 complex. This interaction may result in simultaneous stimulation of CD4- and CCR5-mediated signaling (Signal 1 & 2). In contrast, X4 HIV-1 gp120 binds first to CD4 and then to the CXCR4 receptor. Interaction of the X4 gp120 V3 loop with cellular HSPG may interfere with the association between the CD4–gp120 complex and CXCR4. Consequently, the earliest X4 gp120-induced signaling may be generated mostly by the CD4 receptor (Signal 1). Subsequent binding to CXCR4 may induce signal 2. Thus, R5 and X4 HIV-1 binding to target cells may contribute differently to HIV pathogenesis; on ce the virus is bound to the host cell membrane it is internalized by endocyctosis; https://www.researchgate.net/
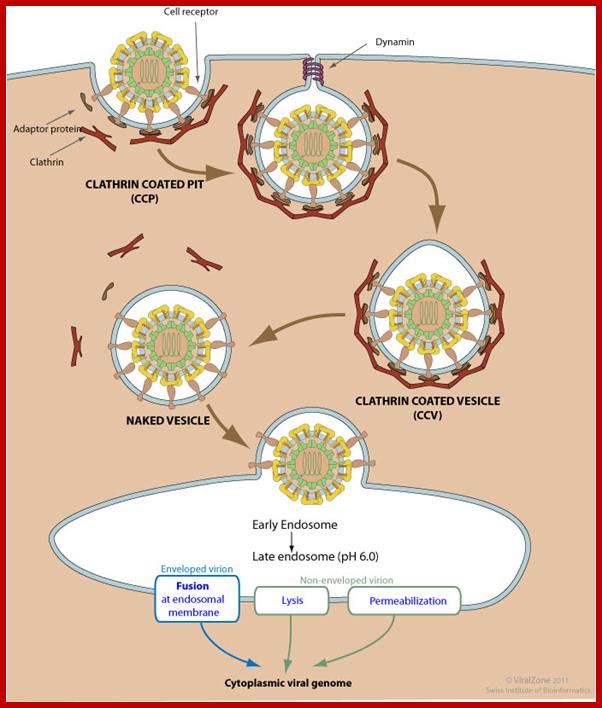
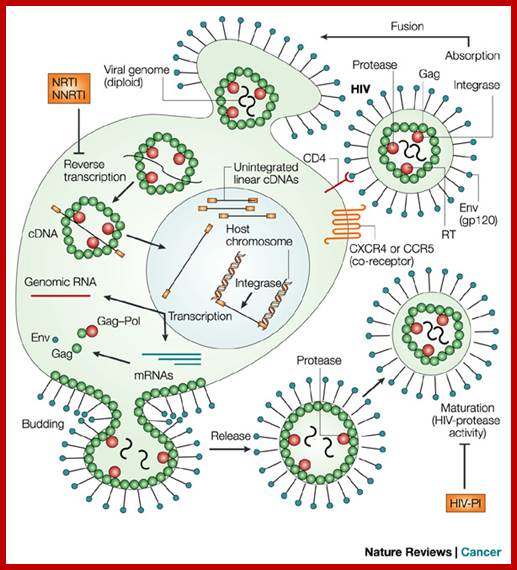
Clathrin-mediated endoycytosis (CME) and dynamin-dependent fusion with endosome; Endocytosis is triggered by the binding of the virion to host receptors. The virion attachment to the host receptor induces the binding of an adaptor protein to the receptor cytoplasmic tail. Endocytosis can occur via clathrin-mediated endocytosis, caveolin-mediated endocytosis or clathrin- and caveolin-independent endocytosis. Adaptor proteins bind to clathrin, and the local concentration of adaptor proteins on the inside face of the plasma membrane allows clathrin to multimerize to form characteristic invaginations or Clathrin-Coated Pit (CCP). Membrane scission proteins DNM1 /Dynamin-1 or DNM2/Dynamin-2-pinch off the CCP from the host membrane thereby releasing the Clathrin-Coated Vesicle with the virus (CCV) into the cellular cytoplasm. The clathrin basket is subsequently released from the vesicle by auxilin and hsc70. The vesicle then delivers its viral content to early endosomes. Endosomal acidic pH and /or receptor binding usually induces structural modifications of the virus surface proteins that leads to genome penetration into the cytoplasm via fusion or permeabilization mechanisms. http://viralzone.expasy.org/

http://www.microbiologybook.org/
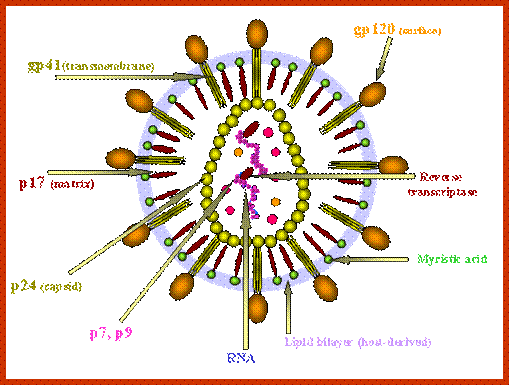
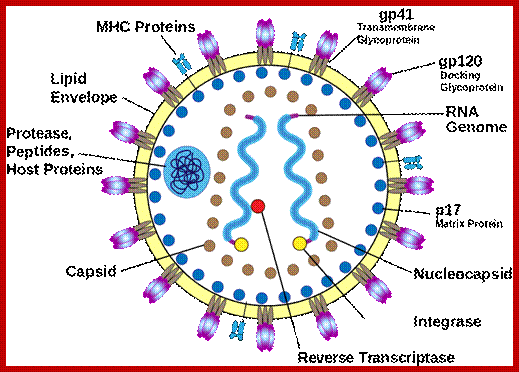
Virus is enveloped with a membrane and many proteins are embedded in it, like Gp41associated with gp120, matrix protein p17; inside one finds two positive RNA strands (9749ntds) associated with Reverse transcriptase and p7/p9; http://www.microbiologybook.org/
· Human immuno deficiency virus (HIV) viruses contain several proteins on viral enveloped membranes, such as glycoprotein Gp120, transmembrane protein Gp41, Human leucocytes antigenic proteins HLA-I and HLA-II. Some times they also carry MHC components.
