Orthomyxo Viridae:
Influenza virus:
- Some viruses with negative sense RNAs may contain only one genome ex. VSV, Rabies, and Ebola. Some may contain two negative genomes ex. Arenaviridae.
- Orthomyxo Viridae, a group that includes Influenza viruses contains 8-10 (-) RNA genomes (multipartite genomes against bipartite and monopartite genomes).
- Like Rhabdo Viridae, Influenza viruses are enveloped viruses Covered by host cell membranes. These viruses have caused disease in epidemic proportion culminating in pandemics across the seven continents; even now it can cause such pandemics.
- In 1918 pandemic, Spanish flu H1N1 it has killed nearly 20 million people, i.e. 1% of the world’s population.
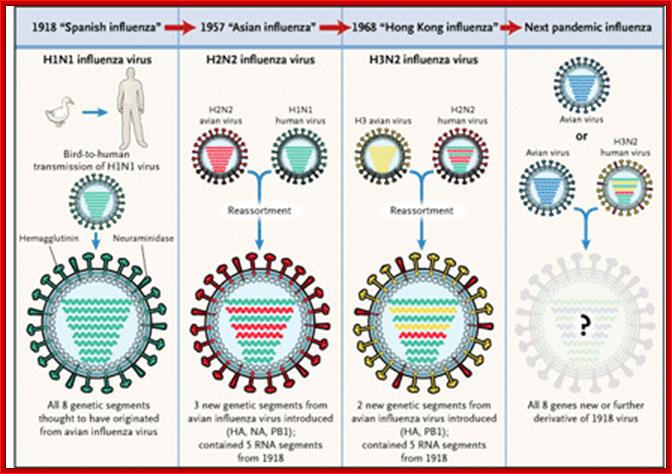
- In 1957- Asian flu H2N2; In 1968- HongKong flu-H3N2, 2007-08 H5N1 showed Pendemic threat, but controlled, Zoonotic H7N7; and endemic are H1N2 Humna flu; all belong to Influenza A; Influenza B exclusively humans and Influenza C –humans and pigs.
- India is the worst hit nation, where entire population of some villages has been wiped out. Only those who medicated locally on garlic, pepper and curcuma were survived, perhaps application of native-indigenous medicine has saved them. The value of such simple medication is still to be validated or the practice is vindicated by practioners of modern medicine. The University of Taxas has found a compound called Curcumin as one of the ingredients in Curcuma longa and found it has some important curative properties. Still in India, whole population use some of these plant products and herbs as curatives for a variety of common diseases; the scientists are yet take cognizance of it and analyze the components and find what compound has the curative properties. The components of garlic and pepper have been identified and their effects are being studied. Garlic contains, among others Allicin (allyl thiosulfates); pepper contains Piperines or piperidines.
- Asian flu in 1957 did not cause much damage. The 1968 epidemic, in spite of efforts to control, killed not less than 700,000 people all over the world.

http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/I/Influenza.html; http://www.iaff.org /http://www.iaff.org/
http://www.brandeis.edu/
Characteristic of flu: Fever (not everyone with flu will have a fever); Feeling feverish or having chills, Cough, sore throat and runny or stuffy nose, Muscle or body aches, Headaches, Fatigue and Vomiting or diarrhea
Epidemiology: http://www.ufrgs.br/
- The most feared out break in 1997-98, just killed only 3 (recorded), because of war footing preventive steps taken by the worlds’ health organization (WHO). The recent (2005-2006) birds’ flu has been reoccurring in various parts of the world and found to be deadly. Only the time will tell when the flu genome mutates and produce deadly strain that can infect humans and cause a pandemic.
- In all the three past episodic events, the origin of the viruses has been traced to china and chine’s pigs and poultry. In 1997 millions of chicks have been sacrificed to save the world, similar to killing of cattles and sheep in European countries, because of foot and mouth disease and Bovine Spongiform Encephalitis (BSE).
