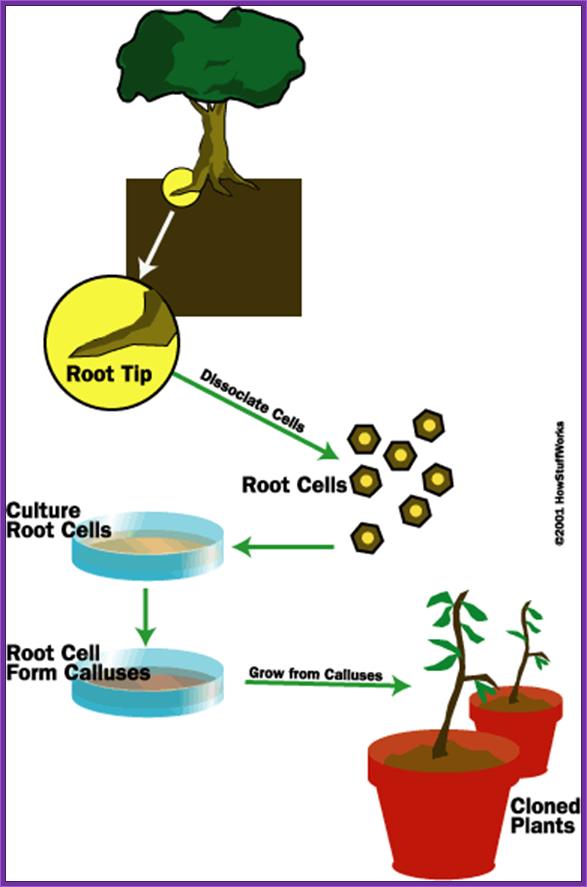
Transformation-Plant Cells :
Agrobacterium a Natural Genetic Engineer:
Agrobacterium tumefaciens now called as Rhizobium radiobacter is a soil gram negative bacteria; its cousin is Agrobacterium rhizogenes. Both have quorum sensing abilities. The former causes crown galls and the later induces hairy roots. Both have some specific oncogenes and they are responsible for the diseased morphology when they infect. Both these bacteria are capable of transferring their specific plasmid DNA into plant cells, which gets integrated into plant chromosomal DNA and causes the disease. Plant scientists are using the bacteria transferring the required gene into plant to obtain Transgenic plants for a specific purpose. Plant genetists have created thousands of such transgenic plants and many of them are in use in various countries. Some countries are against using such transgenic plants for agriculture or for other purpose. Those who are against the use of transgenic plants think they are the most intelligent people than the people use them.

http://www.anselm.edu/
Agrobacterium tumefaciens:
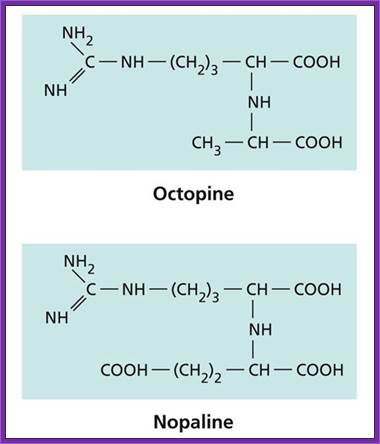
It is a soil bacteria, causes tumors in plants especially in Solanaceae members. These bacteria can also cause tumors in some members of liliaceae and few other monocots. They are gram-negative bacteria; they contain a plasmid, which is capable of inducing tumors, so the plasmid is called Tumor inducing plasmid (Ti-plasmid). Depending upon the kind of tumors they produce they are classified as Nopaline synthesizing plasmids called Nos plasmid or Octopine plasmids.
Agrobacterium New nomenclature:
The Agrobacterium have been renamed as Rhizobium species (Young et. al., 2001), they are listed below, with their new names. See the LPSN for more information on the Agrobacterium. These generally do not form symbiotic root nodules, unless they harbour a Sym plasmid or other transposable element with nodulation genes. In this case, I would define them as rhizobia. Other Agrobacterium have been reclassified as Stappia, Ruegeria, and Pseudorhodobacter.
|
Old names |
New Name |
|
Agrobacterium larrymoorei |
Rhizobium larrymoorei (Young et. al., 2001) |
|
Agrobacterium radiobacter = Agrobacterium tumefaciens |
Rhizobium radiobacter (Young et. al., 2001) |
|
Agrobacterium rhizogenes |
Rhizobium rhizogenes (Young et. al., 2001) |
|
Agrobacterium rubi |
Rhizobium rubi (Young et. al., 2001) |
|
Agrobacterium vitis |
Rhizobium vitis (Young et. al., 2001) |
Kingdom: Bacteria,
Phylum: Proteobacteria,
Class:Alpha Proteobacteria,
Order; Rhobiales;
Family:Rhobiaceae
Genus ;Rhizobium,
Species: radiobacter
(old name: Agrobacterium tumefaciens),
11 August 2011
Agrobacterium used for gene transfer into plants; Bacteria bound to cell surface..
http://www.davidson.edu

Agrobacteria are harmful to plants, but useful to plant scientists; http://www.genomenewsnetwork.org/

Tree tumor: https://www.flickr.com
Kingdom: Bacteria,
Phylum: Proteobacteria,
Class:Alpha Proteobacteria,
Order; Rhobiales;
Family:Rhobiaceae
Genus ;Rhizobium,
Species: rhizogenes,
Updated August11-2011
Old Classification
|
Kingdom: |
|
|
Phylum: |
|
|
Class: |
|
|
Order: |
|
|
Family: |
|
|
Genus: |
|
|
Species: |
A. rhizogenes |
|
Agrobacterium rhizogenes synonym with |
|
|
New name:Rhizobim rhizogenes |
|

Agrobacterium rhizogenes
It is gram negative soil bacterium that produces hairy root disease in dicotyledonous plants. A. rhizogenes induces the formation of proliferative multi-branched adventitious roots at the site of infection; so called 'hairy roots. Agrobacterium rhizogenes doesn’t have Cytokinin genes; instead they have genes like rol-A, rol-B and rol-C. These are responsible for developing hairy roots and they can regenerate plants.

Agrobacterium rhizogenes induced hairy roots. http://www.biotechnet.ch/
Rhizogene induced rootnodules