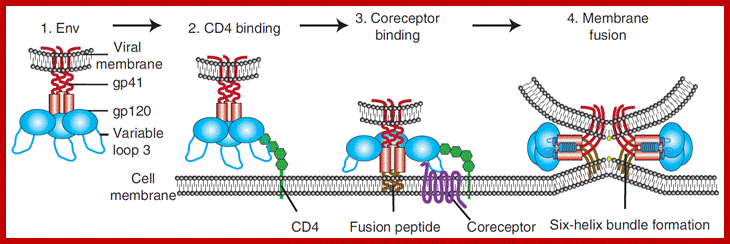
· Infection is through its gp-120 (N-terminal region of the protein) binds to CD4 (55KD) of T-cells or subset of T-cells, helper T-cell or cells responsible inducing immune response.
· Viral gp120 and TM protein form trimeric complexes, where the TM is anchored in the membrane and the Su is noncovalently bound to Tm41 protein. The protein is glycosylated at least at 35 positions at aspartyl residues.
· The gp120 shows 5 domains (motifs from N end to C end of the protein).
· Among the gp120, the domain V3 is important for it contacts CD4 and co receptors.
· The gp-41 helps in the fusion of viral membrane with host membrane, resulting in the entry of the naked viral particle into the cytoplasm.
· Another cellular receptor protein, which helps the entry of the virus, is co receptor CCR5 or CXCR4 domains, (26Kd), which has protease activity. Viral gp-120 has another motif called V3; contact co receptors, it facilitates membrane fusion and internalization.
· Most important reason for immune deficiency is due to rapid dysfunction of T- helper cells, for T-H cells have maximum number of CD4 receptor than any other host cells, so they are incapacitated first.
· Even cytotoxic T-cells with CD8 get destroyed.
· The B-cells become hyperactive.
· HIV patients though carry B-memory lymphocytes, yet opportunistic infections and infectious agents are common. Many of the infected cells are destroyed.
· Monocytes in the infected persons express CD4 receptors, so they also become targets for viral infection.
· Some strains of HIV have greater preference to Monocytes.
· Even macrophages are infected and natural killer cells become defective.
· Infections of cells with a delay lead to the production of viruses and they are released into body fluids by the destruction of cells.
· In some, upon infection, the viral genome gets integrated and such cells expand.
· Detection can be accurately made by immuno diagnostic methods; in fact antibody based assays give more than 98 % accuracy for several different strains. DNA-PCR or RNA-PCR is also used for detecting the disease, but clinically, the technique does not give 100% accuracy.
· Infected cells with integrated genome can be activated by another viral infection or infection of the HIV virus itself.
· Human immuno-deficiency viruses contain some additional genes, which are not present in other retro viruses.
· One of the HIV genes tat-1 codes for proteins that activate the expression of its own genes but also activates few cellular genes.
· Expression of the REV gene of HIV is important for it is this protein that is responsible for the transportation of the full length and unspliced mRNA into cytoplasm for the production of viral particles.
· Most of the retroviruses are 100nm thick and have a membrane envelope, studded with two types of proteins; one is surface Glycoprotein (gp12Kd), which actually recognizes host cell receptors.
· Second one is a transmembrane protein (gp 41Kd), which is cross-linked to Glycoprotein. During internalization process, the transmembrane protein cleaves the glycoprotein at a particular site, thus the viral and cell membranes are brought together and facilitate fusion.
· Another host cell protein, that performs an additional role in fusion, is CD26. This protein has protease activity.
· HIV/HTLV viruses interact with mitochondria and exhibit pro-apoptotic properties, however Mitochondria mediated immune responses might be an evolutionary adaptation by which mitochondria might have prevented the entry of invading micro-organisms thus establishing themselves as an integral part of the cell.
HIV1 and HIV2 show some differences in terms of transmission from persons to persons and HIV2 is found hard to transmit from person to person as HIV1. Genomically HIV1 contains VPU and HIV2 contains VPX. Genomically they are not identical. HIV2 is more close to simian immunodeficiency virus.