- Influenza strains have been grouped into A, B, C and d. It has been named as A-PR8 (A/PR8/34), A-HK (A/hongkong/8/68); A-H5N1 is the most recent one. The reoccurrence of this disease is due to mutation in viral genome, or may be due to recombination among genomes of different strains in a mixed infection or may be due to mixed or mixed segregation of the genomes again in the mixed infection.
- Birds are considered to be the main reservoirs and the source variation of these viruses. Influenza viral epidemic appears once in decade or two; the newer strains are more powerful and more devastating than the earlier once. It is for this reason people are scared; perhaps that is one of the ways the nature controls worlds population.
- Unbelievingly, the term Influenza has its origin from a civilized country, Italy. They, in those days 1918 or so, people used assign a specific god for one specific event or an event of catastrophes. The god-fearing population, when the disease hit their country, opined that the disease is due the “influence” of a god (which God!), so the term “Influenza”, a “Pandemic God” indeed! Influenza normally epidemic in some areas and it can be pandemic –global. In 20th century new strains of flu are recognized as Spanish flu 1918-1919, Asian Flu 1957-58 and Hong knog flu 1968-1969
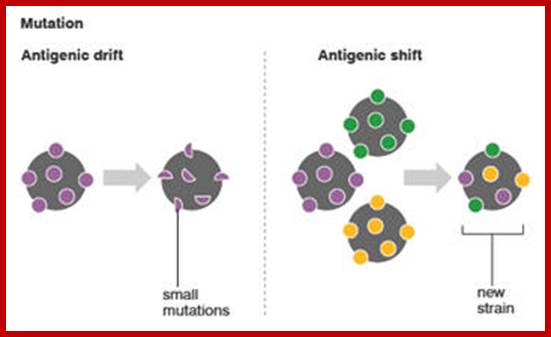
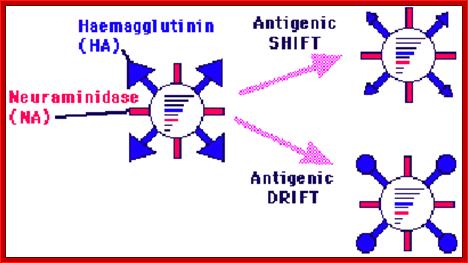
The influenza A virus can mutate in two different ways; antigenic drift, in which existing antigens are subtly altered, and antigenic shift, in which two or more strains combine. Antigenic drift causes slight flu mutations year on year, from which humans have partial, but not complete, immunity. By contrast, the new strain of H1N1 appears to have originated via antigenic shift in Mexican pigs.
Source: BBC News. “US reports first swine flu-death.”
