Picorna Virus:
Pico RNA virus,
And SARS virus,
Picorna virus:
It is one of the smallest animal viruses known. It belongs to a group of viruses containing more than 200 serotypes. It is an RNA virus.
|
Virus |
Serotypes |
Disease they cause |
|
Aphtho virus |
7 |
FMD, Foot-and-mouth disease |
|
Cardio virus |
2 |
Cardiac diseases |
|
Entero virus |
111 |
Polio |
|
Hepto virus |
2 |
Hapatitis-A |
|
Rhino virus |
105 |
Causes common cold |
|
Unassigned |
3 |
|
|
|
|
|
- Picorna viruses are very small animal virus, cause deadly diseases like polio, foot and mouth (FMD), Hepatitis-A and other diseases. The F & M DV disease has devastated European cattle and sheep population and also other country’s economy ex. South America
FMD Epidemics: dates all over the world
5.7 Japan and the Republic of Korea, 2010
- At least 1 billion doses of vaccines are required for South America only.
- In India, this disease frequently appears, and people have native medicine for it; they use Oscimum, Leucas, Garlic and Ginger juice, and administer the same to animals orally. How the plant extracts work is yet a mystery for no scientist has ever tried to look into this aspect of indigenous medicine biochemically. This is of Ayurvadic medicine; it is a common Indian medicine, used for centuries.
- Poliomyelitis is another disease, which causes life long disability. Poliovirus was first isolated in 1909 by Karl Landsteiner and Erwin Popper. In 1981, the poliovirus genome was published by two different teams of researchers— by Vincent Racaniello and David Baltimore at MIT and by Naomi Kitamura and others at the State University of New York, Stony Brook. Poliovirus is one of the well-characterized viruses, and has become a useful model system for understanding the biology of RNA viruses. This virus causes severe disability muscles and infected persons become disabled. Vaccines are available and in some countries people are using public vaccination.
- Hepatitis-A is water borne virus. In third world countries, it is as common as any other water borne diseases and people have learned live with it.
An Egyptian Stele thought to be a polio victim, 18th Dynasty (1403–1365 BC)
Morphology:
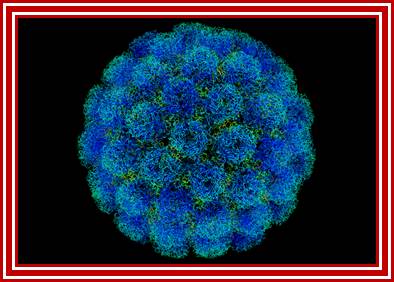
- Polio virus is about 27 to 30 nm in size, isometric (icosahedral), with 60 protomers (20 faces X 3 protomers= 60 protomers) and each protomer is made up of 4 subunits, called V1, V2, V3, and V4. The genome is of plus sense, single stranded RNA of 7433 (2500nm in length) ntds long packed into the capsid of ~30nm along with Na^+ and K ^k and polyamines.
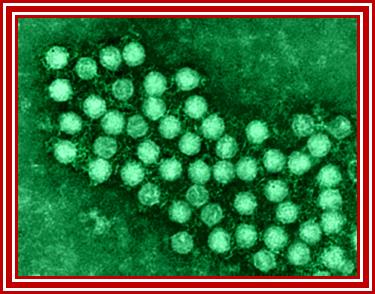
Electron microscopic pictures of Picorna viral particles
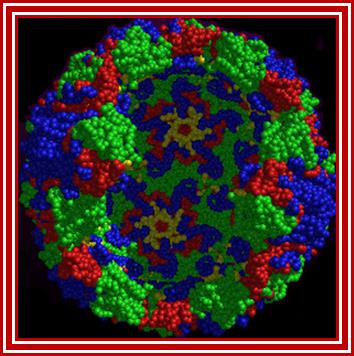
Rhino viral particle colored to show different capsid proteins; http://www.microbiologybytes.com/

FMD virus; http://www.virology.wisc.edu/

Hepatitis A virus, Dr Richard Hunt
http://www.microbiologybook.org
Polio Viruses- morphology; Synthetic vaccine could prevent future FMD disease; http://www.nature.com/
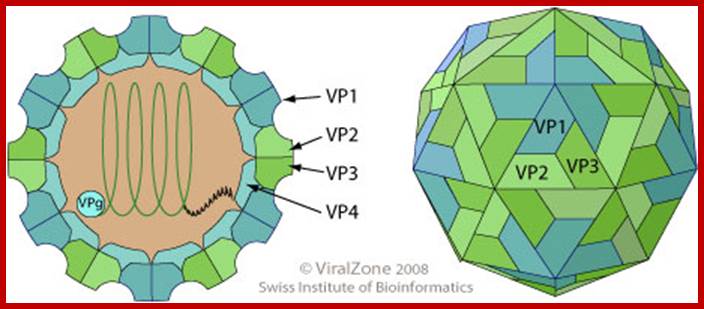
Virus non-enveloped, 30nm, Pseudo icasohedral capsid inside one finds RNA genome, the icasohedral capsid consists of 60 protomers, each consisting VP1,VP2,VP3 and VP4; The VP4 located at the inner layer of the capsid. http://viralzone.expasy.org/
Virus classification:
|
Order: |
|
|
Family: |
|
|
Genus: |
|
|
Species: |
FMD is caused by Picorna viruses, belongs to a family of viruses such as polio and cold viruses. These viruses mutate rapidly, traveling around as a quasispecies cloud. The clouds can be easily divided into seven broad groups, and within the most common serotype (O) there are 8 distinct subgroups (see the map to the right for their geographical distribution).
