Qb Virus:
These are bacteria eating viruses and they belong to RNA phages, which are classified into 3 serologic groups.
RNA Bacteriophages:
|
Viral group |
Specific viruses |
Features |
|
Group-I |
F2, fr, FH5, MS2, R17, M12, |
|
|
Group-II |
Ff, If |
|
|
Group-III |
Q-beta |
|
|
|
|
|
Morphology:
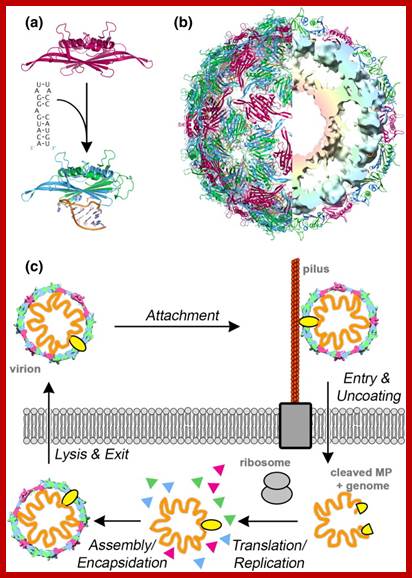
Qb ssRNA phages is a coliphage with 4217 ntds long RNA. They are extremely small viruses, 25 nm thick, isometric, adsorb to specific bacterial sex pili proteins and infect. So they are male dependent viruses. The phage has 20 faces and 12 vertices. Each face is made up of six subunits and each vertices is made of five subunits; in all it has 180 protein subunits. Inside the capsid a single stranded genomic RNA is packed; it is a linear molecule but extensively folded into secondary structures.
Qbeta viruses related to another similar virus called MS2, whose genome is more or less same as that of Qbeta.
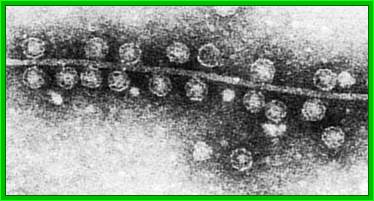
Q-beta phage particles
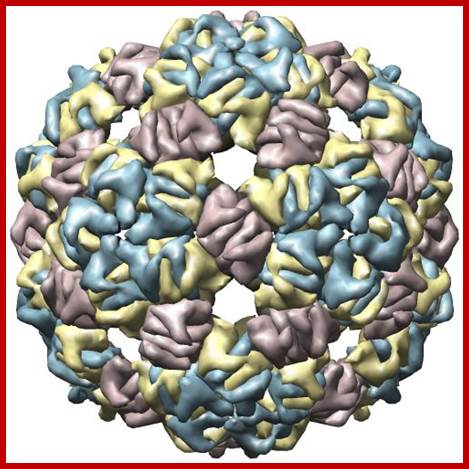
Bacteriophage Qbeta capsid; http://www.rcsb.org/
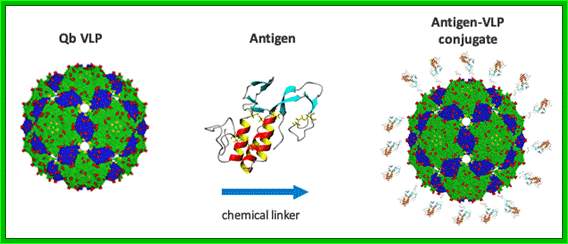
Schematic illustration of Qbeta virus-like particle linked to an antigen using succinimidyl-6-[beta-maleimidopropionamido]hexanoate (SMPH) linker. The Qbeta VLPs are approximately 25 nm in diameter.http://www.cytos.com/; http://www.igert.org/
Infection:
Viruses bind to sex pili in large numbers. Through the tubular sex pili (made up of oligomeric protein Pilin) or F-pili, the RNA genome is transported leaving behind the empty capsid. The pili is about 6-8nm in diameter. Once infected, they can easily replicate and produce about 5000 to 10000 particles per cell in about 60 minutes. Though the infection is through sex pili, the exact mechanism of the genome transfer is not known. The genome is linear, but it has very high level of secondary structures (62 to 80% of the genome), with many hairpin like loops, even at 5’ and 3’ ends. In fact the secondary structure provides protection from cellular exonuclease digestion. The length of the RNA is 4217 ntds and the genome has positive sense and codes for four proteins-A1, A2, CP and qb Replicase. At 5’ end it has ppp5’G-GGU and at 3’ end it has CCA 3’OH, both ends have hairpin structures. The RNA is linear and single stranded; it has positive sense. Replication of the phage F-specific RNA requires fewer than cfu ml^1 E.coli actually it requires 35-40 minutes.