http://news.bbc.co.uk/2/hi/health/8021958.stm
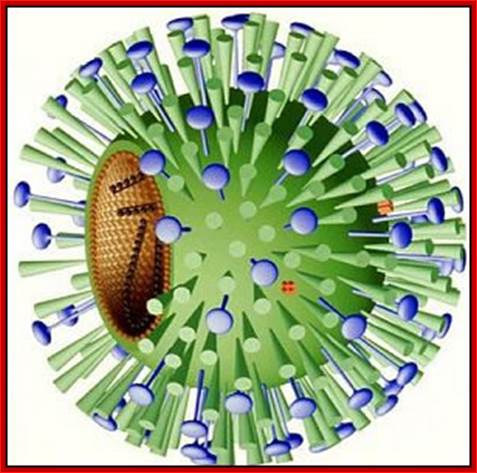
Virus with glycoproteins; http://www.departments.bucknell.edu/
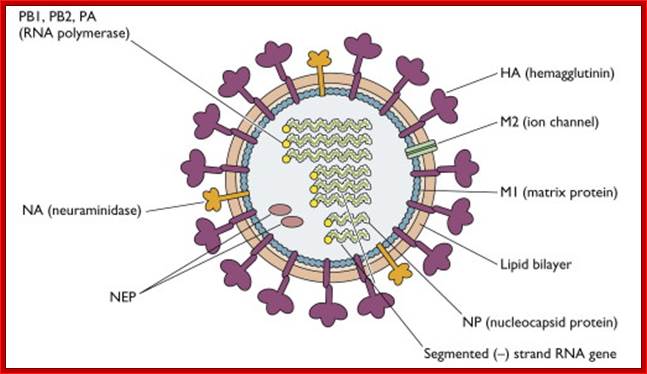
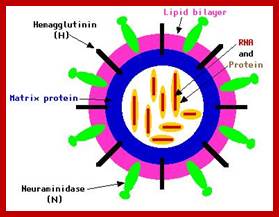
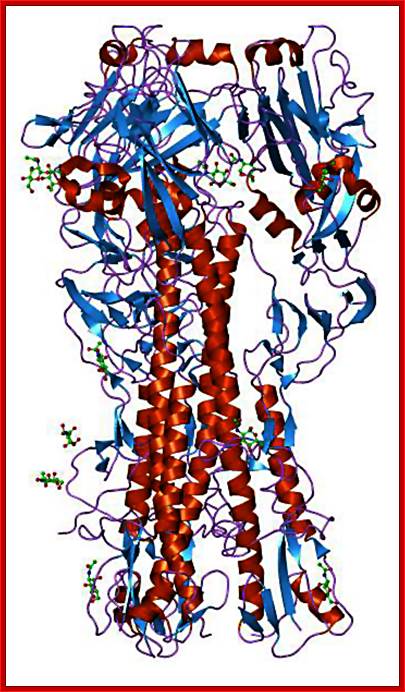
The influenza virion (as the infectious particle) is roughly spherical. It is an enveloped virus – that is, the outer layer is a lipid membrane which is taken from the host cell in which the virus multiplies. Inserted into the lipid membrane are ‘spikes’, which are proteins – actually glycoproteins, because they consist of protein linked to sugars – known as HA (hemagglutinin) and NA (neuraminidase).http://www.virology.ws/
Morphology:
- Structurally influenza viruses exhibit polymorphism, from spherical to ovoid or twisted shapes.
- All these viruses have enveloped membranes studded with two types of proteins, one a glycoprotein consisting of Haem-agglutinins (HA). This protein causes agglutinisation of human RBCs. The HA targets cell receptors, which are Glycophorin- Each virion contains at least 100 such glycoproteins.
Another membrane protein found is Neuraminidase (NA). This hydrolyses the link between terminal Sialic acid groups from Galactose or Galactosamine residues. Breakdown of this link, at the time of infection, facilitates transport of viruses into cells through mucins. About ~100 copies of this protein per virus are present on the viral membranes.
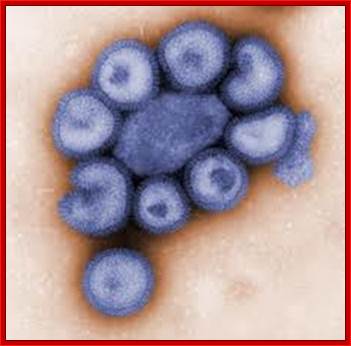
This colorized negative-stained transmission electron micrograph (TEM) depicts the ultrastructural details of a number of influenza virus particles, or “virions." Image: CDC/ Dr. F. A. Murphy; Read more at : http://phys.org/news176055264.html#jCpViral shape; http://phys.org/

Swine flu: http://www.mcb.uct.ac.za/
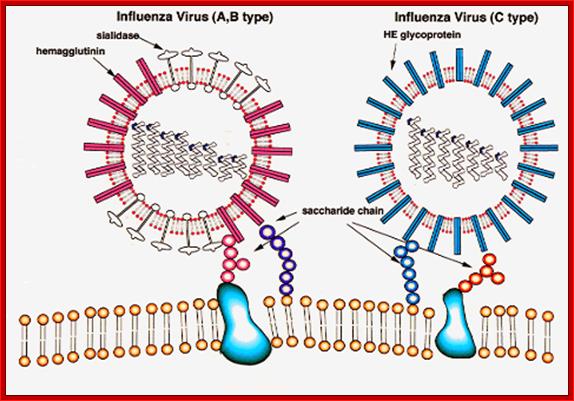