HIV Subtypes:

Within group M there are known to be at least nine genetically distinct subtypes of HIV-1. These are subtypes A, B, C, D, F, G, H, J and K. http://www.avert.org/
Most of the recombinant viruses originated from Africa (Table 1) and substantial majority of them are 12 full length hybrids, include segments of subtype A. This is not surprising, since it appears that the various subtypes of HIV-1 first arose in central Africa where they have co-circulated for many years. Furthermore, in that region, subtype A is the most common. Among the hybrids of African origin, MAL is interesting because it was one of the first African (and, in retrospect, nonsubtype B) viruses characterized. Li et al. first suggested that MAL may be a recombinant virus (1987), but the realization of the widespread occurrence of recombination among HIV-1 subtypes did not occur for about another 7 years. While MAL includes regions that are clearly of subtype A and D origin, there is a long region encompassing the 5' half of the pol gene that has defied characterization. Now it appears that part of that region may be subtype I (see below), but the origin of the remainder has still not been determined Thus, the ancestry of MAL may have involved successive recombination events of sequences derived from three or four different subtypes. Z321B, which also appears to include sequence blocks from three (or four?) different subtypes, is the oldest of the known hybrid viruses, having been isolated in 1976. Clearly, by that date, the group M subtypes had diverged sufficiently that the various segments recombined within Z312B are detectable as having distinct subtype origins. This implies that the subtypes must have begun to diverge substantially earlier than the 1970s, and is consistent with the recent finding that a virus from 1959 appears, from phylogenetic analyses, to have postdated the group M radiation. Among the hybrid viruses isolated outside of Africa are two subtype "E" viruses (CM240 and 93TH253.3) from Thailand. As shown in Figure 1, a large fraction of the genome of these subtype "E" viruses, including the gag and pol genes, appears to be of subtype A origin. Since all Thai "E" viruses appear to be closely related, and since the mosaic structure of their genomes appears to be identical to that of the other full length subtype "E" virus (90CF402.1) from the Central African Republic, it is most likely that a single recombination event occurred in Africa, and that the subtype "E" viruses in Thailand are due to the introduction of a descendant of that recombination event. While the subtype "E" viruses clearly indicate that a subtype E arose in the past, it is possible that full length subtype E viruses no longer exist. In contrast, the recombination event between subtypes B and F to generate 93BR029.4 most likely occurred in Brazil, where both of these subtypes are common. At least three other viruses from Brazil, for each of which only partial sequences are available, appear to represent independently generated B/F hybrids]. The other non-African example, 94CY032.3, was isolated in Cyprus. Initially, partial sequences from this virus formed the basis of the proposition of subtype I. Analysis of a recently determined full length sequence suggests that it is a recombinant of at least three subtypes, A, G and I. Since 94CY032.3 is the prototypic subtype I virus, it is most parsimonious to assume that all segments which cannot be assigned to any other subtype are of a common (i.e., subtype I) origin. Two of the recombinant viruses from Africa, MAL and Z321B, contain short segments of sequence closely related to 94CY032.3, but not similar to other subtypes; thus, these regions have also been designated as being of subtype I origin (Figure 1). It is not clear whether the multiple recombination events in the ancestry of 94CY032.3 occurred in Africa, or in Cyprus, where a number of different subtypes are also found.
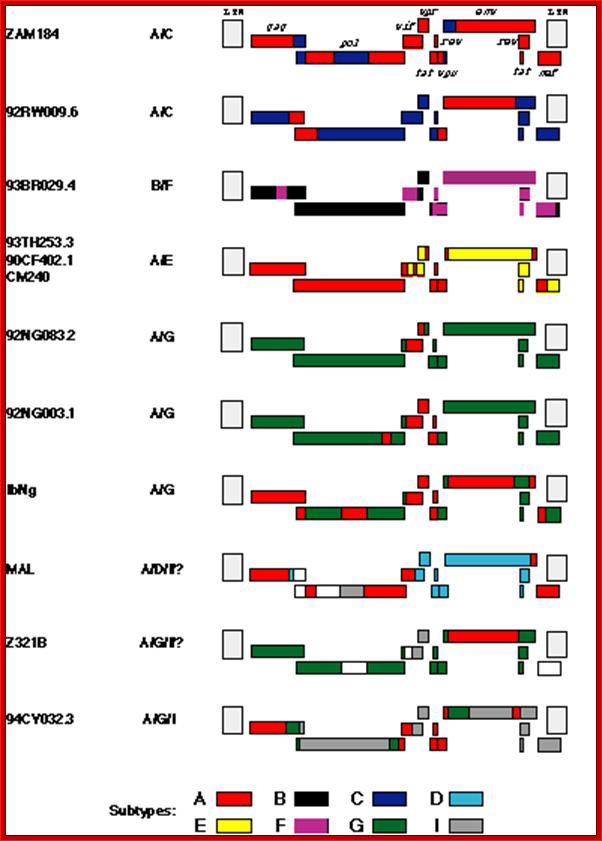
Full-length inter subtype recombinant HIV-1 sequences. The color
code indicates the subtype origin of different segments of the genomes; regions
in white are sequences whose origin has not been determined. The LTRs are
hatched because they have not been analyzed in detail. See Table 1 for more
details. http://www.thebody.com/
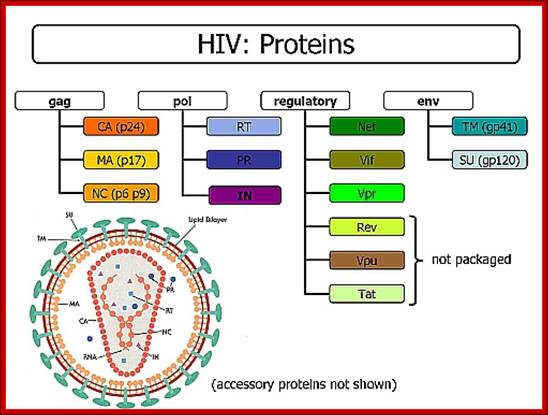
Genome: components: http://www2.gsu.edu/
Viral Proteins:
|
Acronym |
Name of the protein |
Protein Mol.wt in KD |
Gene |
Character |
MA |
Matrix proteins |
17/18 |
Gag |
At inner surface of the envelope, p-17 has nuclear localization signal
|
|
CA |
Capsid |
6 outer 24/25 inner |
Gag |
Abundant proteins, covers inner core |
|
NC |
Nucleo capsid |
7/9 |
Gag-pol |
Essential for the cleavage of gag proteins
|
|
PR |
Protease |
12 |
Gag |
Essential for the cleavage of gag proteins
|
|
|
|
|
|
|
|
RT |
Reverse transcriptase |
51/66 |
Pol |
It is a RNA dependent DNA polymerase
|
|
IN |
Integrase |
32 |
Pol |
Performs cleavage and ligation
|
|
SU (S) |
Surface glycoprotein |
120 |
Env |
Glycoprotein sticking out of Env. Membrane
|
|
TM (N) |
Transmembrane |
41 |
Env |