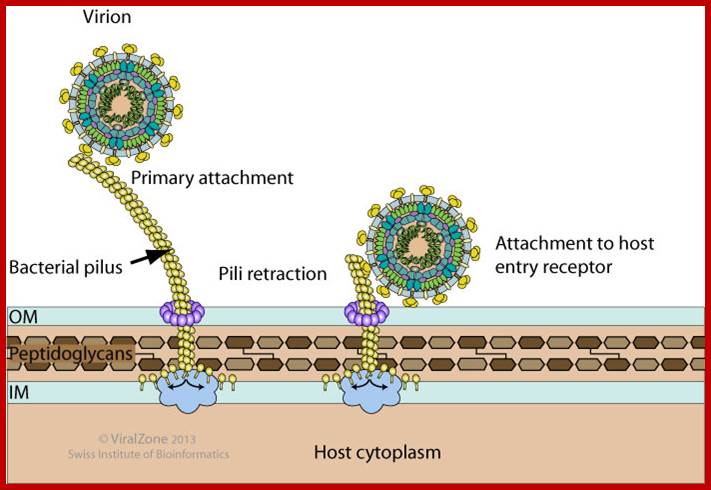
Pili are proteic retractile filaments up to 20 micrometer long that protrude from gram-negative bacteria. There are many types of pili and each bacterial virus binds specifically to a precise type. Some filamentous viruses like bacteriophage fd adsorb onto the tip of F-pili and enter the cell at the pili basal pore. Other filamentous phages can target I-pili (phage If1), N-pili (phage IKe), or type IV pili (e.g. bacteriophages Pf1 and Pf3) . Icosahedral virus usually adsorb onto the sides the pili. http://viralzone.expasy.org/
Some RNA and DNA bacteriophages use pili to attach to the host cell.
· Coat protein = 13.7 KD, there are 180 subunits per phage.
· A1 protein = 38 KD, extension of coat protein. Only one copy per virus, it is required for attachment of the phage to virus. It is a read through product of the coat gene.
· A 2 protein = Maturation protein, also has lytic function. It is required for the maturation of the phage i.e. it is required for the assembly of the particles within the host cell and ultimately leads to the lysis of cells.
· qb1 = Replicase, 65.5 kDa. It resembles to that bacterial beta sub unit, with template site and an active site. But it requires few more proteins from the host cell for its activity. It gets associated with ribosomal S1 subunits 961.2kDa; it also contains host EF-Tu and EF-Ts (subunits g- 43.2 kDa, and d- 30.3 kDa); it also require a host factor Hfq (which acts like a chaperone).
· Coat proteins are produced in greater abundance than the others, the ratio is, 20 coat protein: 5 b protein: 1 A2.
· A2 gene ; the initiator codon is GUG, terminator codon is UAG. It is a maturation protein. A2 functions in host cell recognition. A2 inhibits peptidoglycan formation similar to penicillin.
· Coat protein is coded by RNA with AUG as the initiator, UGA or UAA as terminators, UGA act as read through codon during the synthesis of A1 protein.
· Replicase (beta) = AUG is the initiator, terminator is UAG. The replicase has four subunits, one coded by the qb RNA.
· The RNA polymerase that replicates both the plus and minus RNA strands is made up of four proteins as mentioned above: the beta subunit (replicase) is encoded by the phage, while the other three subunits are encoded by the bacterial genome: alpha subunit (ribosomal protein S1), EF-Tu is gamma subunit and EF-Ts delta subunit.
ppp5’G-GGU—62------1322-1345----1744----2332-2353-----4120-4217-3’ Qb


Monopartite, linear, ssRNA(+) genome about 3.4-4.3 kb in size. The 5' end is capped. Encodes 3 (Allelovirus ) or 4 proteins (Levivirus ). http://viralzone.expasy.org
Coli phage MS2, Monopartite, linear, ssRNA(+) genome about 4 kb in size. The 5’ end is capped. Encodes 4 proteins. http://viralzone.expasy.org/
From 1-62, 5’ end leader sequence.
From 62 – 1322 ntd, A2, maturation protein (75 aa long).
From 1345 – 1744 ntd, coat protein (129 aa long).
From 1345 – 2332 ntd, A1 attachment protein (353 aa long).
From 2353 – 4120 ntd, Replicase protein.
From 4120 – 4220 ntd, 3’ end noncoding terminal sequence.
From 1322 to 1345, from 2332 to 1353 are noncoding spacers.