Viral glycoproteins on the surface interact with host cell receptors and bind; http://employees.csbsju.edu/
A surface protein on the surface of the viral particle-
Hemagglutinin; http://blogs.unimelb.edu.au/
Neuraminidase; http://home.cc.umanitoba.ca/
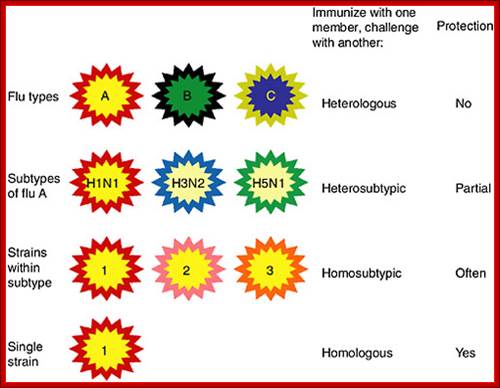
Categories of influenza viruses and immunity they induce. Colors indicate similarity. For example, influenza virus types A, B and C differ for both internal proteins and the HA and NA external glycoproteins. Within the influenza A type, subtypes have major differences in HA and NA but only subtle differences in internal proteins. Within an influenza A subtype, for example, H1N1, the HA and NA differ in more subtle ways shown by the more similar colors. Note that the term ‘heterologous’ immunity is also used to refer to immunity induced by one virus and reactive with an unrelated virus (Selin et al., 1994) but the term will not be used that way in this review.Types ABC and D and subtypes: http://www.cdc.gov/https://www.nap.edu
https://www.cs.mcgill.ca/
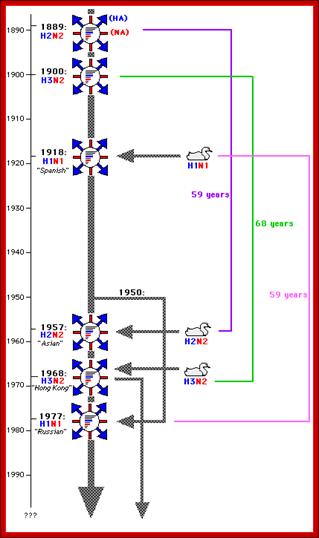
Certain modification of Hemagglutinin has caused disease in pandemic proportion during different time periods in different countries; most of them originated from China. http://www.lehigh.edu/bioS 353.html
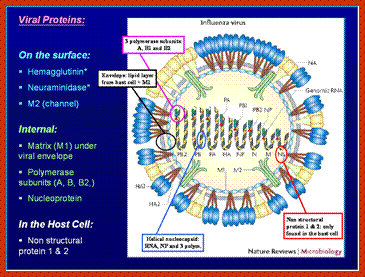
- On the inner surface of the enveloped membrane, a matrix layer of 6 nm thick is found, consisting of the most abundant proteins called capsid or matrix proteins.
- Inside this thick protein enclosure, eight RNA genomic strands of different lengths, are found, where each of them are helically coiled and coated with nucleo capsid proteins (NPs).
- In influenza strain-C contains seven genomic RNA segments (?). The viral core also contains another set of proteins PA1, PB1, PB2, and NS1, NS2 and M2. They are associated with each of the nucleoprotein threads.