Crown galls in Walnut trees’ Juglans regia; http://ucce.ucdavis.edu/
Few Agrobacterium strains used in Gene transfer:
The strains like LBA 4404 have plasmids, which lack tumor inducing genes, but they do contain a restrained Ti plasmid with Vir genes, which can provide components for transferring DNA segments that are found in other plasmid DNA, if located in between the LB and RB. Genes such as Shi, Roi and opine producing genes are deleted. The said strains also lack in endonuclease activity (Rec ^-- RE ^ - and they are sensitive to some antibiotics such as carboxylin.
Plant Oncogenes:
Any gene or genes, which are capable inducing tumors, is called Oncogenes. In Agrobacterium system the genes such as Auxin synthesizing and Cytokinin producing genes when get integrated into host genome, they are activated by cellular factors. These gene products produce Indole acetic acid from Tryptophan, and Isopentinyl adenine is produced from Isopentinyl pyrophosphate and Adenine mono phosphate respectively. The concentration of Auxin and Cytokinins produced is balanced so as to transform the cell into actively dividing undifferentiated cells. This produces a mass of undifferentiated cells, which is called callus, or this can assume the shape of tumor, so the genes responsible for inducing callus are called as plant Oncogenes. It is not unusual to see such tumor as bid as six feet in diameter on various plants.
- Agrobacterium infects plants through external wounds.
- The bacterial infection and compatibility depends upon certain gene products such as Chv-A, B and C and Exo-C. They have a role in the attachment of the bacteria to the host cell. Mutations in the said genes affect the attachment of the bacteria to the host cells. Certain mutations and variation in ViR-A exhibits limited host range (LHR). On the contrary super virulent strains have wider host range capabilities.
- However host range compatibility and incompatibility lies in bacterial factors and host plant factors as well; a situation similar or akin to Rhizobium infection of certain legume members during the development of rood nodules.
- Certain phenolic compounds released to the surface of the host cells during injury act as signals and Agrobacterium attracted to such phenolic compounds and bind to host cells. Some of the glycoprotein on the surface of the bacterial cells may play a pivotal role in the recognition and binding.
Agrobacteriums’ Ti plasmid:
Ti plasmids range in size from ~200 to ~300 kbp. Of this, about 30% is common to Nopaline and Octopine plasmids (Genes VIII, Benjamin Lewin, pp. 525). The Ti plasmids are classified into different types based on the type of opine produced by their genes. The different opines specified by pTi are Octopine, Nopaline, Succinamopine and Leucinopine (Wikipedia). The plasmid has 196 genes that code for 195 proteins. There is no one structural RNA. The plasmid is ~206,479 nucleotides long, the GC content is 56% and 81% of the material is coding genes. There are no pseudogenes.
Types of Ti Plasmids
Two classes of Ti-plasmids exist. The nopaline and octopine Ti-plasmids have T- DNA regions but the structure of these regions differs.
Nopaline T-DNA - single continuous segment of
about 22kb
Octopine T-DNA - three segments; left DNA (TL) = 13 kb; center DNA (TC),
DNA = 1.5 kb; Right DNA (TR) = 7.8 kb; TL DNA contains the oncogenic function and TR contains opine synthesis genes. The opines released out of plant cells are consumed by bacteria bound at externally at the cell surface.
The DNA that is transferred from Ti DNA in the bacteria to plant cell is called T-DNA is referred to as the T-region. T-region is approximately 10 to 30 kbp in size and represents less than 10% of the Ti plasmid. Ti plasmids generally contain one T-region, but some Ti plasmids have multiple T-regions in them. T-region is flanked by T-DNA border repeats. These are 25 bp long and highly homologous in sequence. The T-DNA border sequences flank the T-region in directly repeated orientation. The T-DNA border sequences delimit the T-DNA and are the target of the border-specific endonucleases. The sequences are
TG Pu G ATTGGCAGGATATAT N TGTAANC C T TC
The Ti plasmid consists of a replication origin OriT, It has Opine catabolytic gene, It also contains Virulent genes (Vir), Bacterial chromosomes contain important regulatory genes such as Chv.
A part of TDNA – from the left->
Vir= Virulence regulators left of LB.
LB = Left border.
iaaH = (aux2 and tms 2), Indole-acetamide hydrolase gene which converts IAM to IAA. iaaM = (aux 1, tms 1, Tryptophan-2mono-oxygenase, which converts Tryptophan into indole3-acetamide (IAM).
IPT = Isopentinyl transferase gene.
Nos = Nopaline synthase gene.
RB = Right border.
o/D = Over drive sequence right of RB.
The said plasmid genes are involved in the synthesis of Auxin and Cytokinin.
The Ti plasmid, outside the T-DNA region, also contains several other genes such as opine catabolising genes such as Nopaline catabolising gene (NOC), Octopine catabolising gene (OCC) and Agropine catabolising gene (AgC). On to the left of the T-DNA there is a segment of DNA, 36kbp, consisting of a cluster of genes called ViR (virulence) genes. On the right flank of the RB of T-DNA abutting to it is 25 bp long sequences called Over- Drive (5’TAApuTpyNCTGTpuTNTGTTTTGTTTG3’). This region is recognized by ViR-C gene products and facilitates ViR-D2 gene products to cut the strands in the RB.
Along with there are regions for the replication Origin called Ori-V and Ori-T (Tra) for replication and transfer of T-DNA. There are many other genes located on bacterial chromosome, they are Chv for chromosomal virulence (Chv-A, Chv-B, Chv-D, Chv-E, cel, exoC and Psc I) involved in recognizing the host, which actually determine host bacterial compatibility. Genes such as exo-C and att are involved in the binding of bacteria to host cells.
Vir -LB-I------I--iaaH->I<---I-iaaM—I—Iptà--Iàtml-à--I--ßNos---I-RB-overdrive
The T DNA is constrained by direct repeats called Borders- left and right- LB and RB. The border sequences are There are at least 30 different opines described so far. From the RB, the first gene present codes for a specific Opine. Next to it is a Cytokinin synthesis gene; next there are two genes for Auxin synthesis.
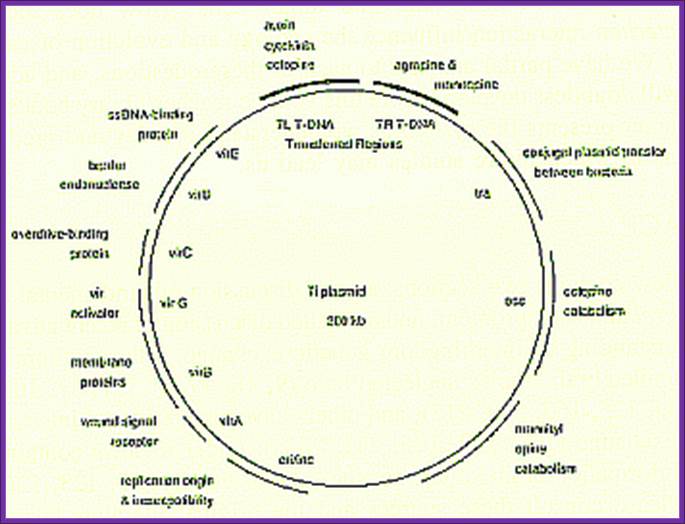
A general view of the Ti plasmid
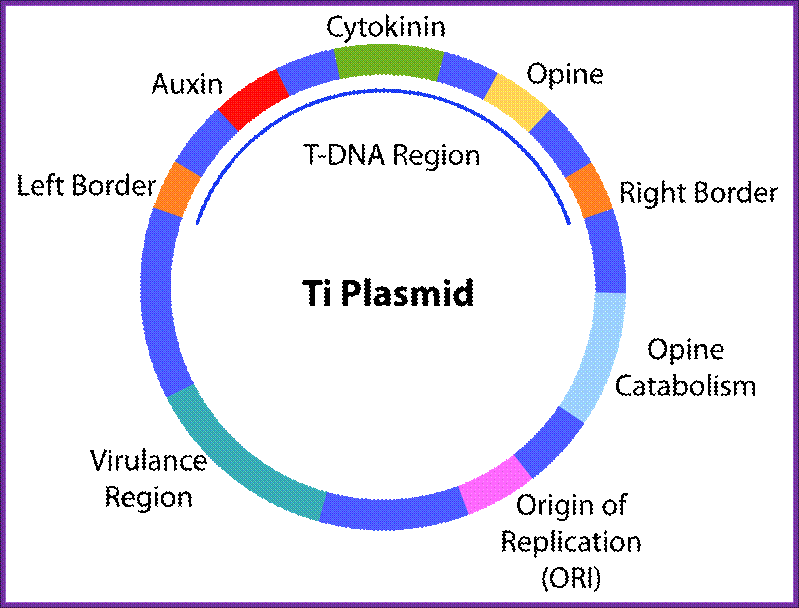
Ti Plasmid; http://en.wikipedia.org/
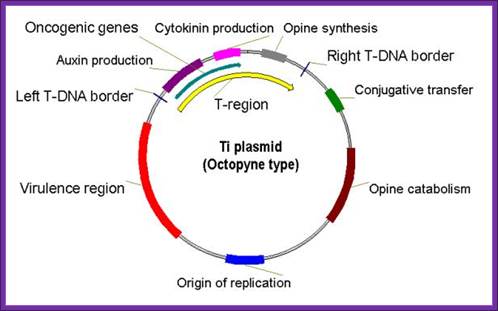
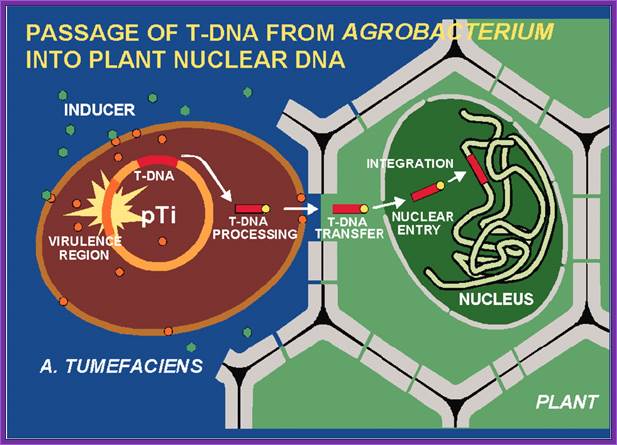
Agrobacterium tumefaciens and the Plant: The David and Goliath of Modern Genetics; http://www.plantphysiol.org/
In response to wounds, plants secrete a variety of Phytochemicals including phenolic compounds. They in turn induce cells to respond. The bacterium now binds firmly to the plant cell wall. The ‘two system’ surface receptors found in the outer membrane bind to the signal ligands. The bacteria have quorum sensing mechanism.