Both negative and positive strands have 5’GG and 3’ CCA
Qb - RNA replication enzyme:
The replication enzyme is called RNA dependent RNA polymerase or RNA Replicase. The replicating enzyme is a complex made up of two virally coded b protein and few host proteins. It replicates from 3’end towards 5’end, no primers are used.
- b- Protein: 60 –61 KD, (544 aa long), virally coded, it is similar to that of bacterial beta subunit, which has a template binding site and a polymerizing site.
- a- Subunit: 70kd, from the host cell, it is from one of the small ribosomal protein (S1).
- ¡- Protein: 45kd, bacterial protein called elongation factor EF-Tu, required for elongation during protein synthesis.
- d- Protein: 35 KD, it is again host factor called EF-Ts, which is involved in recycling of Tu with GTP dependent manner.
- HF1: 72kd protein, another host protein; it is a hexamer. It is associated with ribosomes.
|
Protein |
Mol.wt (subunits) in KD |
Features |
|
Beta protein |
60-61 (544 aa) |
Viral coded, has enzyme properties |
|
Alpha subunit |
70 (host’s small ribosome s10) |
One of the host small ribosomal subunit |
|
Gamma protein |
45 |
Bacterial elongation factor EF-Tu |
|
Delta |
35 |
Bacterial EF-Ts |
|
HF1 |
72 (hexamer) |
Associated with ribosome |
Replication:

Alloleviviral RNA, q-beta-http://www.lookfordiagnosis.com
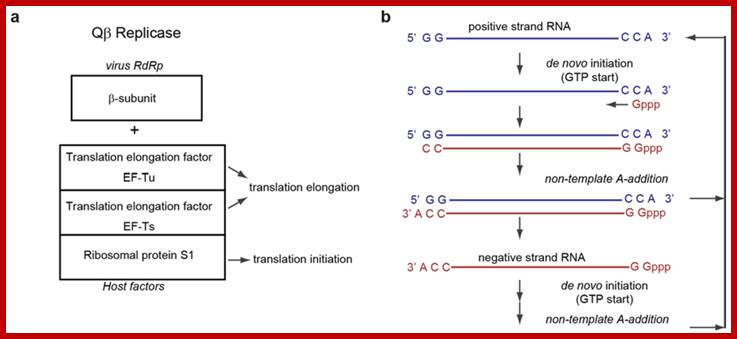
Plus-strand RNA viruses contain RNA elements within their genomes that elements is that of local RNA structures. This perspective, however, is changing due to increasing discoveries of functional viral RNA elements that are formed by long-range RNA–RNA interactions, often spanning thousands of nucleotides. The plus-strand RNA genomes of tombusviruses exemplify this concept by possessing different long-range RNA–RNA interactions that regulate both viral translation and transcription. Here we report that a third fundamental tombusvirus process, viral genome replication, requires a long-range RNA–based interaction spanning ∼3000 nts. In vivo and in vitro analyses suggest that the discontinuous RNA platform formed by the interaction facilitates efficient assembly of the viral RNA replicase. This finding has allowed us to build an integrated model for the role of global RNA structure in regulating the reproduction of a eukaryotic RNA virus, and the insights gained have extended our understanding of the multifunctional nature of viral RNA genomes.A Discontinuous RNA Platform Mediates RNA Virus Replication: Building an Integrated Model for RNA–based Regulation of Viral Processes; mediate a variety of fundamental viral processes. The traditional view of these Baodong Wu, et al; WWW.Journals.plos.org
The genomic RNA, before initiation of replication, undergoes translation. Initiation of translation takes place at an AUG codon found in non-hairpin region.
- This is the only internal ribosomal entry site (IRE) found in at the end of A2 reading frame and at the beginning of coat protein gene for initiation of translation. Ribosomes use internal guide sequence called (IRE) and thread through and initiate translation of the coat protein region.
- As the translation progresses the hairpin structures are opened ahead of the ribosome, this leads to the opening of the gene Replicase initiator region, thus Replicase part is translated. Every time Replicase is translated, it is initiated at coat protein site i.e. IRE site.
- Host factor EF-Tu-GTP in the complex binds to tRNA like sequence CCA3” for initiating complementary strand synthesis; which is similar to the host factor TF-Tu binding to CCA3’ and placing on ribosomal A site for elongation.
- It is to remind, that HIV and other retroviruses use 3’ end of specific tRNAs like structures as primers for replication of their RNA genomes. Perhaps the 3’ end of the genomic RNA, which exhibits tRNA like secondary structure, is used for proper binding of the enzyme complex and initiates complementary strand synthesis, with first nucleotide added is 5’PPPG. Now it is known it uses phenylalanine tRNA as a primer, but this primer less initiation and it is called recursive replication.