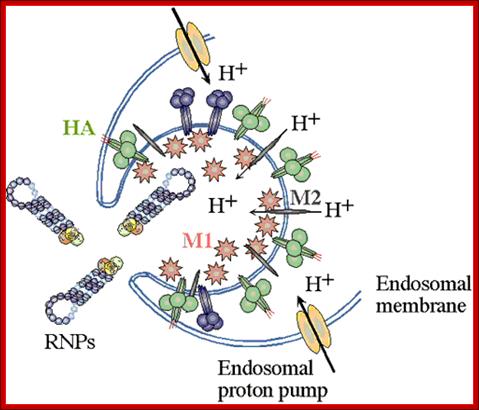
Enveloped viruses gain entry to cells by use of specific viral proteins that have membrane fusion - inducing properties. The first of these viruses for which the entry process was figured out in detail was influenza. Influenza virions (see EM pictures) bind to sialic acid (also known as N-acetyl neuraminic acid) on the cell surface and are endocytosed. The endocytic vesicles become acidified (pH drops from 7 to around 5), and at this pH, the hemaglutinin (HA) trimers in the influenza envelope undergo a structural transition. At some stage the HA monomer is cut into two polypeptides, HA1 and HA2. When the pH drops to 5, the amino terminal end of HA2 flips "upward" from an internal position to become exposed to the aqueous environment. This end of HA2 (called the "fusion peptide") is highly hydrophobic, and it interacts with the vesicle membrane and causes the viral envelope and the vesicle membrane to fuse. (See figure.) This fusion event dumps the viral core into the cell's cytoplasm. (See a figure by Paul Digard, Department of Pathology, University of Cambridge.) ; Flu viral endocytosis and endosomal membranes; http://www.lehigh.edu/
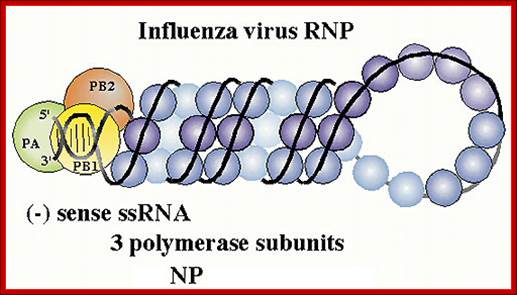
Viral RNPs: http://www.microbiologybytes.com/
Infection and replication:

Host cell invasion and replication; https://www.cs.mcgill.ca
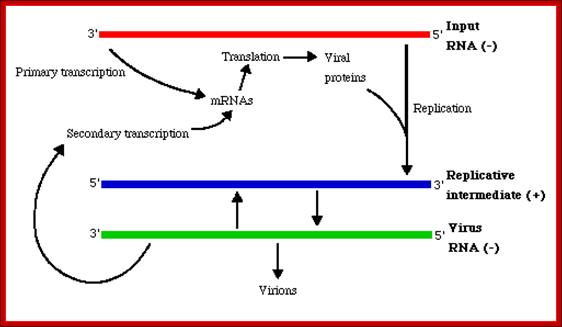
(-) genetic RNA replication; http://www.slangenforum.com/;http://www.slangenforum.com/
Infection and multiplication:
Influenza viruses bind through hemagglutinin onto sialic acid sugars on the surfaces of epithelial cells; typically in the nose, throat and lungs of mammals and intestines of birds (Stage 1 in infection figure). The cell imports the virus by endocytosis. In the acidic endosome, part of the hemagglutinin protein fuses the viral envelope with the vacuole's membrane, releasing the viral RNA (vRNA) molecules, accessory proteins and RNA-dependent RNA polymerase into the cytoplasm (Stage 2). These proteins and vRNA form a complex that is transported into the cell nucleus, where the RNA-dependent RNA polymerase begins transcribing complementary positive-sense vRNA (Steps 3a and b). The vRNA is either exported into the cytoplasm and translated (step 4), or remains in the nucleus. Newly-synthesised viral proteins are either secreted through the Golgi apparatus onto the cell surface (in the case of neuraminidase and hemagglutinin, step 5b) or transported back into the nucleus to bind vRNA and form new viral genome particles (step 5a). Other viral proteins have multiple actions in the host cell, including degrading cellular mRNA and using the released nucleotides for vRNA synthesis and also inhibiting translation of host-cell mRNAs.