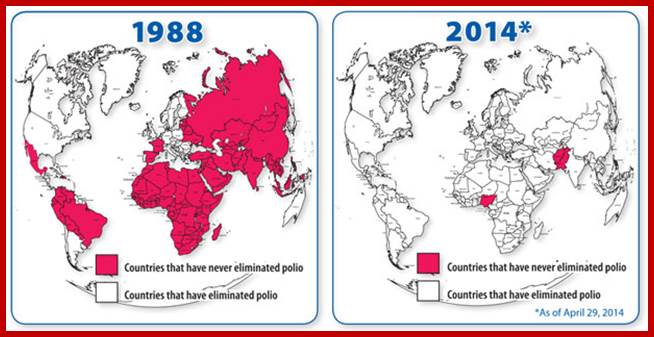
Centers for disease control and prevention; our progress against Polio; the number 35000 in 1988 is reduced to 407 in 2013; http://www.cdc.gov/
The FMD genome is 7741 (8134?) nucleotides long, and the sequence analysis has been used for epidemiology like the 7 different prototypes based on no more than 8% of that length-the VP1 gene, usually. That’s enough to track high-level changes, because of FMD’s rapid mutation rate: The FMD is a virus that affects many hooved animals; it’s not usually fatal, but causes productivity loss. FMD outbreaks are economically devastating, because aside from the productivity loss in many countries, some are free of the disease; people will refuse to take meat or other agricultural products from outbreak areas.
The 2001 outbreak in Great Britain actually came from outside the country. The 2007 outbreak, though, was clearly from a local source: They have a FMD research lab in the Institute for Animal Health (IAH) Pirbright, and Surrey. The epidemiology of that outbreak, surveyed used the genome sequencing to track and predict sites of FMD.
· The FMD viral genome consists of a single strand of positive sense RNA of approximately ~8,134 nucleotides. The RNA is initially translated as a single polypeptide which is subsequently cleaved by viral-encoded proteases to produce the structural and non-structural proteins depicted in the genome map.
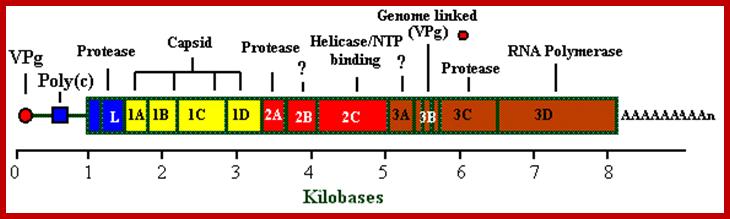
Picorna viral Genome; http://bioinformatica.uab.es/
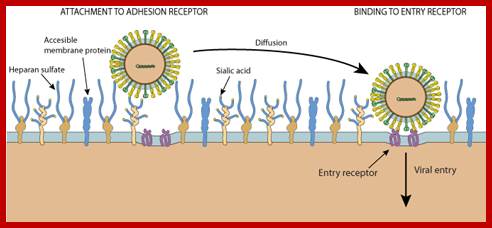
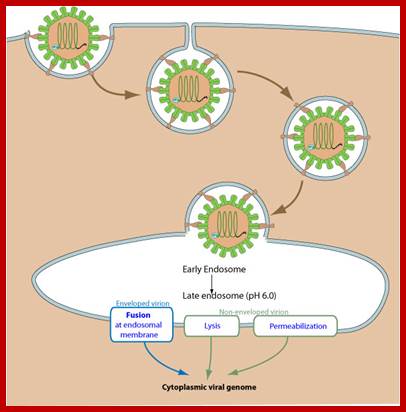
Unusual entry of virus into host cell with out the use of clathrin or caeoin; but depend upon cholesterol, DNM2/Dynamin-2, small GTPases or tyrosine kinase and possibly involve non-caveolar lipid rafts; http://viralzone.expasy.org/

Life cycle; http://www.mcb.uct.ac.za/
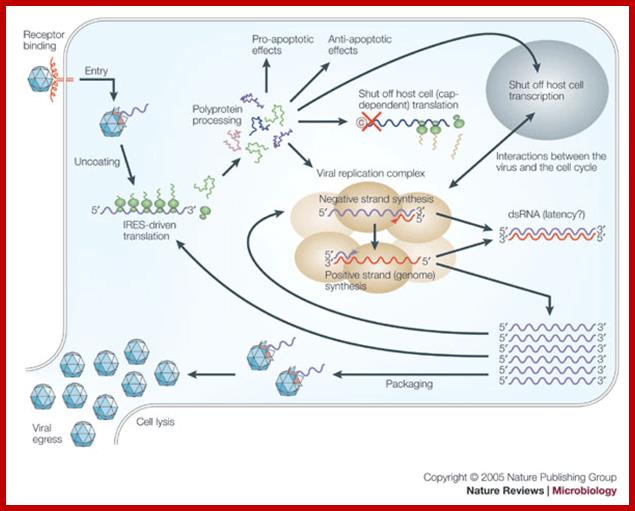
Life cycle of Picorna VirusJ. Lindsay Whitton et al; http://www.nature.com/