- Probably the hexamer might act as the helicase and the enzyme processively progresses to make a new strand, i.e. the (-) strand. As the enzyme moves, hairpins in the (+) strand are opened ahead of the enzyme progressively; at the same time the newly synthesized strand frees from the (+) strand and assume their secondary structure dictated by their complementary sequences. At the end of replication, it is believed both (+) and (-) RNAs are poly Adenylated? Synthesis of minus strand continues till sufficient number is built up.
- The same enzyme complex now initiates plus strand synthesis and uses the secondary structure in the minus strand.
- Synthesis of sufficient number of plus strands helps in the production of coat proteins. When coat proteins level builds up, they bind to the initiator region of Replicase segment and thus prevent the synthesis of Replicase; this is an example of translational control.
- S1 is required only on (+) strand, helps in binding of the Replicase to the template and initiate replication and produce (-) strand. It is not required for the replication of (-) strand on (+) RNA.
- Once adequate number of capsid proteins is generated, they bind to package sequences in the genomic RNA and assemble viral particles, which are further matured into full-fledged viral particles. Then they are released by the lysis of the cell. There are few problems not explained so far.
(+) RNA- 5’ UGG--------------------------------------------->CCA 3’
(+) RNA- 5’ UGG--------------------------------------------->CCA 3’
(-) RNA- 3’ ACC<----------------------------------------------GGU 5’
(+) RNA- 5’ UGG---------------------------------------------->CCA 3’
![]()
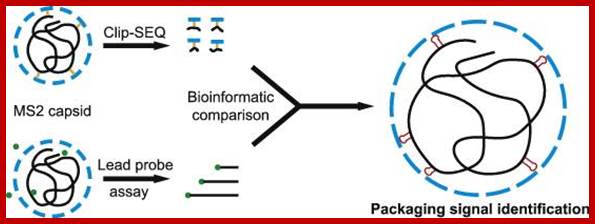
https://www.ncbi.nlm.nih.gov
MS2 packaging.https://www.ncbi.nlm.nih.gov
Most families of these viruses encode single-copy, high-affinity CP-binding sites in their genomes that are thought to act as assembly initiation sites [20], [21]. These RNA sequences form defined secondary structure elements that are specifically recognized by the viral CPs. The best characterized of these interactions is the 19-nt-long stem–loop named TR in the MS2 genome (Fig. 1) that acts as a translational operator for the replicase cistron ;The free energy for this process has been thought to come mostly from favorable electrostatic interactions between the negatively charged RNA and positively charged residues and domains in the CPs ;Packaging of qb viral RNA into capsid is similar to the of its related virus MS2; https://www.ncbi.nlm.nih.gov
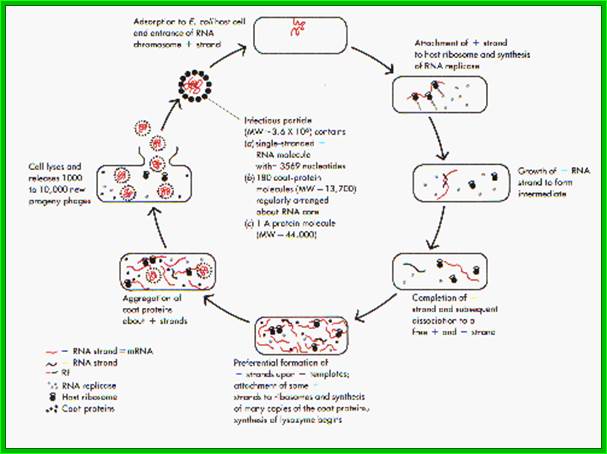
Q beta Life cycle
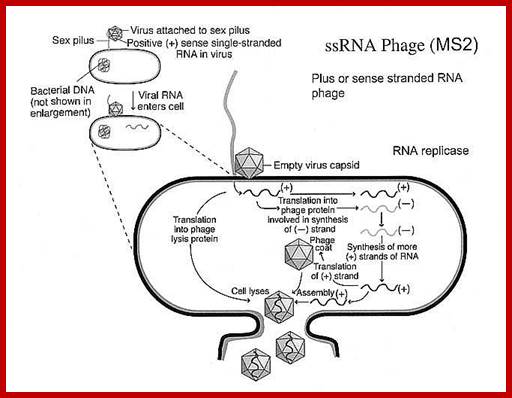
Infection, growth and lyses; Replication of Q-beta RNA, first the +RNA is copied by its Qb-Replicase enzyme, then the –strand is copied to + strand; http://www.lookfordiagnosis.com