Negative-sense vRNAs that form the genomes of future viruses, RNA-dependent RNA polymerase, and other viral proteins are assembled into a virion. Hemagglutinin and neuraminidase molecules cluster into a bulge in the cell membrane. The vRNA and viral core proteins leave the nucleus and enter this membrane protrusion. The mature virus buds off from the cell in a sphere of host phospholipid membrane, acquiring hemagglutinin and neuraminidase with this membrane coat is an important feature of the emerging virus. As before, the viruses adhere to the cell through hemagglutinin; the mature viruses detach once their neuraminidase has cleaved sialic acid residues from the host cell. After the release of new influenza viruses, the host cell dies.
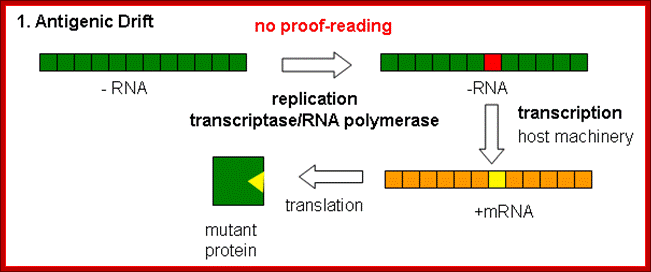
Because of the absence of RNA proofreading enzymes, the RNA-dependent RNA polymerase makes a single nucleotide insertion error roughly every 10 thousand nucleotides, which is the approximate length of the influenza vRNA. Hence, nearly every newly-produced influenza virus is a mutant—antigenic drift. The separation of the genome into eight separate segments of vRNA allows mixing or reassortment of vRNAs if more than one viral line has infected a single cell. The resulting rapid change in viral genetics produces antigenic shifts and allows the virus to infect new host species and quickly overcome protective immunity. This is important in the emergence of pandemics. https://www.cs.mcgill.ca
Dr. Margaret Hunt; http://www.thailabonline.com/

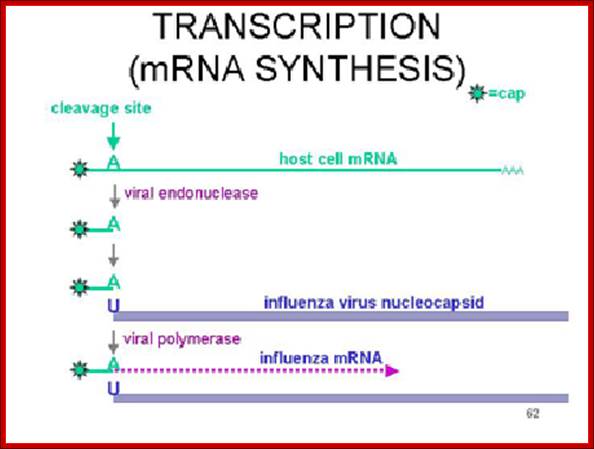
The enzyme that reproduces influenza RNA is known as an RNA-dependent RNA polymerase. This enzyme, which consists of the viral proteins PA, PB1, and PB2, is present in every virus particle. The influenza viral RNA polymerase is a primer-dependent enzyme. The enzyme cannot copy the (-) strand RNA template without a small piece of RNA that aligns on the template RNA and provides a starting point for RNA synthesis. The primers for influenza viral mRNA synthesis are produced from the cell’s own collection of mRNA molecules. The influenza viral RNA polymerase actually cleaves cell mRNAs near their 5′-ends, generating the primers it requires for RNA synthesis. This process has been called cap snatching, because the primers are made from the 5′-ends of cell mRNAs, which have an unusual chemical structure called a ‘cap’ (box labeled ‘c’ in illustration). Cap-snatching is also a feature of mRNA synthesis of other viruses. http://www.virology.ws/
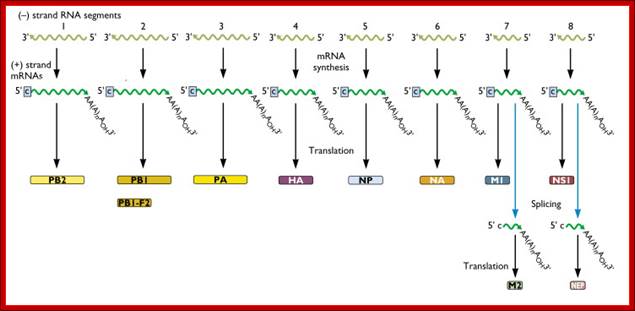
Within the influenza A virion are eight segments of viral RNA. These molecules carry the all the information needed to make new influenza virus particles. These eight RNAs are shown schematically as olive green lines at the top of the illustration. RNAs are chains of four different nucleotides, A, C, G, U. In the case of influenza virus, the eight RNAs are a total of about 14,000 nucleotides in length. The nucleotides make up the genetic code – it is read by the cell’s translation machinery in groups of three, with each triplet specifying an amino acid. Vincent Racaniell; http://www.virology.ws/

Influenza Virus Life Cycle: Adam; http://nursingcrib.com/
Infection:
Entry of viruses is through cell receptor and viral HA binding. Hydrolysis of the link between the Sialic acid and Glycans, leads to internalization of viral nucleo capsid leaving the membrane. The viral membrane and its proteins merge with the host cell membrane.