Understanding of the interplay between the extracellular environment and intracellular decision circuitry of a cell is important but is an arduous goal to achieve since many interacting factors, difficult to measure and control in experiment, are involved. The authors address this problem by means of computational modeling using the example of a bacterial population that cooperatively switches on a common gene expression program if a certain critical population density is achieved. They developed a detailed model of the intracellular control network and demonstrated that it can operate as an “on–off” gene expression switch that is sensitive to environmental control and yet highly robust to intracellular molecular noise. The population-wide transition is further modeled using a novel method in which each bacterium is given a unique copy of an intracellular network. This approach, which allows monitoring of both the dynamics of individual cells and population behavior, provides an explanation for the gradual appearance of the transition to the “on” state that has been observed in experiments, and quantitatively predicts the critical value of the population density at which this transition occurs. Unexpectedly, a comparison of the cell densities required for the transition in different environmental conditions brought about a hypothesis regarding the previously elusive ecological and evolutionary function of this cooperative phenomenon. Transition to Quorum Sensing in an Agrobacterium Population: A Stochastic Model, Andrew B. Goryachev1 etal
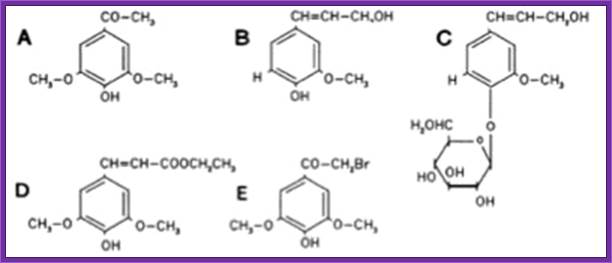
When tested the vir-inducing abilities of several different phenolic compounds using two wild-type strains of A. tumefaciens, KU12 and A6. Authors found that several compounds such as 4-hydroxyacetophone and p-coumaric acid induced the vir of KU12, but not A6. On the other hand, acetosyringone and several other phenolic compounds induced the vir of A6, but not KU12.
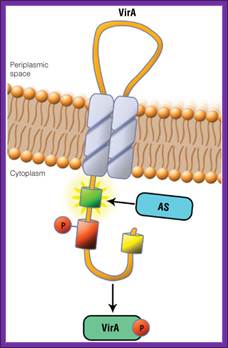
The receptor that responds to signal is VirA. It has autokinase activity and phosphorylates at serine. Then the activated VirA-P phosphorylates VirG found at the cytosolic side of the VirA. Both genes on Ti plasmid are constitutively expressed. The activated G is a transcriptional factor (TF). It activates the rest of the Vir genes.
Schematic showing the VirA receptor location.
In A. thumefaciens, the VirA receptor is located in the inner membrane with a loop structure in the periplasmatic space. The ligand acetosyringone (AS) activates VirA. After autophosphorylation of VirA the phosphate is transferred to the response regulatorVirG.
http://www.promega.com/
Schematic showing the VirA receptor location:
In A. tumefaciens, the VirA receptor is located in the inner membrane with a loop structure in the periplasmatic space. The ligand acetosyringone (AS) activates VirA. After autophosphorylation of VirA the phosphate is transfered to the response regulator VirG.
The VirA receptor consists of 829 amino acids and is a transmembrane protein in the inner membrane of A. tumefaciens (Melchers LS, 1989). VirA spans the inner membrane, with two transmembrane domains, a large periplasmic region, and a large C-terminal cytoplasmic domain (Banta LM, 1994). VirA directly senses the phenolic compounds for vir activation (Lee YW et al., 1996). Therefore the linker domain is essential for induction by phenolic compounds (Chang CH and Winans SC., 1992). The linker region is located in the cytosolic site at position 280 to 414 (Lee YW et al., 1996). This region between the amino acids 283 and 304 was highly conserved in four different strains of Agrobacterium, and therefore likely to serve as the receptor region for the phenolic inducers which are common to all four strains (Turk SC et al., 1994).
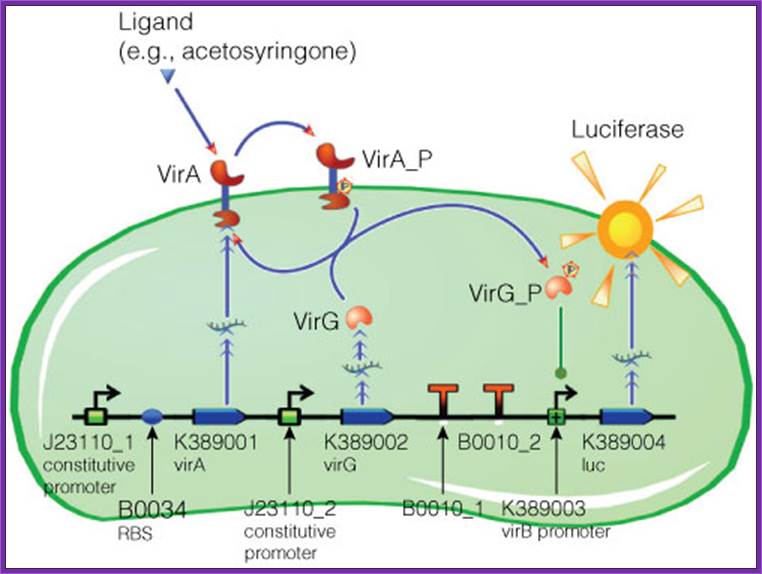
Figure . Model of acetosyringone inducible luciferase expression in E. coli.
Construct contains constitutive expression promotors for the two component receptor system (virA + virG), the two component system as well as the luciferase gene under control of the inducible virB promotor. Ligand binding to theVirA receptor initiates the internal signal transduction cascade through VirG phosphorylation. Activated transcription factor VirG binds to the vir Box containing Promotor virB. Luciferase expression occurs relative to the initial VirA ligand concentration. Gene numbers describing the database reference number of BioBrick database.. http://www.promega.com/
VirA is a transmembrane dimeric sensor protein that detects signal molecules, released from wounded plants (Pan et al., 1993). The signals for VirA activation include acidic pH, phenolic compounds, such as acetosyringone (Winans et al., 1992), and certain class of monosaccharides which acts synergistically with phenolic compounds.
Activated VirA has the capacity to transfer its phosphate to a conserved aspartate residue of the cytoplasmic DNA binding protein VirG (Jin et al., 1990; Pan et al., 1993). VirG functions as transcriptional factor regulating the expression of vir genes when it is phosphorilated by VirA (Jin et al., 1990a, 1990b). The C-terminal region is responsible for the DNA binding activity, while the N-terminal is the phosphorilation domain and shows homology with the VirA receiver (sensor) domain
VirA protein can be structurally defined into three domains: the periplasmic or input domain and two transmembrane domains (TM1 and TM2). The TM1 and TM2 domains act as a transmitter (signaling), TM2 has kinase domain p-lates His742 (autop-lation).
Construct contains constitutive expression promotors for the two component receptor system (virA + virG), the two component system as well as the luciferase gene under control of the inducible virB promotor. Ligand binding to the VirA receptor initiates the internal signal transduction cascade through VirG phosphorylation. Activated transcription factor VirG binds to the vir Box containing Promotor virB. Luciferase expression occurs relative to the initial VirA ligand concentration. Gene numbers describing the database reference number of BioBrick database.
Virulence genes (ViR):
Vir Genes and their Function |
|
Vir Gene |
Function |
|
Vir A, |
A receptor with two component system reacts from wounded plant cells and phenolic compounds such as acetosyringone, syringealdehyde or acetovanillone which leak out of damaged plant tissues. VirA gets activated with the bindng ofd signals and autophosphorylates and inturn phosphorylates VirG
|
|
VirG |
VirG is Transcriptial factor, p-lated VirG induce expression of other virulence genes. |
|
VirD2 |
Endonuclease; cuts T-DNA at right border to initiate T-strand synthesis and also it has an important role in integration of T DNA into host chromosomal DNA |
|
Vir D1 |
Topiosomerase;
Helps Vir D2 to recognise and cleave within the 25bp |
|
Vir D2 |
An endonuclease
nicks the 5’end of the T-DNA at the RB and covalently attaches to the 5’ end
of the T-strand, thus forming the |
|
VirD4 |
VirD4 is the ATP-dependent linkage of protein complex necessary for T-DNA translocation
|
|
Vir C1 |
Binds
to the 'overdrive' region to promote high efficiency T-strand
|
|
Vir E2 |
Binds to T-strand
protecting it from nuclease attack, and intercalates |
|
Vir E1 |
Acts as a chaperone which stabilizes Vir E2 in the Agrobacteriu. It is essential for the xport of E2 into the plant cell and into the nucleus |
|
Vir B & Vir D4 |
Assemble into a
secretion system which spans the inner and outer bacterial |
|
VirBs |
Proposed- VirB7, VirB8, VirB9, and VirB10 are the primary constituents of the T-DNA transport pore. VirB7, a lipoprotein, is anchored to the outer membrane (13), while VirB8 and VirB10 are inner membrane proteins. , and VirB10 participate in the formation of oligomeric complexes. These studies support the proposed role of the VirB7 to VirB10 proteins in transporter assembly. VirB4 and VirB11 are homo- and heterodimmers (Dang and Christie, 1997). The VirB7-VirB9 heterodimmer is assumed to stabilises other Vir proteins d uring assembly of functional transmembrane channel
|
|
Host interacting factors |
AtKAPα, CypA, PP2C, RocA, Roc4 ASK (Skp1-like), VIP1
|