TM protein holds the glycoprotein by s-s bonds
|
|
Regulatory proteins |
|
|
|
Nef, Vif,Vpr,Rev,Vpu and Tat |
|
|
|
|
|
|
www.lookfordiagnosis.com/mesh_info.php
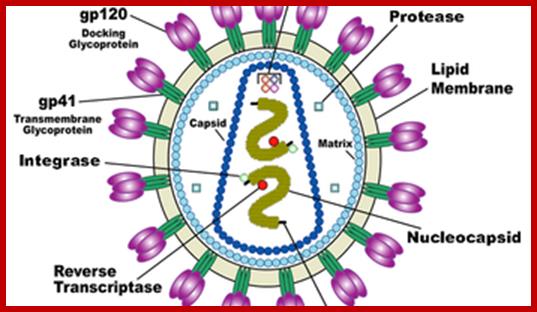
HIV image showing membrane embedded with gp41 and gp120; inner nucleocapsid and the two genomes. http://www.innovationtoronto.com/
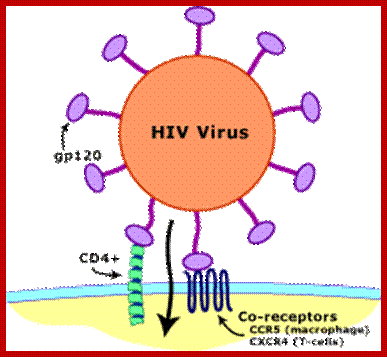
In 2001, Dr. Robert Gallo's group identified such a factor, which appears to be a combination of chemicals (called chemokines): MIP-1-alpha, MIP-1-beta and Rantes. The fact that CCR5 not only allows HIV entry into CD4 bearing cells, but is also a receptor for these chemokines explains two important aspects of the interaction between the immune system and the virus. When the CD8+ cells effectively make a large quantity of the chemokines, they may fill up and block the "doorway" for infection provided by the CCR5 protein. Conversely, when levels of the chemokines are low or absent for any reason, the virus is free to more easily infect cells because the CCR5 receptor protein is readily available to it.
Collectively, the back-to-back discoveries of the role of the chemokines and the CCR5 receptor site shed important new light on how HIV infects cells and may explain why the disease process differs from person to person. It should also be noted that another CD8+-derived antiviral factor (CAF), documented first by Dr. Jay Levy (co-discoverer of HIV), has been shown to inhibit HIV replication, but the origin of this factor remains unidentified. Chemokines block the binding of HIV gp120 to bind to cell receptors.
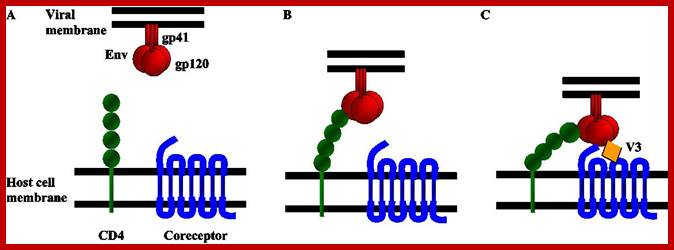
Schematic diagram of the first steps of the HIV entry process. (A) HIV entry into target cells is mediated through the interaction of the viral envelope glycoprotein (Env) with host cell surface receptors. (B) Surface gp120 first contacts CD4 and then (C) a chemokine receptor, typically CCR5 or CXCR4, triggering molecular rearrangements in the gp41 that result in membrane fusion and the initiation of infection. http://www.intechopen.com/
HIV entry mechanism is considered to be a good target for various drugs and many have been designed to inhibit different entry steps, however, as mentioned previously resistance to these drugs arise faster than we are able to create any new ones. This summer I’m also working on such entry-inhibitor molecule. Biochemistry department in Zurich University have created a libraries of DARPin (=designed ankyrin repeat proteins) molecules that have different structures and can target and inhibit activity of different molecules. One of these libraries has been made for DARPins that can bind V3 region of gp120 and therefore, inhibit HIV binding to its coreceptor. Antibodies that target V3 are quite rare in HIV patients and are not very effective (I would guess mostly because V3 is kind of *hidden* inside gp120 and so the ‘bulky’ antibodies can hardly access it). DARPins, on the other hand, are quite small (15-18kDa) and can access V3 and other hidden structures much more readily then the antibodies. Moreover, commercial DARPin production is much more viable as it can be made in prokaryotic systems (by simply expressing DARPin sequence containing plasmid in E.coli), whereas antibody production requires much more resources and eukaryotic systems. Obviously, because of the incredible HIV diversity (i.e. there is diversity among V3 regions as well) many different DARPins need to be tested for their activity against different HIV strains. So during my time here I’ll be introducing mutations in HIV gp120 and trying to inhibit the virus using different DARPins. The project involves making HIV with different V3 region sequences and making infectivity and neutralization assays to analyze how sequence variation affects DARPin effectiveness as well as general HIV infectivity.; http://questiongene.com/researching-hiv/
· At the inner surface of the envelope, there are matrix proteins, which contact both envelope membrane and also several coat proteins and core proteins.
· The capsid protein is believed to have two layers, outer and inner. They form a cylindrical structure, inside which genome is found.
· In the central core region there are two positive sense genomic RNA molecules, but they are held to each other at 5’ region by hydrogen bonds.
· Each of the RNA molecules is bound by a number of nucleo capsid proteins, to give an appearance of chromosomal feature to RNA- proteinaceous thread.
· Associated with this or bound to RNA genomes is reverse transcriptase enzyme.
· Each of genomic RNAs carry specific lys-tRNA, hydrogen bonded to primer binding site (PBS).
· Also found in the core are integrases, protease and any other factors of unknown nature.
· When the virus is delivered into cytoplasm as provirus, it is already activated, and the whole process of unique replication mechanism takes place in cytoplasm and with in the viral capsid core. It is virtually reminiscent of Reo viruses.
· In the activated viral core (one layered coat proteins) contains all the required components to produce new viruses of the same kind in large numbers.