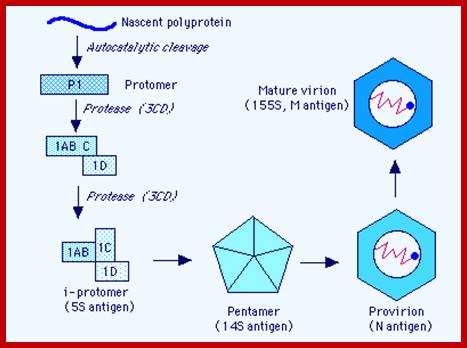
RNA is believed to be packaged into preformed capsids, although the molecular interactions between the genome & the capsid responsible for this process are not clear. Empty capsids (defective) are common in all Picornavirus infections. The capsid is assembled by cleavage of the P1 polyprotein precursor into a protomer consisting of VP0,3,1 which join together enclosing the genome:
Maturation (& infectivity) relies on an internal autocatalytic (?) cleavage of VP0 into VP2 + VP4. Release (in most cases) on the virus from the cytoplasm occurs when the cell lyses - probably a 'preprogrammed' event which occurs a set time after the cessation of 'housekeeping' macromolecular synthesis at shutoff. (Hepatitis A virus is relatively non-lytic & sets up a more persistent infection).
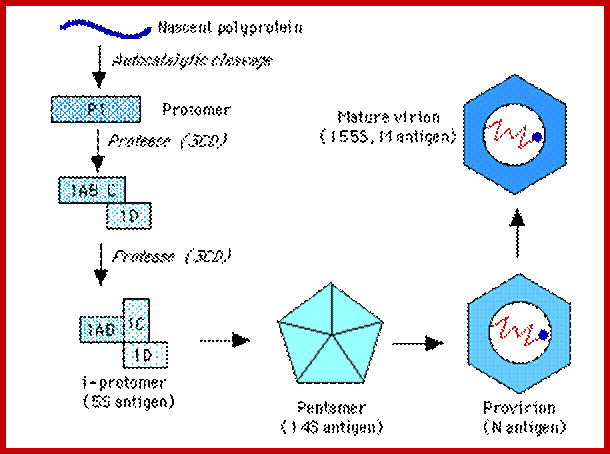
The Nascent Polypeptide chain is processed to generate individual polypeptides which have their own functions; http://www.mcb.uct.ac.za/
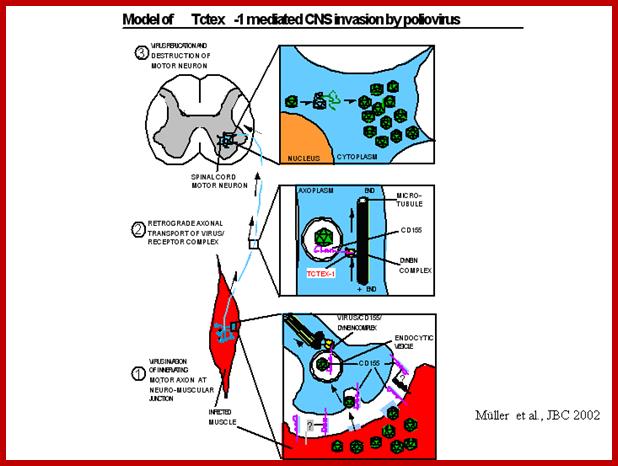
Polio Vrus-Infection:
- Infection is through lymphoid cells associated with oropharynx and gut region; cellular receptors recognize V1 coat protein and internalize viruses by endocytosis.
- Uncoating releases RNA strand into cytoplasm, where the RNA replicates and produces a large number of viral particles numbering 10^5 (?) in about 8 to 10 hrs after infection.
- The released viruses enter blood stream and finally land up in CNS, particularly motor neuronal cells in the anterior horns of spinal cord and brain stem.
- Virus multiplication ultimately leads to cell lysis, which leads to plaque formation. Inflammation is mainly due to over reaction of the immune system to virus particles. Only one percent of the infected population get paralytic stroke, but the rest of the people experience asymptomatic infections. Death may be due to respiratory failure by paralysis of inter costal muscles and Diaphragm.