CHV Genes(Chromosomal Virulence): They are located on bacterial chromosomes. There are several loci and their functions are varied. The loci chvA and chvB, involved in the synthesis and excretion of the b -1,2 glucan (Cangelosi et al., 1989); the chvE required for the sugar enhancement of vir genes induction and bacterial chemotaxis (Ankenbauer et al., 1990, Cangelosi et al., 1990, 1991); the cel locus, responsible for the synthesis of cellulose fibrils (Matthysse 1983); the pscA (exoC) locus, playing its role in the synthesis of both cyclic glucan and acid succinoglycan (Cangelosi et at., 1987, 1991); and the att locus, which is involved in the cell surface proteins (Matthysse, 1987).
Role of Chv genes :
|
Gene
|
Function |
|
Chv-A |
Codes for inner membrane protein essential for the transport of b-1, 2 glucan from bacterial cytoplasm into periplasm |
|
Chv-B |
An inner membrane protein likely to be involved in the synthesis of b-1, 2 glucan |
|
Chv-D |
Required for expression of Vir genes |
|
Chv-E |
Required for the expression of Vir genes |
|
Exo-locus
Exo-C |
Exo-biosynthesis of attachment polysaccharides Exo-C is involved required for the synthesis of b-1, 2 glucan |
|
Cel |
Cellulose fibril synthesis to enable bacterial cells to be firmly adhere to plant cell walls |
|
Psc-I |
|
Agrobacterium Ti plasmids are used for reconstructing the plasmid for dual purposes. One for using T DNA to transfer the required a foreign gene into plant cells. Another, as Ti plasmid contains Vir genes, Ori and a marker gene, can be used as helper plasmid.
The T DNA containing part of the Ti plasmid has been cloned into suitable plasmids found in E.coli. These plasmids are reconstructed in such way they contain TDNA borders with over drive at the RB. They have required Ori for replication in E.coli and also in Agrobacterium. They have antibiotic markers one for T DNA and another for E.coli. Auxin and Cytokinin synthesizing gens have been removed. In their place a strong antibiotic gene is placed at the left border of the T DNA under strong opine promoters. Another construct contains a strong and universal promoter such as 35s CAMV with few cloning sites. This plasmid also contains its own Ori and marker gene. Such vectors are called Binary vectors; originally they were used as Bac vectors.
- If one wants the plasmid to replicate in plant cells as episomal DNA, then one requires an origin similar to that of free replicating plant viruses or plasmids.
- Require a selection marker gene.
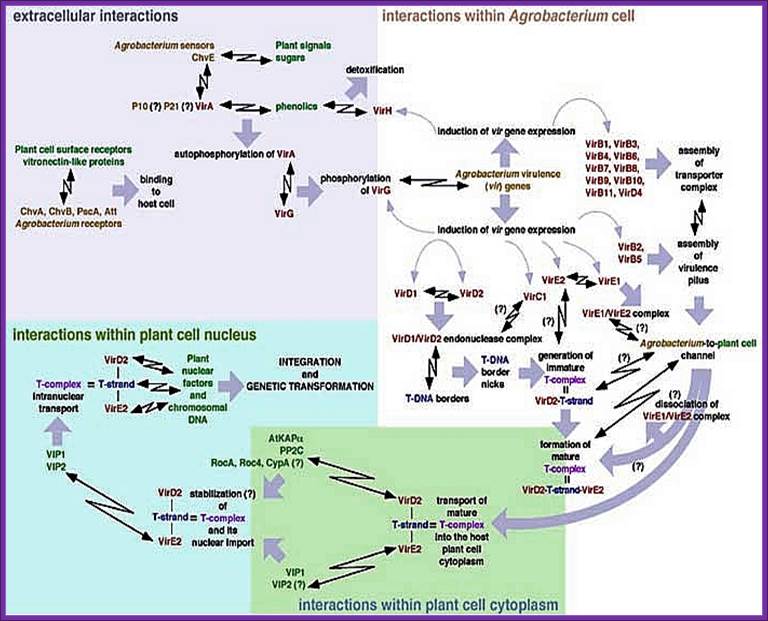
Bacterial virulence system:
The T-DNA transfer is mediated by products encoded by the 30-40 kb vir region of the Ti plasmid. This region is composed by at least six essential operons (vir A, vir B, vir C, vir D, vir E, virG ) and two non-essential (virF, virH). The number of genes per operon differs, virA, virG and virF have only one gene; virE, virC, virH have two genes, while virD and virB have four and eleven genes respectively.. VirA and Vir G are constutively expressed. Others are expressed by the activity of VirG.
Vir Gene Promoters: In the -10 region, the vir promoters share a consensus sequence that is homologous to a DNA sequence found in the same region of E. coli promoters i.e. TATAATA. In contrast, the -35 region sequences are variable. Several vir genes contain two common hexanucleotide sequences, 5'CGAGTA3' and 5'GCAATT3'. Translation initiation codons for all vir genes, except virG, are preceded by sequences homologous to the ribosome binding site sequences called Shine Delgarno (SD) found in E. coli.
ViR Genes:
It is a cluster of several genes and each of which have their own hexa nucleotide boxes in the promoters. Each of the operons is inducible. They are involved in activation and transfer of the T-DNA from the bacterial cell into plant cell. The ViR region consists of seven operons called ViR-A (2kbp, 1 gene), ViR-B (9.5kbp, 11 genes), ViR-G (1kbp, 1 gene), ViR-C (2kbp, 2 genes), ViR-D (4.5kbp, 4genes), ViR-E (2kbp, 2genes) and ViR-F (? 1 gene).
Vitaly H. Citovsky; http://www.mindfully.org/
Organization of Vir-genes from left to right:
-//--Vir-Hà-ViR-Aà--ViR-B->-ViR-G->-ßViR-C---ViR-D-à-Eà-F->//
ViR-A: It is constitutively expressed at low level. The protein is localized on the plasma membrane of bacterial cell and it acts as a receptor. It has an autokinase activity and phosphorylates ViR-G. With the activation of ViR-G receptors are produced in large numbers. It acts in conjunction with Chv-E (a sugar binding protein).
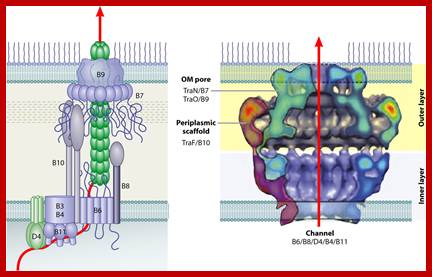
ViR-B: It consists of 11 genes called ViR-B1 to ViR-11. Their location is plasma membrane. They perhaps organize into channel like structure and involved in the transport of T-DNA in single strand form. The B-11 has ATPase activity.
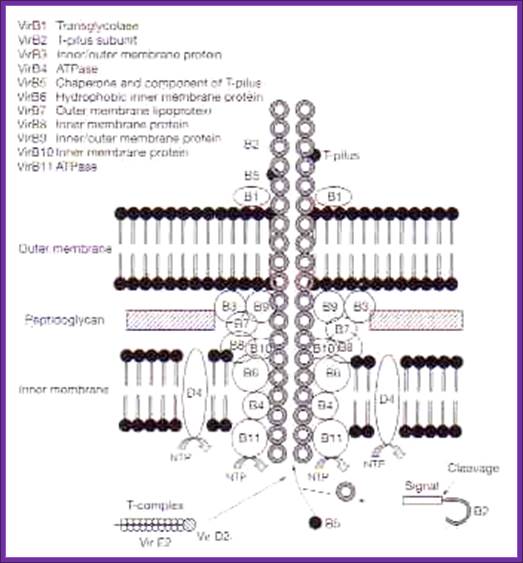
VirB protein complex (12 proteins) act as transporter of T DNA from bacteria to plant cells; This process is deemed as T4ss type
ViR-G: Its location is cytosol. Before activation the gene products are at low level, but once they are activated, they in turn activate its own gene and also activate all other ViR genes. In super virulent strains this product is over produced. When this gets phosphorylated by activated ViR-A it becomes a dimmer and now it acts as a DNA binding protein. Thus all ViR gene operons can be considered as a regulon for they all get activated by ViR-G
ViR-C: This consists of two genes called viR-C1 and viR-C2. Location of these proteins is cytosol and they bind to over drive region at the right end of the right border of T-DNA, acts as a helicase and helps in unwinding the dsDNA. It also helps in activating cleavage of DNA at the right border. They help ViR-D products in this process.
ViR-D: It consists of viR-D1, -D2, -D-3, and D-4. ViR D1 is considered as Topoisomerase and it binds to the right border of the T-DNA. The vir-D2 is considered as an endonuclease. They recognize the RB sequence ACC and cuts at that sequence to generate – ACC 5’
The 5’ of the nicked T-DNA gets covalently bound to D2. Binding to the 5’ end it prevents 5’ exonuclease activity. Perhaps this protein is also involved in recombination events in the host cells.
ViR-E: It consists of two genes called E1 and E-2. They act as single strand binding protein (ssBPs), which in turn bind to the peeled of single strand of T-DNA region from right border to the left border. This protects from the exonuclease or endonuclease digestive activity. They may also help in transporting the DNA across the channel produced by ViR-B products. This protein is also believed to be involved in recombination in the host.
Vir-F: It consists of few sub groups of F genes. The ViR-F genes help in DNA transfer. It is believed that some ViR-F-pin genes activate several host cell genes.
ViR-H: Not much is known about this gene. Two accessory vir operons are vir F & vir H., vir H operon consist of two genes that code for Vir H1 & Vir H2 protein. These Vir protein are not essential but could enhance the transfer efficiency, detoxifying contain plant compounds that can effect bacterial growth.
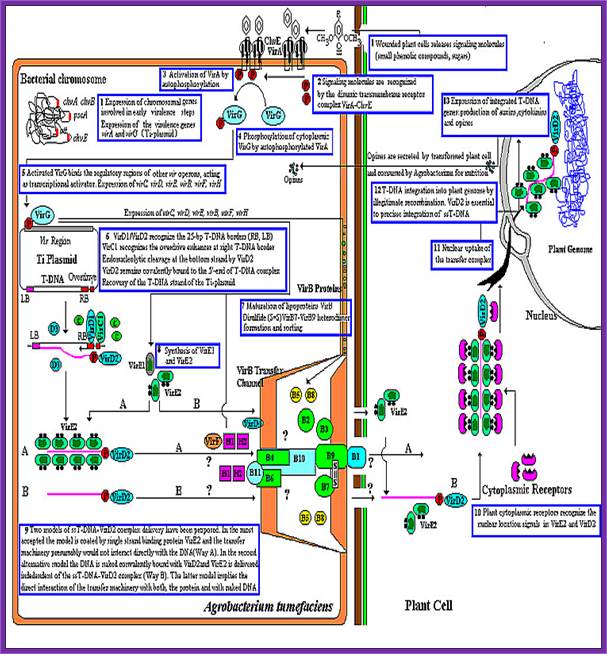
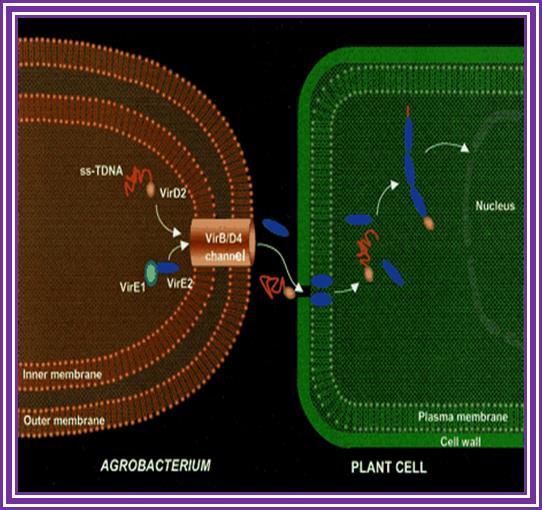
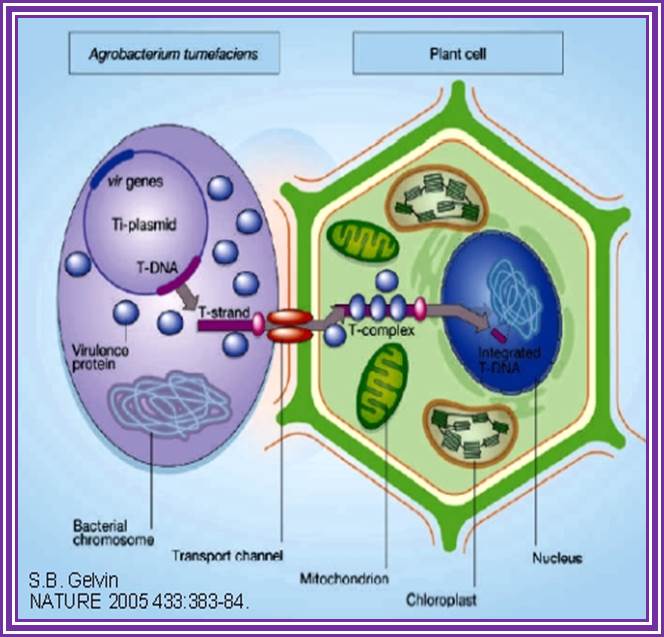
This is an overall view of the components involved in transferring the DNA from the bacteria into plants. One has to read carefully to understand the process. Basic steps in transformation of plant cells by Agrobacterium tumefaciens. The T-DNA transfer is represented according to updated knowledge on this process, although many of involved mechanisms have not been elucidated yet and the experimental results only allow hypothesize about it. Entering of T-DNA complex into the plant cell is almost completely uncharacterized and experimentally only the VirB7-VirB9 disulfide bound heterodimers have been evidenced. The most important events are briefly mentioned in chronological order (boxes 1 through 13). Each step is described in the text http://www.ejbiotechnology.info