· Besides the genomic RNA, the core of the pro-viral component contains, reverse transcriptase enzyme; more to it, all the required dNTPs can easily enter into the core.
· Reverse transcriptase, in a unique mechanism produces first a complementary cDNA strand and then it generates ds cDNA.
A List of Viral Genomes and their Primer tRNAs:
|
Virus |
Size of the genome (ntds) |
Primer tRNA |
|
|
RSV |
~9300 + 3’ Src gene |
Trp-tRNA |
|
|
MoMuLV |
~8300 |
Pro-tRNA |
|
|
ALV |
~7300 |
tryp -tRNA |
|
|
HTLV-1 & 2 |
~8500 |
Pro-tRNA |
|
|
HIV-1&2 |
~9749( ?) |
Lys-tRNA |
|
|
ALV |
~7300 |
Trp |
|
|
MLV mammalian C type |
8300 |
pro |
|
|
MMTV |
8600 |
lys |
|
|
HFV , H foamy virus (spuma) |
11200 |
lys |
|
|
Fish- WDSV |
12300 (13kb) |
his |
|
HFV = human foamy virus
WDSV = Wally dermal sarcoma virus
MMTV = mouse mammary tumor virus.
MoMuLV = Melony murine Leukemia virus
Structural Organization of the Genomes:
· All genomic RNAs are plus strands, capped at 5’ end and polyadenylated at 3’ end, a typical feature of eukaryotic mRNA, however it does not contain apparent introns.
· Next to 5’ end there is a block of sequence named as R, 20 to 250 (10 to 80ntd in some strains) ntds long, the same sequence is also present at the 3’ end as direct repeats, so the term R (R= Repeats; can be called Terminal Repeats).
· Next to R from 5’ end, another sequence called U5 is present, meaning untranslated 5’ end region. The size of it varies from 80 to 250 ntds.
· Next to U5 is 18 ntd long primer binding site (PBS). It is the site at which 3’ end of a specific tRNA is base paired. The free 3’OH group acts like a primer for the synthesis of complementary strand; it is reminiscent of primers used in DNA synthesis. Primers, in general, are not used in RNA synthesis, but replication of DNA requires primers; here the enzyme uses RNA to produce a complementary copy of DNA, hence this genetic RNA requires primers (reasonable explanation).
· Next to PB is sigma, a region of 100 –450 ntds long extending into 400 or more ntds into 5’ end of GAG gene. This region is very important for it is used as packaging sequences.
· Then the GAG gene, Polymerase gene and Envelope envelop gene, of 2000, 2900 and 1800ntds long respectively, are present in same order.
· Each of the genes has translation termination sites at named as t1, t2 and t3 respectively.
· They have splice donor sites one at the 5’ end of the UTR region, located between PB and packaging signal sites and the other at the end of Pol segment.
· Similarly, one splice acceptor site is located at the end of Pol or say at the beginning of the Env. The second one is at the end of the Env gene.
· Next to Env gene is another untranslatable region called U3 of 200 to 1200 ntds, then there is another direct repeat segment of 20 to 250 ntds long R region.
· Between or at the junction of Env and U3 there is a 8 to 16 ntds long polypurine rich tract present, consisting of A s and G s, which is of very importance for it acts as the primer during complementary (+) strand DNA synthesis.
· The 3’ R region has a sequence called poly (A) signal, AAUAAA, located about 30 ntds upstream from the start of Poly (A) tail.
· The poly (A) tail is about 200 to 250 ntds long.
· Basically all Retro viral genomes have the said characteristic features such as 5’ and 3’ ends and 3 genes.
5’-U5-->--> PB--> sigma-->gag-->pol-->env-->ppt-->U3->- (A) n-3’
|
Virus |
Size and Organization of Genes in viral genomes
|
|
RSV: |
5’cap- UTR—tRNA-trp--gag—pol—Env—src—UTR—3’ (A) n [>9,3kb]. |
|
ALV: |
5’cap--UTR—tRNA-pro--gag---pol---Env---UTR3’ (A) n(7.3kb) |
|
MuMLV: |
5’cap--UTR---tRNA-pro--gag---pol---Env---UTR—3’ (A) n [8kb] |
|
MMTV: |
5’ Cap-UTR---tRNA-pro gag----pol—Env-X—UTR3’ (A) n(8.6kb) |
|
MPMV: |
5’ cap-UTR—tRNA-pro--gag---pol—Env—UTR3’ (A) n . |
|
HTLV |
5’cap--UTR---tRNA-pro--gag---prot-pol-tax-rex-env--UTR (A) n [9.2kb] |
|
HSRV: |
5’ cap-UTR---tRNA-? ------Gag—pro-env-x-y-z-UTR3’ (A) n. |
|
HIV1: |
5’cap--UTR—tRNA-lys---gag—pol-vif-vpu-vpr-tat-Rev-nef -3’UTR (A) n [9.749kb] |
|
HIV2: |
5’cap--UTR—tRNA-lys---gag—pol-vif-vpx-vpr-tat-Rev-nef -3’UTR (A) n [9.749kb],Note in HIV2 VPU is absent, in its place VPX is present |
Size (approximate) of U3, R and U5 segments of some retroviruses:
|
Retroviruses |
Size of RNA |
U3 size |
R size |
U5 size |
|
RSV |
9300 |
230 |
20 |
80 |
|
ALV |
7300 |
230 |
20 |
80 |
|
MLV |
8300 |
450 |
70 |
80 |
|
HIV-1 |
9200=/- |
450 |
100 |
80 |
|
m-PMV |
7900 |
220 |
30 |
100 |
|
MMTV |
8600 |
1200 |
- |
- |
|
HTLV-1 |
8500 |
350 |
230 |
220 |
|
WDSV |
12300 |
440 |
80 |
70 |
|
HFV |
11200 |
910 |
190 |
160 |
|
Fly |
7200 |
190 |
50 |
240 |
|
|
|
|
|
|
· Gag and Pol together produce about 7 proteins such as matrix protein, inner and outer capsid proteins, nucleo capsid proteins, protease, Integrase and reverse transcriptase.