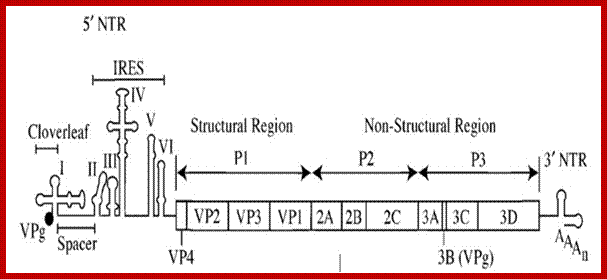
Organisation of the poliovirus genome showing 5′ and 3′ non-coding regions (green), structural proteins (red) and non-structural proteins (blue). VPg is a small virus-encoded protein covalently linked to the 5′ terminal nucleotide of the genome,PD Minor, NIBS,UK
 The polio genome is
linear 7.0 to 8.9 kb, poly adenylated positive strand; the genome consists of
single coding genome encoding polypeptide sequence. The viral genome is RNA
contains VPg with IRES at its 5’ end. It has internal ribosomal entry site; it
has a 3’ UTR important for (-) strand synthesis. The L at 5’ end has additional
N-terminal leader protein sequence. As the genome is positive strand, it allows
translation to generate polyproteins. The poly protein is produced by viral
proteases into precursors and final proteins to yield replicase, VPg and many
other proteins;
http://viralzone.expasy.org/
The polio genome is
linear 7.0 to 8.9 kb, poly adenylated positive strand; the genome consists of
single coding genome encoding polypeptide sequence. The viral genome is RNA
contains VPg with IRES at its 5’ end. It has internal ribosomal entry site; it
has a 3’ UTR important for (-) strand synthesis. The L at 5’ end has additional
N-terminal leader protein sequence. As the genome is positive strand, it allows
translation to generate polyproteins. The poly protein is produced by viral
proteases into precursors and final proteins to yield replicase, VPg and many
other proteins;
http://viralzone.expasy.org/
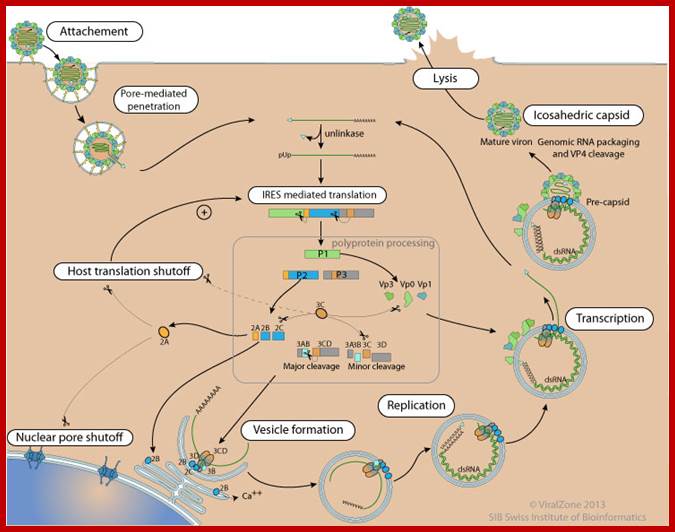
Polio Viral Repl;ication Cycle: http://viralzone.expasy.org/
Infectious Myelitis; http://quizlet.com/
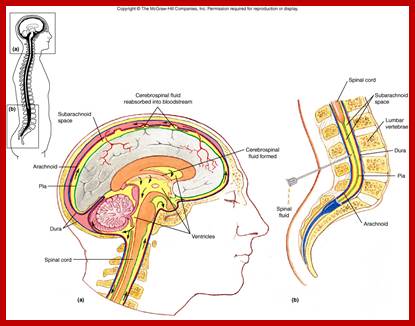
Final location i.e. CNS where polio is manifested; http://www.uark.edu/
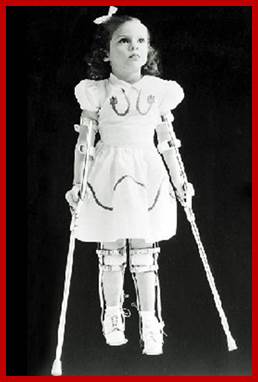
Affected Child; http://www.steadyhealth.com/
Replication:
The genome is 7741-42 ntds long, the first 740 ntds act as 5’UTR leader sequence and the last segment from 7361 to 7441nts act as 3’UTR,
- The genomic RNA shows distinct features; the 5’ end is capped by VP-G, which is a 22a.a long protein, which is covalently bound to the terminal 5’‘U’ nucleotide through Tyrosine residue, a similar feature is also found in adenovirus DNA genome, where a 50KD protein is covalently bound CMP at 5’ end of the DNA.
- The 3’ end of the RNA is made up of a poly- (A) tail, with a stem loop structure. Poly-A is a characteristic feature of eukaryotic mRNAs.
- Cellular factors like PAB-I that bind to poly-A tail prevent digestion from exonucleases, so also the 5’ end is protected by the presence of VP-G protein. The genomic RNA acts as mRNA.
- The 5’ leader sequence shows six stem loop structures.
- At the 5’ end the leader sequence has eight AUG initiator codons, but only the eighth one is used for it is in proper context, i.e. A/GCC AUG G, and it is in proper reading frame.
- Another feature that greatly helps in initiation of translation is stem loop secondary structures (at least 6 of them) at 5’ end. The first one appears like a tRNA structure. Ribosomes enter this site and initiates translation. This site is called Internal Ribosome Entry Site (IRES). The stem loop structure at 392 ntds is has *GCCAUGG* which acts as the initiator codon, for it is in proper context of ORF.