· They are then transported to the nucleus, via transport proteins. In the nucleus, perhaps the nucleo-protienaceous threads unfold.
· Viral Proteins such as PB1, PB2, PA, NS1 and NS2, which are bound to genomic RNAs, play an important role in replication of the genome.
· To begin with, PB1 and PB2 complex bind to the cap region of host’s mRNA (newly made ones), and cut 12 –13 ntds down stream from the cap either at A or at G, because G or A can pair with U. Then the Cap-leader sequence is transposed and laid on the 3’ end of the genomic RNA in such a way, G or A can be paired with 3’U of (-) genomic RNAs. The G or A provides priming 3’OH group.
· Proteins PB1, PB2 and PA in association with nonstructural proteins such as NS1 and NS2 act as Transcriptase complex which extends the primer into a complementary copy of (-) RNA into (+) RNA. Immediately, these transcripts are added with poly- (A) tail at the 3’ end by host enzymes, how? The answer is not clear. Even the process of poly-Adenylation cannot be explained, for the 3’ end of the (+) RNA does not show any polyadenylation signal sequences. It is expected that the host enzymes bring about poly-(A) addition.
· Synthesis of (+) RNAs can considered as transcription (?) for it produces translatable RNAs.
A List of Influenza Genomic RNAs, Theirs Features:
|
Genomic-RNAs |
Length in bases |
Name of the RNA |
Function
|
|
1 |
2341 |
PB2 |
Produces-Transcriptase, cap binding |
|
2 |
2341 |
PB1 |
Transcriptase, elongation |
3 |
2233 |
PA |
Transcriptase! |
4 |
1778 |
HA |
Hem-agglutinin, trimer, 135^Ao |
5 |
1565 |
NP |
Nucleoprotein, |
6 |
1413 |
NA |
Neuraminidase, tetramer, 60A^o |
7.a |
1027 |
M1 |
Matrix protein, major viral proteins |
7.b |
“ |
M2 |
Ion channel, integral membrane protein |
8.a
8.b |
890
“ |
NS1NS2 |
Nonstructural, nuclearNuclear and cytoplasmic |
- Translation of each of the transcripts leads to the production of respective proteins. Transcripts of genomic segment 7 and 8 undergo alternate splicing, thus each of them produce two different proteins.
- As the proteins are synthesized on ER surface, some of them are transported and processed in Golgi apparatus and the same proteins are used as viral envelope membrane proteins; even matrix proteins also go through the same process.
- When viral proteins are produced to certain concentrations, the same protein complexes might be used to cut and prime to produce full length (-) RNAs.
- All the species of RNAs, eight in number now associate with their respective nucleo capsid proteins and each of the eight RNAs are assorted as one group, with out making mistake, and finally the viral particles are budded off with host membrane as the envelope containing the viral coded proteins. How the genomic RNAs are assorted and bundled into a set and budded off; this is yet to be elucidated. There should be some packaging signal sequences to perform this grouping the individual RNAs in to a perfect group.