· Alternate splicing, read through and protease digestion at specific sites and at specific time achieve synthesis of all these proteins.
· Alternative splicing of the env gene transcript produces envelope proteins.
· This is a fine example of how a single reading frame of m viral RNA can produce 8 different proteins of different characters and functions; an incredible achievement.
· Though, all retroviral genomes code for the above mentioned gene products, some viruses have additional genes for viral specific functions.
· In the case of HTLV virus, it has tax and rex as extra genes at the end of the genome but overlaps the 3’ end of the Env gene. They are transcriptional activators, similar to adenoviral E1A proteins.
· In the case of HIV, several extra genes are found (they are incorporated during evolution or developed by alternative splicing to generate different proteins; vif-vpu/vpx-vpr-tat-Rev-nef segments. All of them are located after pol gene, some times overlapping pol and Env or overlapping Env and LTR regions at the end.
A List of Extra Genes Found in HIV Genomes:
|
Genes |
Probable functions |
|
Vif |
23kd, Promotes viral infectivity |
|
Vpr |
Weak transcriptional activator. |
|
Vpu |
16kd; Required for virion budding, found only in HIV-I, responsible for CD4 destruction; absent in HIV-II. |
|
Vpx |
Found in HIV-II and absent in HIV-I,
|
|
Rev |
16kd; Regulator of Virion protein expression: structural gene expression, especially it performs the transport of the unspliced or single spliced mRNAs from the nucleus into cytoplasm. The protein binds to rev response element (RRE) found in the transcripts, and facilitates transport.
|
|
Nef |
27kd; Negative regulatory Factor: Uncertain of its function, but perhaps required to maintain high load of SIVs may lead to down regulation of C4s in infected cells, decreased HIV production |
|
Tat |
72-102 aa; Trans Activator of Transcription: Activates the expression of viral genes, because of T-cell activation, which enhances the production of more tat. It is a transcriptional activator; it is also essential for replication.
|
|
RRE |
Rev response element to which REV protein binds during whole length viral mRNA is transported. |
Note: Rev Response Element called RRE is found in the region at the second exon of Rev gene of the HIV RNA.
· Important difference between HIV-I and HIV-II is the presence of Vpu in HIV-I and absence of Vpx.
· In HIV-II Vpx is present and vpu is absent. What is the role of Vpx? It is not known (I don’t know).
· HIV-II in comparison to HIV-I spreads with less efficiently, and takes longer time to manifest disease symptoms.
· Between the RNA of HIV-II and HIV-I, the homology is 42%, and between HIV-II and SIV the homology is 75%.
· ELISA tests developed for HIV-I also detect 80 % of HIV-II.
HIV GENOME
<------Gag---------><--------Pol------------><------Env----------->
MA CA NC PRO PRO.RT.RNase.IN GP120 .GP41
5’UTRIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII UTR 3’
5’G------I---------------------------------------------I---I---(A)--n3’
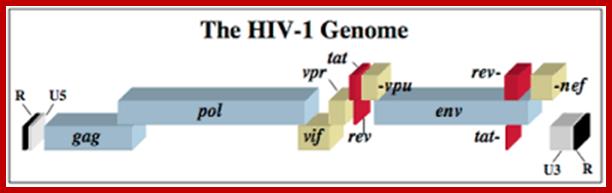
https://microbewiki.kenyon.edu/
HIV coded proteins:
|
Name of the protein |
Protein subunits |
Mol.wt (KD) |
Gene |
Function |
Matrix proteins |
MA |
P17, p18 |
Gag |
Found at inner surface of the envelope, p17 has nuclear localization signal |
|
Capsid protein |
CA |
P24, /p25 |
Gag |
6 outer, 24/25 inner, abundant, covers inner core |
|
Nucleo capsid |
NC |
p9 |
P9-Gag, 96-7-gag-pol |
Bound to RNA |
|
Protease |
PR |
P 10 |
Gag-pol |
Essential for cleaving gag proteins |
|
Reverse transcriptase |
RT |
P51-66 |
RT Pol |
RNA dependent DNA polymerase (N-terminal of RT) |
|
Integrase |
IN |
P 32 |
C-terminal of RT |
Responsible for integrating ds cDNA into host chromosome |
|
Rnase-H |
RH |
P15 |
Middle of RT |
Nicks RNA of RNA-DNA hybrid |
|
Surface glycoproteins |
SG |
120 |
Env |
Sticking on envelope, linked to transmembrane protein |
|
Transmembrane protein |
TM |
P41 |
ENV |
Holds SG by S-S bonds |
|
Extra |
Vif |
|
Vif (Pol-vif-vpr) |
|
|
Extra |
Vpr or Vpx |
|
Vpr (vif-vpr-tat) |
|
|
Extra |
Tat |
|
Tat (vpr-tat-env) |
|
|
Extra |
Rev |
|
Rev (rev-tat-env) |
|
|
Extra |
Vpu |
|
Vpu-env |
|
|
Extra |
Nef |
|
Nef-last of env |
|
|
Human Leukocyte Antigens |
HLA-1 &2 |
Host gene coded |
Host proteins |
|
|
Major Histo Complexity proteins |
MHC-1 & 2 |
Host gene |
|
|
|
Central core contains: |
Two (+) RNAs base paired at R region, tRNA-lys integrase, |
|
|
|
Infection:
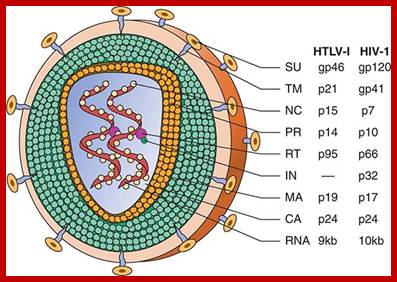
Schematic structure of human retroviruses. The surface glycoprotein (SU) is responsible for binding to receptors of host cells. The transmembrane protein (TM) anchors SU to the virus. NC is a nucleic acid–binding protein found in association with the viral RNA. A protease (PR) cleaves the polyproteins encoded by the gag, pol, and env genes into their functional components. RT is reverse transcriptase, and IN is an integrase present in some retroviruses (e.g., HIV-1) that facilitates insertion of the provirus into the host genome. The matrix protein (MA) is a Gag protein closely associated with the lipid of the envelope. The capsid protein (CA) forms the major internal structure of the virus, the core shell. http://clinicalgate.com/
Infection depends on the cell type and the kind of receptors it has. Based on this relation three modes have been noted.