- Using IREs (Internal Ribosome Entry sites), ribosomes translate full length (+) RNA to produce a polypeptide chain, called Nascent Cleavable Viral Protein (NCVP) of 240 KD (polyprotein) consisting of 3 segments when cleaved it generates 12 segments including endoproteases, VP coat proteins, VpG the cap protein and RNA dependent RNA polymerase. .
- There are few internal sequences with in the polypeptide chain endowed with endo-protease activity. First cleavage is by protease p1A producing P1 and P2P3 segments. A further cleavage is by 3C protease. First is autocatalytic cleavage, which releases few proteases and they in turn further cleave the polypeptide chain in sequence specific manner. The sites used are gln-gly, tyr-gly, asn-ser and tyr-gln.
- One of the protease 2A produced in the cleavage process acts on eukaryotic hosts’ E IF-4G, an mRNA’s cap binding protein and digests it. This shuts off host cells’ protein synthesis.
Polio virus with its IREs. http://commons.wikimedia.org/
The long viral RNA translation is initiated using internal ribosomal entry site (IRE). The Poly-Aat 3’ end is used for producing (-) RNA placing VpG-U as primer. The +RNA is translated to generate a nascent cleavable polyprotein that undergoes a series of protease mediated and site-specific cleavage to generate different sets of capsid and Replicase proteins.
Mechanism of Replication:
- For replication, translation of genomic RNA is essential.
- Translation leads to the formation of VpG and Replicase proteins.
- The VP-g protein gets covalently linked to a Uridine nucleotide through tyrosine.
- The VP-g-U with its 3’ OH free, now acts as the primer. The primer is laid on plus RNA at 3’ end which has poly- (A) s.
- Using the primers’ 3’OH end, the RNA Replicase extends a complementary chain on genomic RNA to generate a (-) RNA strand.
5’VpG-UUAAAACAG----------//---------AAAAAAA3’ (+) RNA
3’AAUUUUGUC<----------------------------- UUUU-gPV5’ (-) 5’VpgUUAAAACAG------------------------------>AAAA3’ (+) RNA
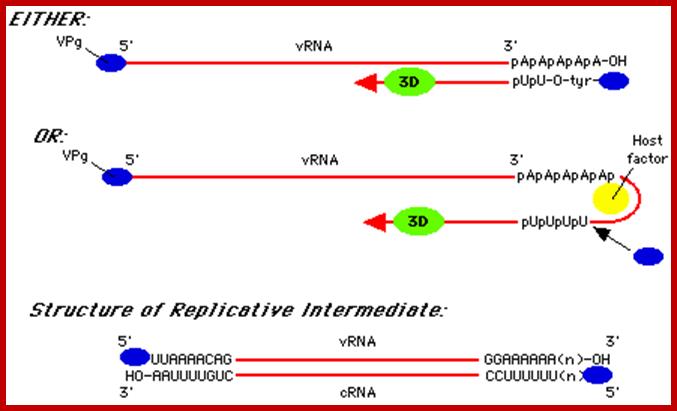
The figure shows alternative forms of replication; http://www.mcb.uct.ac.za/
- The (-) RNA strand is used for the synthesis of plus strand. VpG-U protein primer is added to the 3’ end on ‘A’s and the Replicase extends the chain to produce plus RNA.
- The plus RNA is used for producing proteins as well as replication components like Vp-G and Replicases.
- Once these components are produced, the coat protein, yet not completely digested and released into their respective components, start assembling into star-like structures made up of only auto cleaved P1 proteins. Once they form the structure, they use the 5’ end leader sequence as the viral assembly site and start assembling the viral particle, where RNA is compacted into viral particle.
- As multiplication of the viruses ensues, the cell becomes vacuolated and later the cell gets lysed, how? It is not clear (at least for me). Please refer to other web sites for the information.