Plant transformation: A pilus in Agrobacterium T-DNA transfer; Christian Baron, Patricia C ZambryskI; Agrobacterium tumefaciens transfers a protein–DNA complex to plant cells in a process similar to bacterial conjugation; the mechanism of transfer is beginning to be unravelled by biochemical, genetic and electron microscopic studies.
Thirteen steps or more involved in transferring the T DNA from bacterium to plant cells to Effect:
1. Wounds in plants release signal compounds-phenolic and sugars, When plants are wounded they secrete a variety of substances in response to injuries as defensive mechanism. Some of the compounds like Aceto Syringone and Hydroxyl Aceto Syringone if secreted and in a place where the bacteria happens to be present, Syringones bind to ViR-A receptors found on plasma membrane.

Acetosyringone; https://en.wikipedia.org
- Tight binding between the bacterial and host cells is very important for cell-to-cell communication. Host cell signals may precede the attachment, because the signals binding to bacterial receptors may activate certain bacterial genes located in the bacterial chromosome and their activation may facilitate or actuate the process of binding.
- Actively dividing cells at surface of the wound is an important contributory factor for the ensuring the binding of the bacterial cell to the host.
- However incompatibility may be due variety of host responses such as- hypersensivity of the host cells to bacteria, host cells inadequacy in forming callus at the wounded surface, inhibition of the gene activity of the T-DNA may be due to methylation, or and not producing signaling molecules as inducers. Not all plant types are compatible for the bacterial infection. Some families of plants such as Solanaceae are high susceptible than others, which means the hosts also contribute some chemicals for the bacteria to bind to their surface area.
Bacterial chromosomal genes such as Chv are also expressed. Signaling molecules bind to dimeric transmembrane VirA and ChvE receptors.
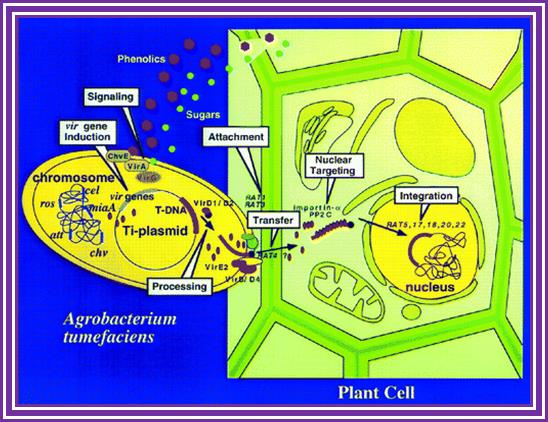
The phytopathogenic bacterium Agrobacterium tumefaciens genetically transforms plants by transferring a portion of the resident Ti-plasmid, the T-DNA, to the plant. Accompanying the T-DNA into the plant cell is a number of virulence (Vir) proteins. These proteins may aid in T-DNA transfer, nuclear targeting, and integration into the plant genome. Other virulence proteins on the bacterial surface form a pilus through which the T-DNA and the transferred proteins may translocate. Although the roles of these virulence proteins within the bacterium are relatively well understood, less is known about their roles in the plant cell. In addition, the role of plant-encoded proteins in the transformation process is virtually unknown. In this article, I review what is currently known about the functions of virulence and plant proteins in several aspects of the Agrobacterium transformation process; Gelvin, S.Annu, Annual Rev.Plant Physiology.
VirA activated by auto P-lation, VirA phosphorylates VirG.
VirG-p, a transactivator activates several vir Genes-Vir C,D,E,B,D,F and H,
VirD1 and D2 recognize RB (25bp), VirC1 recognizes over drive sequences at RB. VirD2 cuts at RB and binds to 5’end of the T-ssDNA strand.
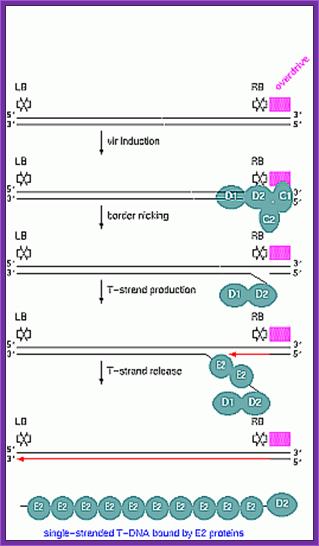
Once the Ti plasmid genes are activated, the VrC1 binds to the RB over drive sequence; this leads to the assembly of VirD2 and endo nuclease assemble on to the 25bp long RB and cuts DNA at 5’ end releases only one strand and the VirD2 covalently bind to the 5’ of ss DNA strand.
VirB forms disulfide bonds, VirB7-9 form heterodimers organize into DNA transfer channels? Vir B channel consists VirB1, B7, B9, B10, B4 and B6 from the outer membrane towards the inner membrane.