Ectotropic: Say a particular virus infects only species specific animal cell ex. HIV infects only Human cells and not any other cell type.
Xenotropic: Infect only non-human cells.
Amphitropic: Infect both human and non-human cells.
Entry of the virus is mediated by viral and host cell protein-protein interaction, this leads to the entry of the viral particle, de-coating and release of naked viral particle, which undergoes partial uncoating to release active core particle. Entire replication process takes place in cytoplasm and with in the active viral core.
Harrison Principle of Internal Medicine; http://clinicalgate.com/

Organization of the genome of the HIV provirus together with a summary description of its 9 genes encoding 15 proteins. (Adapted from WC Greene, BM Peterlin: Nat Med 8:673, 2002.); http://clinicalgate.com/
http://summer2012microbiologyuncg.blogspot.in/
<--t5’
5’cap------I------I--I--------//----------I-------I-------IAAAAA (n) 3’ <---I-------I--It5’
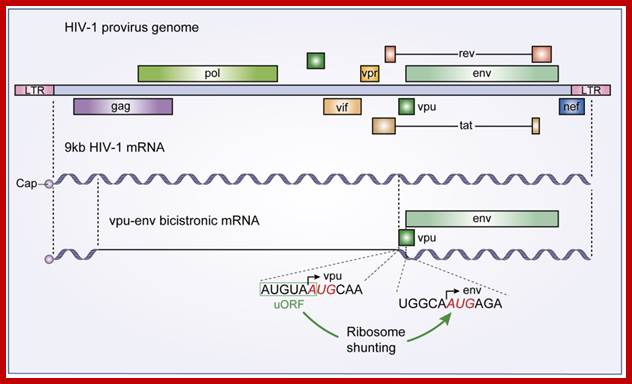
The HIV-1 vpu-env bicistronic mRNA: A schematic diagram of the HIV-1 provirus genome is presented along with its primary 9 kb mRNA transcript. The vpu-env bicistronic mRNA is formed after splicing of the 9 kb mRNA. Start codon sequences of both vpu and env ORFs are presented and the upstream ORF that stimulates Env translation is boxed in green.; Santiago Guerrero et al; http://www.mdpi.com/
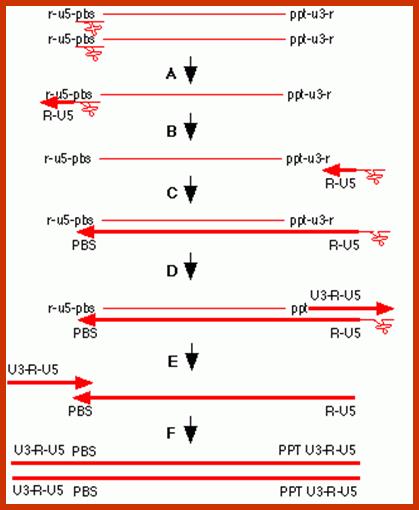
The line drawing shows stepwise events leading to the production ds c-DNA, which is actually little longer than the original length of the genomic RNA.
The genomic RNA contains one tRNA-lys at primer binding site of 18 ntds long. Reverse transcriptase, which is a RNA dependent DNA polymerase, is found bound to each of the RNA genomes, initiates replication in an unique way and performs the replication in more than one unique ways; these are excellent systems.
Reverse Transcriptases:
- Reverse transcriptases are unique enzymes because they use RNA as the template and produce ds DNA that is little larger than the size of the RNA.
- It is, trivially called reverse transcriptase; it is actually RNA dependent DNA polymerase.
- Reverse transcriptase is coded for by the viral genomes.
- Even bacterial and higher organisms do produce their own reverse transcriptases. Ex. Telomerase is a reverse transcriptase, required for the replication of chromosomal DNA ends.
- Occasionally higher systems produce their own Reverse transcriptases and use them, where few cellular RNA transcripts are made into cDNA, which are then incorporated into genomic DNA Ex. Some Introns.
- In bacterial systems and higher systems, retrospons produce reverse transcriptase for copying the transcript and for integration of the same into the genome.
- Copia of Drosophila, IAP of rodents, Hapdna viruses, all produces reverse transcriptase.
- Quite number of processed genes in eukaryotes is due to the reverse transcription of the mRNAs.
Maloney Murine Leukemia Viral (MuMoLV) RT:
- The MuMoLV RT enzyme is 80kDa and it is a monomer.
- More than half of the polypeptide chain from the N-terminal end contains polymerase activity. The C-terminal region has Rnase-H activity; no apparent 3’ >5’ exonuclease activity has been detected.
Avian Myeloblastasis Viral (AMV) RT:
- The functional enzyme is a dimer. The 95kDa protein is called b subunit. Subunits soon become homodimers.
- The 95 KD proteins, at N-terminal part (a small segment), have endonuclease activity.
- At the c- terminal end, it has RNase-H activity.
- The rest of the region contains polymerase activity.
- One of the two b units undergoes proteolytic cleavage at the N-terminal end and produces two fragments.