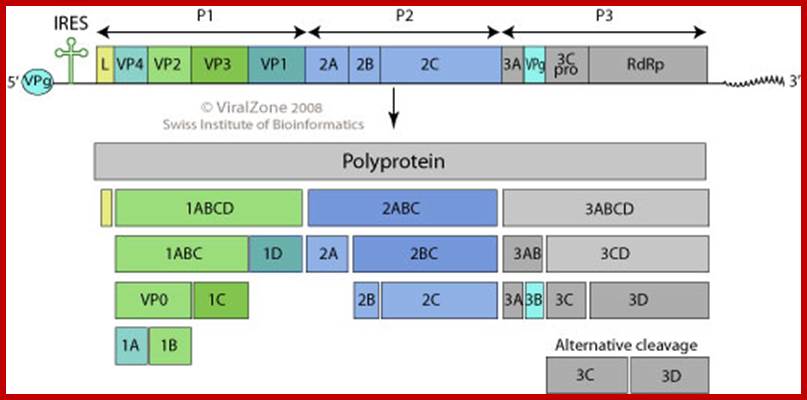
Organisation of the enterovirus genome, polyprotein processing cascade and architecture of enterovirus capsid The genome of enteroviruses contains one single open reading frame flanked by a 5′-and 3’ untranslated regions (UTR). A small viral protein, VPg, is covalently linked to the 5′ UTR. The 3’UTR encoded poly(A) tail. The translation of the genome results in a polyprotein which is cleaved into four structural proteins (dark gray) and seven non-structural proteins (light gray and yellow). The sites of cleavage by viral proteinases are indicated by arrows. The four structural proteins adopte an icosahedral symmetry with VP1, VP2 and VP3 located at the outer surface of the capsid and VP4 at the inner surface. The single strand genomic RNA is located inside the capsid. http://www.intechopen.com/
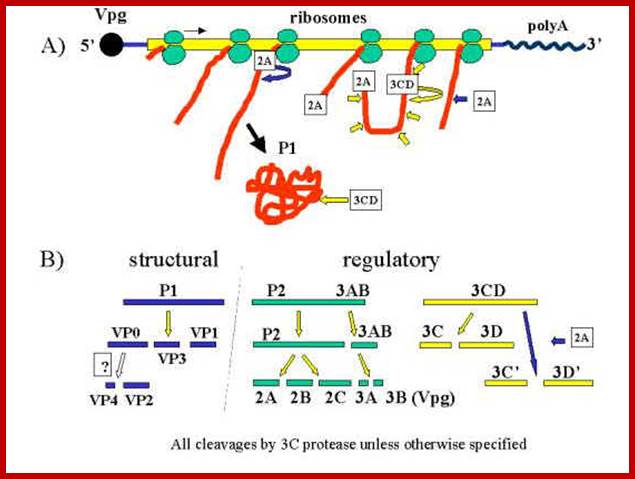
Polyprotein produced by the linear RNA is cleaved first by its own endopeptidase and then the protein is cleaved into specific fragments. The P1 generates all its Vps The 3B produces the cap called VpG and the 3D produces RNA dependent RNA polymerase. www.en.ppt-online.org
[VP1 acts as the binding protein to cell receptor.
· Vpo—>cleaves into VP4 and VP2.
· VP2ABC---->cleaves into 2A, 2B, 2C; all are proteases.
· VP3ABCD---->cleaves into 3A-B and 3CD; then 3A-B cleaves into 3A, the 3B a useless segment.
· The 3-A is VPg protein.
· The 3CD are cleaved into 3C and 3D.
· The 3C is a protease, which cleaves most of the sites, and 3D is large protein and acts as RNA dependent RNA Replicase.]
Human Corona Virus:
Another (+) sense large sized RNA genome.
Introduction:
This virus has most talked about in recent years for it caused Severe Adult Respiratory Syndrome (SARS) in humans, and warning has been flashed all over the world of its new Avatar, found to cause very severe respiratory and enteric diseases. Though more than 8000 people reported be infected only few have died. This is more effective in children than in adults. The strains of this virus also infect murines, porcines, canines, bovines and chicks and devastate animal populations.
This viral disease was first isolated in 1937 from chicks. Late in 2002 this syndrome was observed in Guangdong province of China. It is also in china that pandemic Flu started in late nineteenth century, which affected the entire world. In India those who consumed garlic, pepper and turmeric concoction have survived, the chemical principles responsible for cure are not known. Nearly 65% of the population ascribed to colds still is ascribed to known the SARS. However in 1965 Tyrrell and Bynoe used human cell cultures and isolated first human corona virus; there are more than 13 species. The symptoms are high fever (38-60^c), headache, body aches and can cause liver damage. This disease outbreak was peaked in the early 2003. Though more than 8000 cases have been reported but death was reported to be 3-30% (775 reported).
This virus belongs to Group: IV (+ ssRNA), Order; Nidovirales (nidus = nested), Family: Coronaviridae, Genus Corona virus.
Morphology:
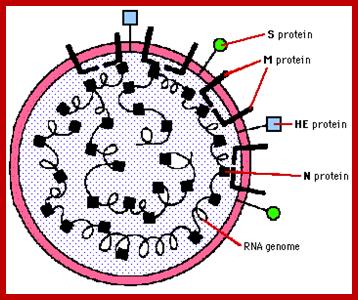
It looks more or less spherical but with little distortions; covered with a membrane, studded with spike like structures, ending in blob or spherical structures. It is because of the surface looks crown like; hence the viruses are called Corona viruses. The size of the virus varies from 65nm to 110nm. The membrane contains tripartite fibers called S proteins, which are called ‘peplomers’ (like the structure found in a blouse at the waistline).