Models of type IV secretion (T4S) system-mediated substrate translocation. Agrobacterium tumefaciens.
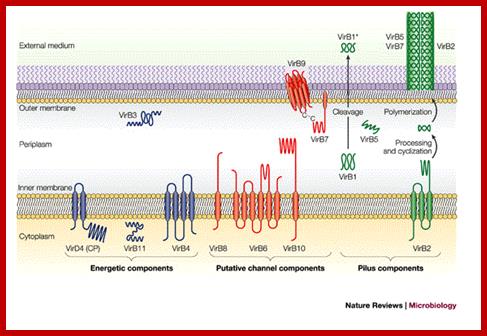
The versatile bacterial type IV secretion systems;Vir B proteins associate to form a channel like structure. The coupling protein (CP) VirD4 and the mating-pore-formation components (VirB1–VirB11) are represented according to their proposed functions: energetic (blue), channel (red) or pilus (green) components. Several proteins are post-translationally modified in the periplasm. Signal sequences of VirB1, VirB2, VirB5, VirB7 and VirB9 are cleaved by signal peptidases. VirB1 is processed to form VirB1*, which is exported across the outer membrane. VirB2 undergoes a novel head-to-tail cyclization reaction, and polymerizes as the T-pilus. VirB7 is modified as a lipoprotein that associates with the T-pilus and also forms an intermolecular disulphide crosslink with VirB9, a possible SECRETIN113. The VirB and VirD4 proteins are postulated to assemble as a supramolecular structure composed of a transenvelope channel and an extracellular pilus. Eric Cascales & Peter J. Christie; http://www.nature.com
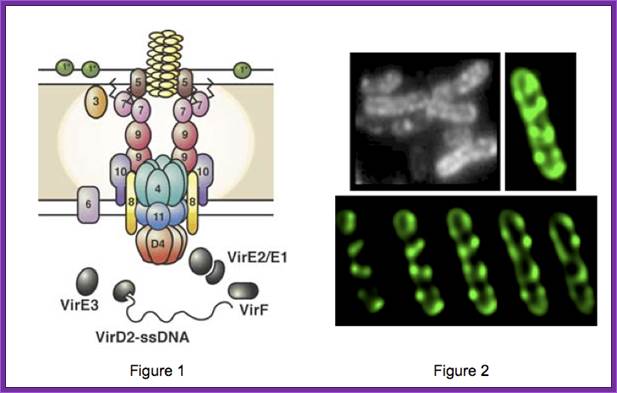
Patricia C. Zambryski http://pmb.berkeley.edu/
T4SS system; The above mentioned channels are a paradigm for type IV secretion systems (T4SS).
A model of how the 12 proteins (VirB1-B11, VirD4) essential for T4SS form a membrane-spanning channel is presented in Figure 1. In addition, 4 proteins, VirD2, VirE2, VirE3, and VirF are exported by the T4SS to plant cells.
This localization pattern was determined by deconvolution fluorescence microscopy of GFP fusions to T4SS components and substrates. Figure shows several optical sections through the bacterial cell. This localization pattern was confirmed by immuno-fluorescence microscopy of native T4SS proteins. This localization pattern likely facilitates binding of Agrobacterium to susceptible plant cells. Indeed, if one tags the T4SS with GFP, one can see Agrobacterium binding laterally along their lengths to single plant cells.
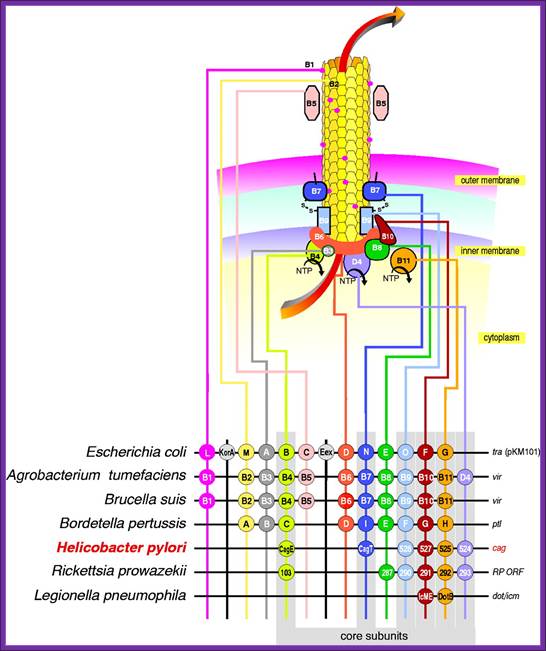
Type IV secretion system (T4SS or TFSS)
It is homologous to conjugation machinery of bacteria (and archaeal flagella). It is capable of transporting both DNA and proteins. It was discovered in Agrobacterium tumefaciens, which uses this system to introduce the T-DNA portion of the Ti plasmid into the plant host, which in turn causes the affected area to develop into a crown gall (tumor). Helicobacter pylori uses a type IV secretion system to deliver CagA into gastric epithelial cells. Bordetella pertussis, the causative agent of whooping cough, secretes the pertussis toxin partly through the type IV system. Legionella pneumophila, the causing agent of legionellosis (Legionnaires' disease) utilizes type IV secretion system, known as the icm/dot (intracellular multiplication / defect in organelle trafficking genes) system, to translocate numerous effector proteins into its eukaryotic host.[4] The prototypic Type IV secretion system is the VirB complex of Agrobacterium tumefaciens.[
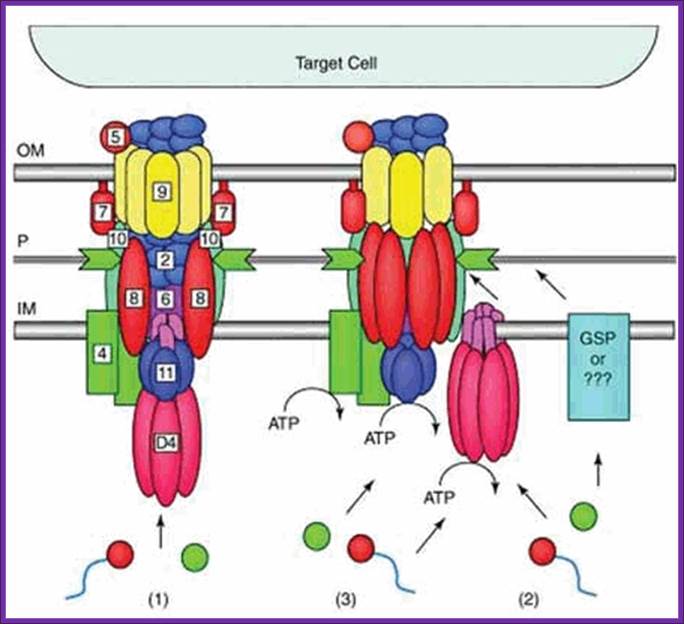
Figure. Possible architectures and substrate transloction routes for the T4SS. Numbers refer to VirB/VirD proteins of the Agrobacterium T-DNA transport system. Three working models describe the possible machine architectures and translocation routes: (1) a one-step model using a transenvelope channel, (2) a two-step model using the T4CP or alternative translocase for substrate transfer across the inner membrane (IM) and the mpf complex for outer membrane (OM) translocation, and (3) another two-step model, the “shoot and pump model”, whereby theT4CP recruits substrates and transports DNA across the IM and delivers protein substrates to the mpf protein export machinery. Blue line,T-strand; red circle, relaxase bound to theT-strand; green circle, protein substrate; P, periplasm. Reprinted with permission from: Ding Z, Atmakuri K, Christie PJ. Trends Microbiol 2003; 11:527-535.©2003 Elsevier.
An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells
Hypothetical model for T-DNA transfer from the bacteria into the plant cell. In the bacterial cell, the VirE1 chaperone prevents binding of VirE2 to ssDNA, as has been suggested (10–12), and possibly prevents VirE2-dependent channel formation in the bacterial membrane. VirE2 is transported through the VirB-VirD4 channel and subsequently inserts into the plant plasma membrane, allowing the transport of the ssDNA-VirD2 complex. The way in which the VirE2 molecules enter the cytoplasm is unclear. However, once in the cytoplasm, VirE2 protein molecules coat the complex, permitting its transfer to the nucleus (1–3). For simplicity, the pilus and its involvement in T-DNA transfer are omitted from the scheme.
Transporter complexes- as depicted and explained above transport ssT DNA (E2?) complex with VirD2 head on and few others into plant cell.
a.VirE2 bound DNA translocate through the B complex, DNA VirE2 complex and Vid2 heading enters the nucleus. When the ssTDNA enters it is coated with VirE2 and cytosolic receptor proteins.
b.With VirD2 bound to 5’end and VirE2 bound to ssT DNA move through the speculative channel. Entry into the nucleus is facilitated by the NLS sequences found in VirD2 and VirE2. Furthermore, the host VirE2-Interacting Protein 1 (VIP1) directly binds to VirE2 and facilitates the nuclear import of T-DNA as well as its subsequent targeting to the host genome (Tzfira et al., 2001; Li et al., 2005; Lacroix et al., 2008).
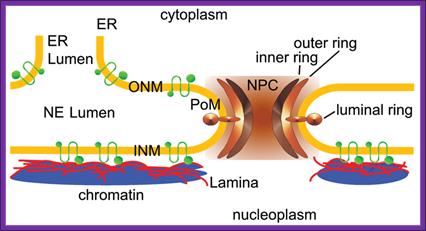
Is there any role for plasmodesmota in the transport of T DNA across the cells? So far this question has not been answered.
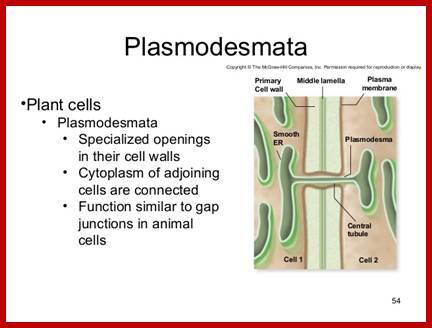
https://www.slideshare.net