- Based on antigenic characters, influenza viruses are grouped into A, B.C. and delta types; each of them have their own subtypes.
- The group A strains have caused pandemics. It is known that migratory birds are carrier of different strains across the world. Escape from the immunity defense against the viruses is due to changes in HA and NA amino acid sequences by what is termed as antigenic shift or antigenic drift.

http://www.microbiologybytes.com/virology/303
http://www.microbiologybytes.com/
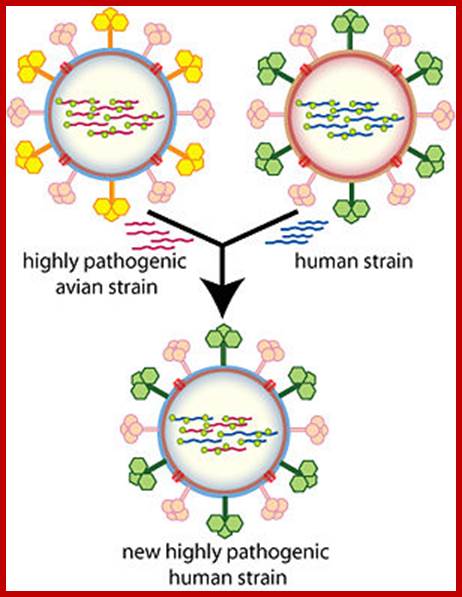
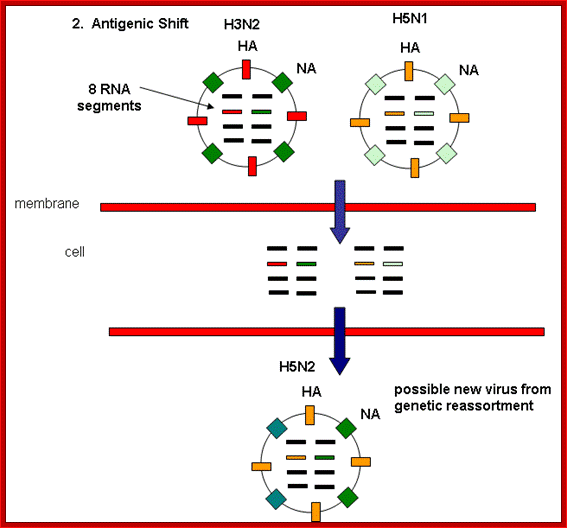
Antigenic shift: It means that the gene coding for HA is totally replaced by another gene from another source i.e. replacement of HA of one species with another species from other source. This can easily make human species not to recognize the earlier Flu infections. By the time human immune system responds and raise and mount its immune attack; the damage to persons is considerable. This is possible by mixed viral infection.
http://employees.csbsju.edu/
Antigenic drift: It means where the antigen gene has undergone point mutation. In a course of time it generates a different amino acid sequence in such a way, human immune system fails to recognize and attack.
Evidences suggest that before 1918, the major strain of influenza was H3N8. This was replaced in the 1918 pandemic when an H1N1 virus arose. In the next pandemic (Asian,1957) three genes from avian influenza (PB1, HA, and NA) replaced their counterparts in the H1N1 strain, presumably through re-assortment as shown above, to give the pandemic strain H2N2. Again through a similar process, the PB1 and HA genes from the prevalent strain where replaced by avian genes to produce the Hong Kong H3N2 pandemic strain.
In addition, the sequence of viruses can be compared and phylogenetic tree can be created, not unlike family trees of inheritance. The 1918 virus appears most related to human and swine influenza virus, not avian, but it does clearly have avian features (such as the gene for HA described above). It may have emerged from avian reservoir before 1918, for which no specimen has yet been found to compare. That virus may have then acquired enough mutations to make it infective in mammals. There is yet another possibility: the 1918 virus came directly from a completely different avian source (somewhat like SARS which emerge from civets). http://employees.csbsju.edu/