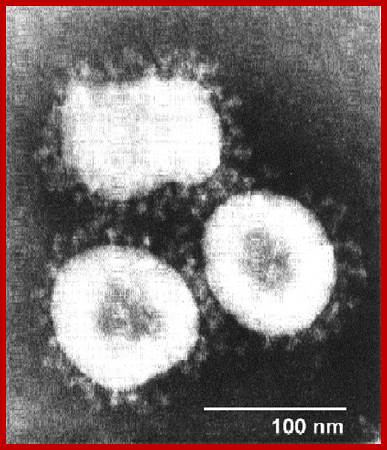

http://sofia.medicalistes.org
Corona virus
The membrane is also studded with transmembrane proteins called Matrix proteins called M. Along with these, one can find another kind of membrane bound proteins called HE. Nucleo capsid proteins found inner to the membrane are bound to long coiled RNA. There is no well defined capsid with any symmetrical organization. The genome in the case of h Sars coV is a plus sense RNA of 29736 nucleotides long, perhaps one of the longest RNA genome found among this class of viruses. The sequence of the genes in human corona viruses is more or less similar to other members of corona viruses. The virus doesn’t enclose any of Replicase.
Viral Proteins:
RNA replicase: RNA dépendent RNA polymérase. The gene for it occupies nearly 21220 ntds from the 5’ noncoding leader sequence; it almost covers 2/3 of the genome. Though this region codes for two proteins, it is translated as an overlapped reading frame. The molecular wt of this protein 1004.5 KD (most unlikely) theoretically, but in vivo the size of it is not known; actually the first part of this longish protein has replicase activity.
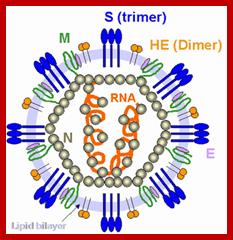

Virus particles range from 70 to 120 nm in diameter and are surrounded by characteristic spike-shaped glycoproteins, as shown in the figure; Coronaviruses contain the largest single-stranded, positive-sense RNA genomes currently known, which range from 25.5 to nearly 32 kb in length. Rachel L. Graham et al; http://www.nature.com
Spike protein: It is a glycoprotein of 150 KD, exist as triple homodimer with spherical conformation at its terminal end and the N end is buried in the membrane; it is a glycoprotein and act as major antigen acts as receptor binding protein, involved in infection.
HE: This is one more envelope protein studded on the surface of the membrane consists of 9-12KD small subunit and other 65kd protein. They form dimers. This is haemagglutinin protein with esterase activity.
M protein: It can be called matrix protein of 31.5-40 KD. It is a trans-membrane protein having an outer domain and an inner domain, contacts N proteins which are bound to RNA genome.
N protein: It is a major protein inside the membrane envelope, bound to 20-27kb long RNA, its Mol.wt. is 60KD, it is this protein found next to membrane, that provides a structural feature, but with no discernable symmetry.
Genome:
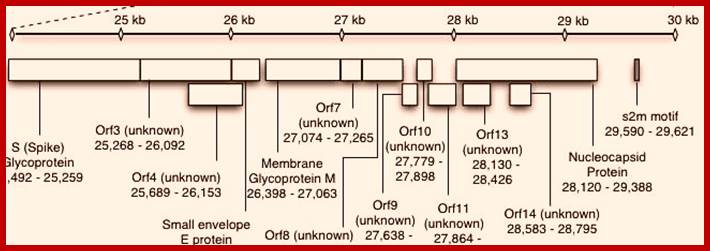
The human corona viral genome is (+) ssRNA, 29736 ntds long. It is packed into to the capsid enveloped by the membrane. At the 5’end it is covalently linked to 7’CH3 G-5p-p-p5’A-.
This large sized RNA genome is unique. At the 5’end it has 265nts long UTR; similarly it has 61 ntds long 3’UTR; the sequence is used for viral packaging. In between the genome has many at least 13 ORFs. The products of some are known and others are yet to be identified. Out of 29717 nucleotides 21220 ntds from 5’ UTR end, the genome is encoded with two cistrons, together generate a huge sized replicase enzyme.
The enzyme has a very poor proof reading. The rest of the genome from 21486 to 29621 contains 5 known ORFs and the other 9 ORFs is not known, so they can be deemed as URFs (unknown reading frames).
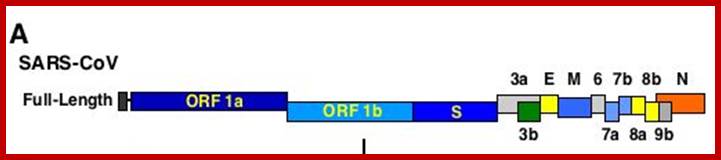
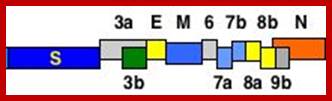
5’Cap-5’UTR-----265-------Replicase----21485-S--E--M--N---3’UTR-29717-(A)n

![]()

Life cycle
Life cycle; http://www.sciencedirect.com/
RNA codes for the following proteins:
5’-265-----13398 =Replicase 1A, 500 KD
--13399---21485 = Replicase 1B, 500 KD
--21492---25259 = Spike- a glycoprotein and a homotrimer (150 kD) trans the membrane.
--25268---26092 and 26117—26347 (E) = HE protein, a membrane, glycol-with esterase (E=19 and HE=65KD).
--26398—27063 = M a Trans membrane protein, it is a glycoprotein (31.5KD).
--2820—24388 = N protein, acts as nucleocapsid protein binds to RNA genome to form RNAsome (60KD).
--8120--29388 = s2m motif.