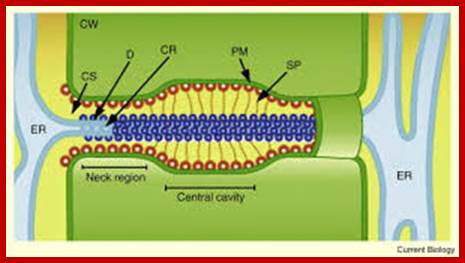
http://www.sciencedirect.com
The structural features and the functional features of plasmodesmota indicate that there is a role for plasmodesmota in locking the Agrobacterium transporter complex with that of Plasmodesmota and transport the TDNA into the cell?
- Another contributory factor perhaps is plasmodesmota sites. It is known that several viral genomes are transported why even the entire viruses are transported across the cells via plasmodesmota channels aided and augmented by “Transport “proteins, which also possess ATPase activity.
Biological Diversity of Prokaryotic Type IV Secretion Systems; http://mmbr.asm.org/
This is another TDNA transport complex showing different subunits organized at the base of inner membrane extending to the outer membrane and beyond. Model for assembly of the Agrobacterium T4SS. Protein interactions determined in this work are depicted in A and B. Previously determined interactions are shown in C. The double lines at the top and bottom of each panel represent the inner and outer membranes, and the shaded region represents the periplasmic space and peptidoglycan. (A) VirB8 functions as a “founding member” of the T4SS and recruits VirB1 to the site of assembly. VirB1 locally remodels the peptidoglycan (represented by decreased shading of the periplasm). (B) VirB1 activity allows recruitment of other T4SS components such as VirB4, VirB7, VirB9, VirB10, and VirB11 by clearing the peptidoglycan or by recruiting components via direct interactions. VirB7–VirB9 heterodimers are critical for stability of T4SS proteins. (C) As the assembly matures, the remaining components are recruited, including VirB3, the pilus (VirB2, VirB5), and inner membrane components (VirB6, VirD4). VirB1* may function extracellularly, and its loose association with the surface is indicated. VirB7 and VirB5 contribute to pilus assembly. VirB4, VirB6, VirB11, and VirD4 all have been postulated to create channels for substrate secretion (blue arrows). (D) A model for assembled T4SS based on interactions described in A–C. VirB7–11 and VirB4 form a functional core of the T4SS that spans both bacterial membranes. Known substrates for export include VirE2 and, perhaps, its specific chaperone VirE1, VirF, and the T-complex (VirD2–ssDNA). VirD4 is likely the recognition protein coupling VirD2 export to the T4SS and may function as a DNA-helicase to liberate the T-strand from the Ti plasmid. VirB11 forms a pore and may facilitate substrate export.
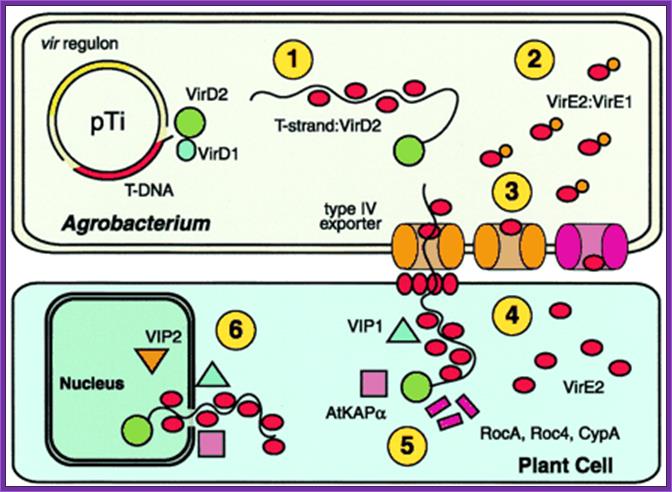
The six functions of Agrobacterium VirE2 :
The six functions of VirE2. The vir regulon, encoding the major loci virA-E and virG-H, is expressed on detection of plant wound signals. VirD2 and VirD1 liberate the T-strand and VirD2 remains covalently bound to the 5′ end. 1: VirE2 coats the T-strand, protects it from degradation, and maintains it in a transportable conformation. 2: VirE2 associates with VirE1, required for VirE2 export. 3: VirE2 exits Agrobacterium via the type IV exporter independently, or as part of the T-complex. Alternatively, VirE2 may exit by an alternate pathway (pink; ref. 4). 4: VirE2 forms a pore in the plant plasma membrane allowing passage of the T-complex and coats the T-strand in the plant cytoplasm. 5: VirD2 and VirE2 interact with plant cytoplasmic chaperones (RocA, Roc4, and CypA). Other factors (AtKAPα, VIP1) may target the T-complex to the nucleus. 6: VirE2 interacts with nuclear factors (VIP2) that mediate interaction with chromatin and facilitate integration of the T strand. http://www.pnas.org
- Once the DNA is in the cytoplasm, it will find its way into the nucleus. Before the DNA enters into the nucleus whether the ssDNA gets circularized or it is converted into double stranded and circularized and then enters into the DNA is not clear.
- Some of the retro viral RNA genomes generate dsDNA, which contain LTR sequence at the ends and then they are circularized by integrase before they enter into the nucleus.
- Even in the case of T-DNA integration of circularized DNA is preferable than the linear DNA. In the nucleus it gets integrated into host genomic DNA, perhaps using T-DNA s border sequences for the some of the border sequences are invariably found in the host DNA.
- Using the left and right border sequences of the T-DNA, it is now known that the DNA gets circularizes before it enters, as the left and right border sequences are well preserved within the host DNA. Especially the right border sequence i.e ---ACC5’ and the left border sequences are found. Right border ACC5’ is very well conserved, but not all the left border sequences conserved.
- The site of Integration of the T-DNA into host cellular DNA is random and there is no sequence bias and again depends upon cells mitotic activity. Yet the site of integration is more or less AT rich.
- Replicating DNA is more amenable for the T-DNA integration. Mitosis provides nuclear membrane barrier free access to host chromosomal DNA.
- Integration of T-DNA at multiple sites is very common and the integrated DNA is very stable, and it is propagated along with its host DNA.
T DNA integration:
TDNA integrates into host DNA, which is greatly facilitated if the cell is in the mitotic stage. VirD2 plays an important role in recombination.
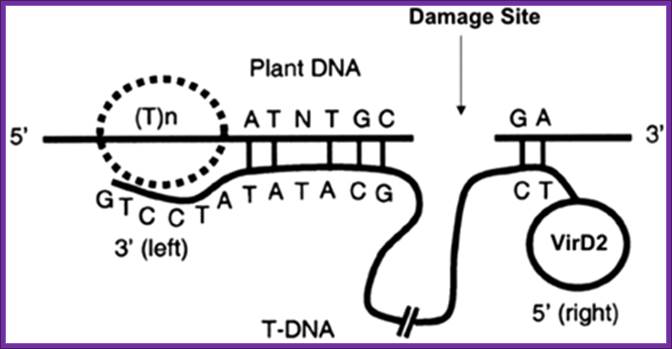
Figure. The current model of T-DNA
integration. The T-rich region [(T)n] and microsimilarities that were
identified as being important for the integration mechanism are depicted.
Modified from Brunaud et al. (2002). http://www.plantphysiol.org/
Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus; B Tinland, B Hohn, and H Puchta.
After transfer from Agrobacterium to plant cells, T-DNA molecules recombined much more efficiently if the homologous sequences were of opposite polarity than if they were of the same polarity.
Single-stranded DNA molecules of opposite polarity can anneal directly, whereas single-stranded DNA molecules of the same polarity first have to become double stranded to anneal. Judging from the relative amounts of single- to double-stranded T-DNA derivatives undergoing recombination, we infer that the T-DNA derivatives enter the plant nucleus in their single-stranded form.
The best model to date compart’s T-DNA integration to illegitimate recombination (Gheysen et al., 1991). It is suggested that the VirD2-bound 5’ end of the T-strand joins a nick in plant DNA. The plant TINA may further unwind to form a gap, and the 3‘ ertd of the T-strand may pair with another region of plant DNA close by. Plant repair and recombination enzymes inay then function to covalently join the 3’ end to plant DNA.
Illegitimate recombination in plants:
A model for T-DNA integration-http://genesdev.cshlp.org/subscriptions Godelieve Gheysen^
Short homologies between the T-DNA ends and the target sites, as well as the presence of filler sequences at the junctions, indicate that T-DNA integration is mediated by illegitimate recombination and that these processes in plants are very analogous to events in mammalian cells. We propose a model for T-DNA integration on the basis of limited base-pairing for initial synapsis, followed by DNA repair at the junctions. Variations of the model can explain the formation of filler DNA at the junctions by polymerase slipping and Green light for gene targeting in plants.
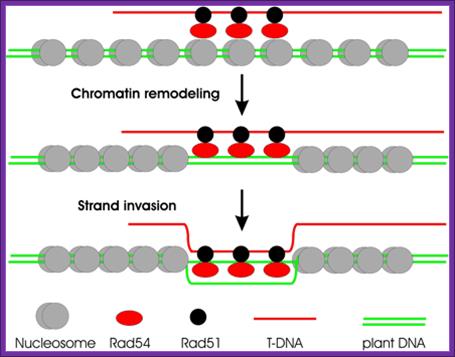
Hypothetical model for Rad54-aided chromatin remodeling and GT. Shown is the chromatin remodeling activity of Rad54 stimulated by the presence of a filament consisting of single-stranded DNA and the single-stranded DNA-binding protein RAD51. In vitro data suggest the following hypothetical scenario for GT in Arabidopsis: T-DNA, coated by the plant ortholog RAD51, recruits yeast RAD54, which, by its capacity to interact with nucleosomes, promotes remodeling of chromatin until homology is encountered. This homology would lead to strand invasion and subsequent formation and resolution of Holliday junctions. It is not known, however, whether the yeast protein would interact with the endogenous RAD51 protein, and species specificity in this interaction has indeed been demonstrated. The activity of the yeast Rad54 protein in dislocating Rad51 from double-stranded DNA may be essential for the later stages of the recombination reaction. (Image courtesy of T. Hohn, Botanical Institute, Basel.
T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites ; Véronique Brunaud1, etal 2002
Abstract
A statistical analysis of 9000 flanking sequence tags
characterizing transferred DNA (T-DNA) transformants in Arabidopsis
sheds new light on T-DNA insertion by illegitimate recombination. T-DNA
integration is favoured in plant DNA regions with an A-T-rich content. The
formation of a short DNA duplex between the host DNA and the left end of the
T-DNA sets the frame for the recombination. The sequence immediately downstream
of the plant A-T-rich region is the master element for setting up the DNA
duplex, and deletions into the left end of the integrated T-DNA depend on the
location of a complementary sequence on the T-DNA. Recombination at the right
end of the T-DNA with the host DNA involves another DNA duplex, 2–3 base pairs
long, that preferentially include a G close to the right end of the T-DNA.
Involvement of F-box proteins in Agrobacterium infection:
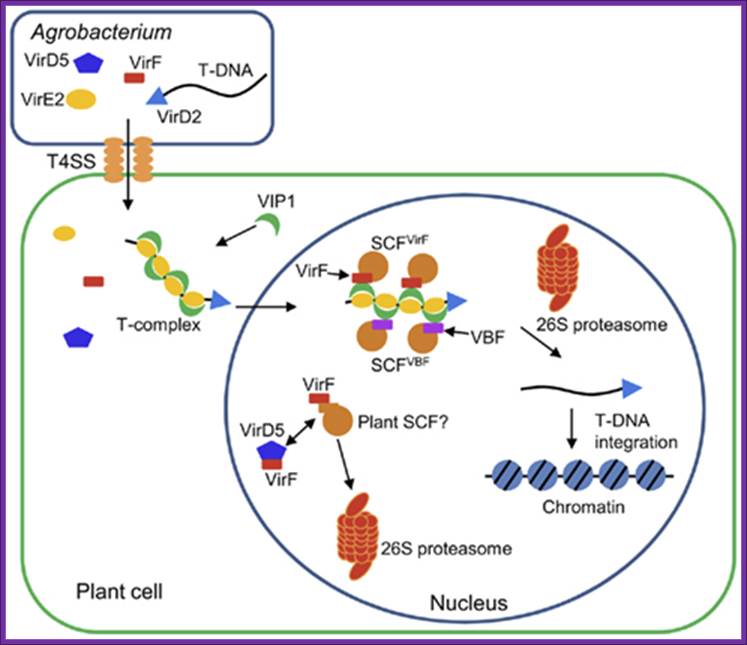
Figure .Hijacking of the host SCF ubiquitin ligase machinery by plant pathogens
Agrobacterium exports a single-stranded copy of T-DNA (T-strand) as well as virulence (Vir) effector proteins into plant cell. Within plant cell, T-strand is assembled into a nucleoprotein complex (T-complex) in which one VirD2 molecule is attached to the 5′ end of the T-strand and multiple VirE2 molecules coat the entire length of the T-strand. In addition, the plant factor VIP1 directly interacts with VirE2 and guides the T-complex into the host cell nucleus. Once the T-complex enters the nucleus, it is presumably disassembled by an SCF complex containing VirF as an F-box component. The SCFVirF complex mediates polyubiquitination of VIP1, thereby targeting VIP1 as well as its associated VirE2 for 26S proteasome-dependent degradation. In plant species that do not require VirF for full virulence, Agrobacterium most likely also utilizes the host F-box protein VBF for the T-complex uncoating. As a defense strategy, the host plants destabilize VirF via the ubiquitin/26S proteasome system (UPS), presumably using an as yet unidentified plant SCF complex. Another exported effector, VirD5, counteracts this host-induced degradation of VirF by directly binding to and stabilizing VirF. Shimpei Magori* and Vitaly Citovsky
http://journal.frontiersin.org/
Most of the said genes are silent when the bacteria are in Free State. But once the said oncogenes are transferred into the host cell and when they get integrated into host cellular genome; the said i.e. Auxin and Cytokinin genes are activated by host cellular factors. The other genes such as OCS, NOS or Agropine gene products are secreted out of the host cells. The Agrobacterium feed on these opines and it is an important nitrogen and carbohydrate source for the bacteria.
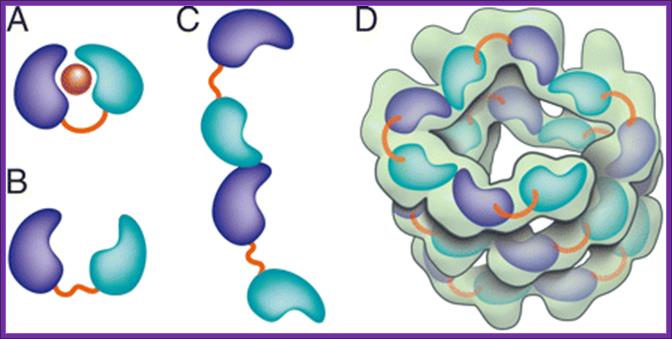
Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners
Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners.
Schematic representation of VirE2 showing how its interdomain flexible linker permits structural rearrangements in complex with its different partners. The two domains of VirE2 are shown in purple and cyan linked by their interdomain flexible linker shown in orange. (A) In the presence of VirE1 (red), the two VirE2 domains are locked by their interaction with VirE1. (B) In the absence of VirE1, the domains of VirE2 are unlocked and free to rotate around the flexible interdomain linker. (C) In the unlocked form, VirE2 has a strong tendency to self-assemble forming N- to C-terminal interactions. Because of the flexible linker, the two domains of VirE2 can adopt a range of orientations resulting in irregular filaments. (D) On addition of ssDNA to the filaments, an ordered solenoid assembly is formed (gray envelope: a tracing of the EM model with 4.25 VirE2 units per turn (26). The ssDNA should wrap along the inner wall of the protein structure, limiting the degree of freedom in the linker and thereby imposing a favorable VirE2–VirE2 arrangement. http://www.pnas.org/
Design of Ti based vectors for plant transformation.
Agrobacterium T-DNA constructs and components for replication in E.coli :
To introduce any foreign gene into plants one has to construct a plasmid, which has the ability to transfer the DNA and ability to integrate into host genome. Generally such vectors are binary vectors. They contain the following features:
Transgenic crops: how genetics is providing new ways to envision Agriculture; http://www.scq.ubc.ca/
- Ti plasmid containing RB and LB and an overdrive sequence at the RB.
- A suitable promoter with regulatory sequences such as response elements or enhancers.
- Contain cloning site and a transcription terminator signal.
- Contain the desired gene and a selection marker gene towards RB or LB.
- A selection marker gene like fluorescent protein gene can be introduced.
- For manipulation in both E.coli and Agrobacterium it should contain origin and a gene for antibiotic resistance.
- The desired genes can be protein coding genes or desired antisense RNA (RNAi) genes can be introduced to knock out specific host gene expression.
- Such constructs can be transferred into plant tissues through Agrobacterium or through gene gun.
- For transferring the gene through Agrobacterium one has to get a bacterial strain that is disabled, it means the entire T-DNA found in between the LB and RB is deleted and all other features are retained in the Ti-plasmid.
- Require a strong and inducible promoter with regulatory elements.
- Require a cloning site or sites.
- Require T-DNA left and right borders, it is with which selection marker gene and expression cassette is included.
- Require a reasonable small sized plasmid having all the above features.
- These plasmids act like shuttle vectors for they can function in E.coli and Agrobacterium.
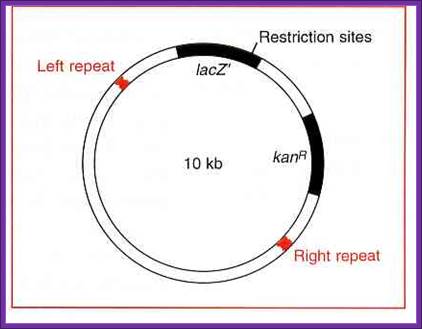
Ti Plasmid 10kb for inserting a recombinant DNA; www.whatisthebiotechnology.com
Few plant vectors used:
pBI 101.
pBI 103.
pBI 121.
pBIN.
pGA 472.
pGK 100.
pGPTV.
The strain should be Rec^- and RE minus. And it should be compatible for the host cells.
Co-integrated Vectors : T-DNA clone disarmed and made into a vector for Bacterial transformation with a selectable marker and a gene of interest.