- One of them, the 63kd is called a subunit and it has Rnase-H activity. The other small N- terminal fragment is called B-subunit; it has endonuclease property, and it may act as integrase also.
Human Immunodeficiency Viral RT:
- The enzyme is a dimeric protein and each subunit is 63 KD called b subunit.
- The beta subunit has N- terminal polymerase function and c- terminal Rnase-H activity.
- However one the 66 KD proteins undergo protease digestion, which removes N- terminal region to generate a 51 KD, fragment called a subunit and it has just Rnase-H function.
<---------------------------66 kd ------------->
NH3^+I=====================I==========ICoo^-
Polymerase RNase-H
· All Reverse transcriptases basically fold conformationally like DNA pol-I, with a folded palm like structure. Like the palm, fingers and the thumb, the protein folds into polymerase. The palm like region has sites for template binding and an active site for extension of the primer thus shows 5’—>3’ polymerase activity.
· The connection between the palm and the thumb, called connecter has presumably the RNase–H activity.
· Here the template first used is RNA and later it uses DNA as the next template.
· In both cases, it requires primers. It can use RNA as primers and it can also use DNA as primers.
· Though it has no apparent 3’—>5’ exonuclease activity, that means it lack proof reading function, yet mutation rate is not that high as expected of such enzymes.
· The RNase-H part of the enzyme is unique in the sense; it removes RNA only when it is hybridized to DNA. It can cut the RNA randomly and displace the RNA fragments. Under in vivo conditions it can chew up RNA either from 5’ end mostly and also from 3’ end. RNase-H means, it shows RNase activity only when it is hybridized to DNA.
- The enzyme has strong affinity to ssRNA and least affinity to dsDNA. The order of affinity is- ssRNA > ssDNA > dsDNA > Poly d (A).
- It exhibits strand displacement synthesis, uses sliding and switching mode.
- It has more insertional errors under in vitro conditions than in vivo environment.
- However the HIV enzyme, incorporation errors is one in every 2000 to 4000 ntds in vitro conditions.
- It uses RNA template very efficiently for c-DNA synthesis, it can also use ssDNA strand for ds DNA synthesis.
- It requires Zn^2+ ions for its effectiveness.
- The enzyme uses both RNA and DNA primers efficiently.
- Its processivity is weak, 150 to 200 ntds long.
- Its fidelity is questionable under in vitro condition.
- Rate of synthesis is 100ntds per second.
Recently scientists have obtained Reverse transcriptase enzymes with 5’--->3’ polymerase activity, a 3’—5’ exonuclease activity, and 5’->3’ and 3’—>5’ RNase-H activity, which provide higher processivity and higher fidelity. https://www.researchgate.net/

http://cbc.arizona.edu/
Plant Retroviruses-called SIRE: by Howard M. Laten, PhD,
Plants are no exception to harbor retroviruses like particles. Plants contain a large number of transposons and retrotransposons. The retrotransposons virtually behave like plant retroviruses. Its genome is 9.3-9.8kb. They contain all the elements that a retrovirus possesses; such as LTRs, gag, Pol and Envelop genes and their ORFs are well defined.
My, (Howard M. Laten) investigations are focused on the characterization and evolution of a unique family of plant retroelements that resemble mammalian retroviruses. Retroelements are a class of DNA and RNA genetic entities that utilize the enzyme reverse transcriptase to sponsor the replication of their small genomes.
The best known retroelements is the Human Immunodeficiency Virus (HIV), the causative agent of AIDS. Non-infectious retrotransposons constitute a second major class of retroelements and are found in virtually every eukaryote. While their genomes are similar to those of the retroviruses, they lack genes necessary for intercellular transfer. The proliferation of both classes of retroelements involves the integration of reverse transcribed DNA copies into the hosts' chromosomes. These copies become permanent residents (endogenous) and record present, past, and even ancient encounters with retroelements.
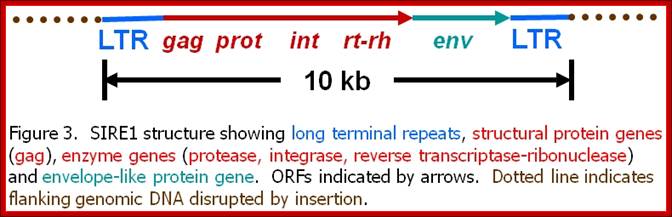
Once considered by many to be pesky genetic parasites, these endogenous elements were dubbed "selfish DNA" because they can proliferate within chromosomes. However, they may actually be key players in the modification of host genomes during critical periods of large-scale evolutionary changes.
The retroelements we discovered, SIRE-1, was first encountered in the soybean genome as a family of a thousand structurally conserved members with the standard complement of retroelement genes. But unlike other plant retroelements known at the time, SIRE-1 also codes for a protein unique to retroviruses -- an envelope-like protein. In retroviruses, the envelope protein confers infectivity. We have now confirmed the presence of SIRE-1 relatives in several other plant species, and expect it was once and may still be a highly successful retrovirus transmitted to plants by aphids or some other invertebrate vector. Our attention is focused on three primary questions: 1) Is SIRE-1 an active plant retrovirus - do host cells make SIRE-1 mRNAs, proteins, virions? 2) Can invertebrate vectors be found? And 3) Can molecular systematic approaches reveal the precise evolutionary relationships among retroviruses and retrotransposons?
Cellular Retro transposons and Retroviruses:
Cellular insertional elements are varied in their structure and composition. Many of them are processed genes (called Pseudo genes), SINES and LINES and retrotransposons. Among them Retrotransposons have features similar to Retro viral genes such as gag and pol and LT sequences are found on either side. Such elements are found both in plants and animals.
If one such element acquires an envelope segment, it can produce viral particles. Perhaps the plant retrovirus called SIRE belongs to this category.