From 21486 to 29388 the genome generates five known proteins and nine URFs, though they synthesize the said proteins, their exact features and functions are yet to be known.
The genomic RNA also contains intergenic sequences such as UCUAAC at five positions after replicase coding region. This sequence is used for transcribing minus strand in nested form.
Infection:
Infection is through S protein and the host cell membrane take the virus into through endocytosis, where all viral membrane and proteins enter into host membrane leaving nacked RNA free in the cytoplasm. As the 5’end is capped and the 3’end with poly(A) tail soon it is covered by poly(A) binding protein I and cap binding proteins, so both ends are protected. As the genome is a plus strand it directly acts as translatable mRNA of a huge size.
Translation process:
The leader sequence facilitates the binding of ribosomes for initiation and elongation, and the translation ends at replicase cistron. Though there two subunits, the translation continues from Replicae A into replicase B. Whether the translation continues to the end or not is not clear, but some authors express the translation continues up to the end and the long polypeptide chain is cleaved by autolysis. But others have different opinions.
Monopartite, linear ssRNA(+) genome of 27-32kb in size (the largest of all RNA virus genomes). Capped, and polyadenylated. The leader RNA (65-89 bp) at the 5' end of the genome is also present at the end of each subgenomic RNAs.
;http://viralzone.expasy.org
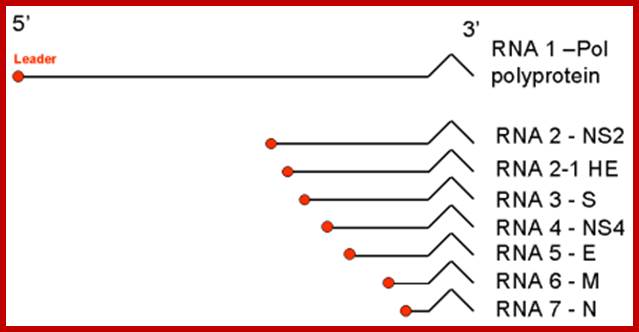
Once the Replicase is produced, it binds to the 3’end and transcribes to generate full length (-) strand which acts as the template to generate mRNAs for translation. But the transcription is nested in the sense; it is transcribed in segment starting from a fixed position, determined by the UCUAAC sequences located at the intergenic region.
The Replicase first transcribes the first 72 nucleotides from the 5’end and replicase dissociates from the leading strand with its 72 ntds long leader places at the first intergenic region and transcribes the (-) RNA up to the end. The mRNA produced is poly cistronic and it is immediately capped at the 5’end poly-adenylated at the 3’end.
Similarly, using the 72ntds long leader and UCUAAC intergenic sequence, the replicase transcribes the other four segments to generate five or more mRNA transcripts that can be translated. Each of the mRNA’s length is successively shorter and generates specific polypeptides, which can under go proteolytic cleavage and generate their respective proteins. Once sufficient amount of proteins are produced, the minus RNA is used to generate full length plus strand and repeatedly the process continues to generate more and plus strands and more and more of proteins.
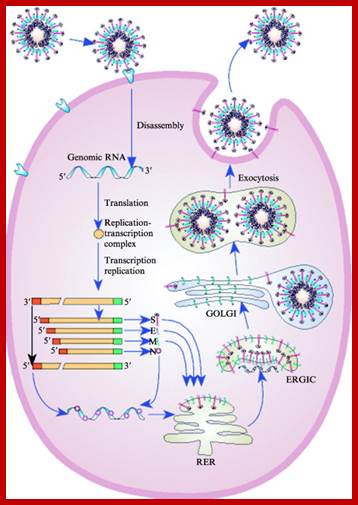
While translation is going on, translated viral membrane proteins, such as S, M and E, are transferred to endoplasmic reticulum; there they are pulled into lumen and processed and packed to cis Golgi, where the viral proteins are modified and localized.
At this point the N protein initiates the binding to 61 nucleotide sequences at 3’ end of the full length plus RNA, and the assembly continues to produce a nucleocapsid thread. With viral membrane proteins assembled in Golgi membranes they bind to Nucleocapsid thread and finally encapsulate the nucleocapsid and budded off as viral particles. The life cycle takes about 24hrs after infection. This spreads to other parts in few days and cause severe symptoms.
CYTOPLASMIC
- Attachement of the virus to host receptors mediates endocytosis of the virus into the host cell.
- The capsid undergoes a conformational change and releases VP4 that opens a pore in the host endosomal membrane and the viral genomic RNA penetrates into the host cell cytoplasm.
- VPg is removed from the viral RNA, which is then translated into a processed polyprotein.
- In entero-, rhino-, and aphthoviruses, shutoff of cellular cap-dependent translationthrough the cleavage of translation initiation factors by viral protease.
- Replication occurs in viral factories made of membrane vesicles derived from the ER. A dsRNA genome is synthesized from the genomic ssRNA(+).
- The dsRNA genome is transcribed/replicated thereby providing viral mRNAs/new ssRNA(+) genomes.
- New genomic RNA is believed to be packaged into preassembled procapsids.
- Cell lysis and virus release.
- Maturation of provirions by an unknown host protease.