Co-integrated vectors or hybrid Ti plasmids, these vectors were among the first types of modified and engineered Ti plasmids devised for Agrobacterium -mediated transformation, but are not widely used today. These vectors are constructed by homologous recombination of a bacterial plasmid with the T-DNA region of an endogenous Ti plasmid in Agrobacterium. Integration of the two plasmids requires a region of homology present in both.
A resulting co-integrated plasmid assembled by in vitro manipulation normally contains: They contain - Vir genes, the left and right T-DNA borders, an exogenous DNA sequence between the two T-DNA borders, and plant and bacterial selectable markers.

Helper plasmid; A helper Ti plasmid, harbored in A. tumefaciens, which lacks the entire T-DNA region but contains an intact vir region and Ti ori. Vir provides factors for DNA transfer.
Binary vectors:
They are extensively used in Agrobacterium mediated transfer of genes into plant cells.
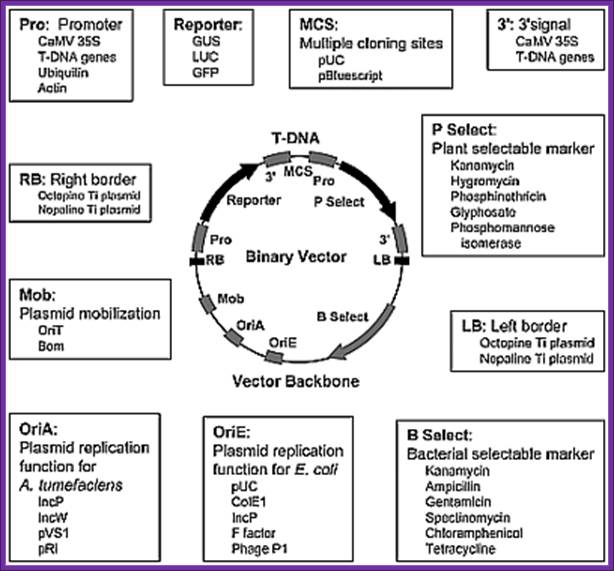
- Binary vectors based on the bacterial artificial chromosome (BAC). Two United States patents and a European application assigned to Cornell refer to this topic. The binary vector that contains the T-DNA region has
- origins of replication for E. coli and Agrobacterium, wherein the plasmids are maintained as a single copy, and
- a unique restriction site for insertion of an exogenous sequence located between a right and a left T-DNA borders
Although binary BAC vectors are devised for cloning long fragments of DNA (around 150 Kb), the designed vectors have a fairly broad scope except that the origins of replication are very specific, that is, they must maintain plasmids as a single copy.
The discovery that the vir genes do not need to be in the same plasmid with a T-DNA region to lead its transfer and insertion into the plant genome led to the construction of a system for plant transformation where the T-DNA region and the vir region are on separate plasmids.
In the binary vector system, the two different plasmids employed are:
- a wide-host-range small replicon, which has an origin of replication (ori) that permits the maintenance of the plasmid in a wide range of bacteria including E. coli and Agrobacterium. This plasmid typically contains:
- foreign DNA in place of T-DNA,
- the left and right T-DNA borders (or at least the right T-border),
- markers for selection and maintenance in both E. coli and A. tumefaciens,
- a selectable marker for plants.
· The plasmid is said to be "disarmed", since its tumor-inducing genes located in the T-DNA have been removed.
- a helper Ti plasmid, harbored in A. tumefaciens, which lacks the entire T-DNA region but contains an intact vir region.
In general, the transformation procedure is as follows:
- The
recombinant small replicon is transferred via bacterial conjugation or
direct transfer to
A. tumefaciens harboring a helper Ti plasmid, - The plant cells are co-cultivated with the Agrobacterium, to allow transfer of recombinant T-DNA into the plant genome, and
- Transformed plant cells are selected under appropriate conditions.
Advantages
Compared with co-integrated vectors, binary vectors present some advantages:
- No recombination process takes place between the molecules involved.
- Instead of a very large, recombinant, disarmed Ti plasmid, small vectors are used, which increases transfer efficiency from E. coli to Agrobacterium.
This vector system is most widely used nowadays. Different types of binary vectors have been devised to suit different needs in a plant transformation process.
Types of Binary vector:
1.a.pGA- series vectors, which contain:
- an ori derived from RK2 for replication in E. coli and Agrobacterium,
- a tetracycline resistance gene,
- the cis-acting factor required for conjugal transfer,
- the right (RB) and left (LB) T-DNA borders,
- a Neomycin phospho-transferase (nptII) gene, which confers resistance to kanamycin and G418 in transformed plants, and
- a polylinker site (multicloning site).
Specific vectors in this series are designed for cloning large fragments (colE1 origin of replication and phage l cos), analyzing promoters (multiple cloning site immediately upstream of a promoterless cat gene), and expressing a gene of interest (polylinker site between a plant promoter and a terminator).
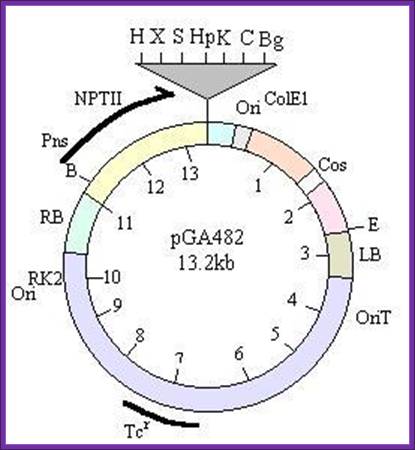
-//--RB-NptII—Mcs-Ori--cos—LB—oriT-Tet---//-
1.b.pGK100 vector: This is derived from pGA472 (Galvin’s vector). This vector was provided by Prof. Veluthambi from Madurai Kamaraj University, Dept. Plant Biotech, Madurai, and Chennai State.
This was modified by Dr.G.R.Kantharaj in USDA, Maryland, USA (1985-86), used for developing Tomato Transgenic plants for resistance to ToMV and ToLCV.
Plasmid size is same as pGA472,
The size of the plasmid is 15500,
It Consists of RB and LB with an overdrive sequence at RB.
It has Neomycin phospho transferase (npt) II gene with promoter towards LB,
It has an ORI and RE sites Pst1, Sca1 (cos cluster).
It has a cloning site Bglll towards RB, At this position CaMV (Cauliflower mosaic promoter is added in between HindIII and BglII.
-//--RB-—Mcs-Ori—cos- NptII — LB—oriT-Tet---//-
Other features are same as that of pGA472. The reason for changing this configuration is that the cloned X gene should enter the plant cell before the npt II gene; this ensures that the cloned gene has entered the cell before the marker gene. The cell resistant to the antibiotic has the cloned x gene.
- pCG series vectors, which contain:
- the origin of replication of the Agrobacterium rhizogenes root -inducing plasmid pRiHRI, which confers more stability in Agrobacterium than the ori derived from RK2, and a ColE1 origin of replication from the vector pBR322 for maintenance in E. coli.
- pCIT series which contain:
- the hygromycin (hph) resistance gene for plants,
- the lambda cos site for cloning long fragments.
- pGPTV (glucuronidase plant transformation vector) series , which have:
- different plant selectable marker genes close to the left T-DNA border. This design overcomes problems inherent with the preferential right to left border transfer of T-DNA and improves the chances of having the gene of interest transferred to the plant cell in cells expressing the selectable marker gene.
- pBECK2000 series, which contain:
- Synthetic T-DNA borders and a bar gene, which confers the plants resistance to the herbicide phosphinothricin. Also, the vectors use the phage P1 Cre/loxP site-specific recombinase system, which permits the transfer and integration of a target and marker genes as a single T-DNA unit into the plant genome or as two independent T-DNAs within a single Agrobacterium. It also allows site-specific excision of marker genes from the plant genome after transformation.
- Binary-BAC (BiBAC) vector
- based on a bacterial artificial chromosome (BAC) vector and is suitable for Agrobacterium-mediated transformation of high-molecular-weight DNA
- comprises low-copy number origins of replication for both E. coli and Agrobacterium to ensure replication of the plasmid as a single-copy in both bacteria; and
- a helper plasmid carrying additional copies of vir-genes in order to clone very large T-DNAs (up to 150 kb) into the plant genome.
- pGreen series, small plasmids of around 3.2 Kb containing:
- a broad host range replication origin (ori pSa) and a ColE1 origin derived from pUC,
- a pSa replicase gene (rep A) that provides replication functions in trans and is located in a compatible plasmid (pSoup) in Agrobacterium, and
- multiple cloning sites based on the pBlueScript vector, which allow any arrangement of selectable marker and reporter genes.
Although binary BAC vectors are devised for cloning long fragments of DNA (around 150 Kb) or more size it has been modified for different uses.

Binary derived BAC vector

The vector contains wide range replication Ori.
Some promoters used for
expression in plants:
CAMV 35S (Cauliflower mosaic virus):
PRS (Pathogen Response Sequence).
Osmotin: stress controlled.
Chl a/b (chlorophyll A and B gene): Light controlled.
RUBISCO light chain (Ribulose bi phosphate light chain gene): light induced.
NOS (Nopaline synthase gene) :wound induced.
OS (Octopine synthase gene) : wound induced.
Hirudln promoter. Anticoagulator gene,
Hordien promoter,
Glutein.
Legumin.
Cecropin: response to bacterial infection
Promoters can chosen for tissue specific or stage specific expression in embryo- early or late, cotyledons, leaves, anther, leaves, ovary, fruit and other tissues
CaMV Promoter:
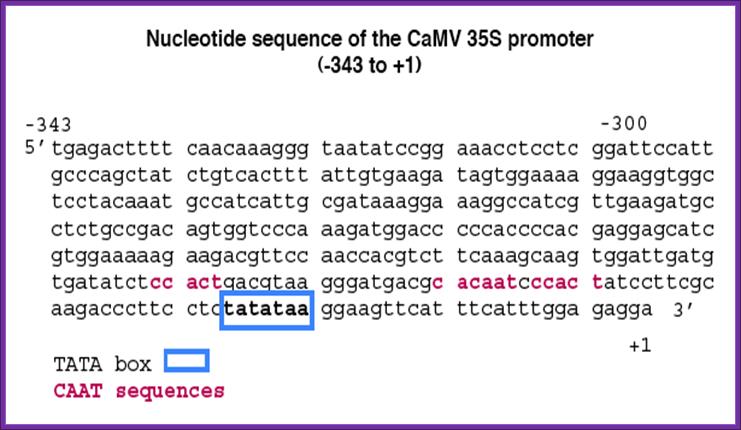
CAMV provides a versatile promoter and it is active all the times.
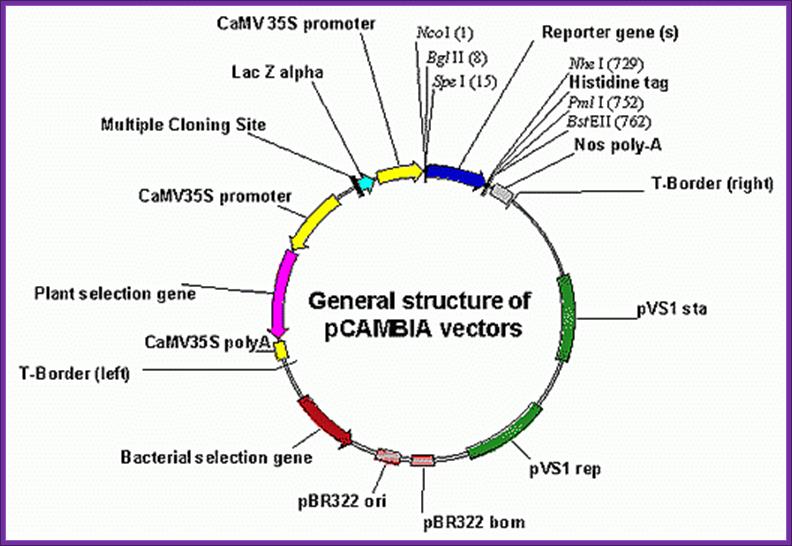
CaMV derived BIA vector called pCAMBIA
![]()
http://www.patentlens.net/
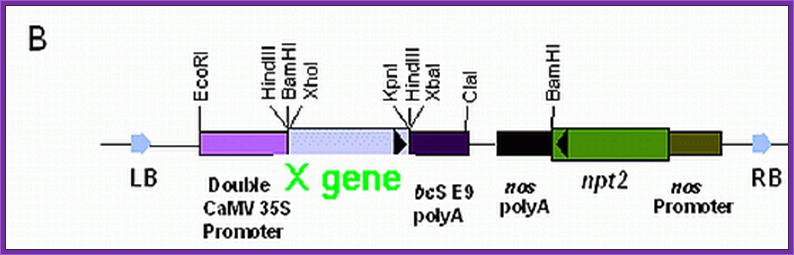
Selection marker genes:
Glyphosate (herbicide resistant gene).
Phosphonithricin (inhibits glutamine synthesis).
Some of the reporter genes used are Glucorunidase and Luciferin, and Protein-fluorescent green (PFG).
--LB—P/osmotic--X- gene---Ttr---P/Nos—Hygromycin—Ttr-RB-O/Drive—(circular mode)
P/O = Promoter Osmotin.
LB = Left border.
RB = Right border.
O/Drive = over drive.
Nos = Nopaline synthase gene promoter.
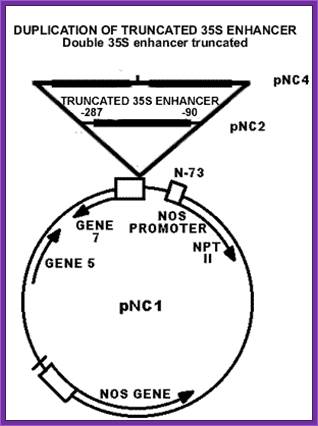
Nos-Plasmid (Nos):
Nos plasmids synthesize Nopaline, an Arginine dérivative. Nopaline is N-alpha dicarboxy ethyl-L Arginine. This provides both Nitrogen and carbohydrate as a source for the bacteria. This Ti-plasmid produces teratomas, which can differentiate into roots and shoots.
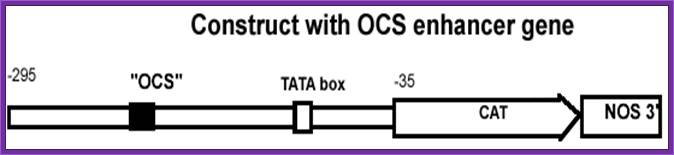
Octopine Plasmids (OCS):
They generate Octopine- N-2-1-3 dicarboxy propyl-arginine. The tumors don’t develop differentiated organs such as roots and shoots.

Agropine Plasmids (Ag S):
They also produce substance called Agropine. They produce poor tumors and they don’t differentiate.
Agrobacterium Rhizogene Plasmids:
Ri plasmids: The entire genome of the Ri plasmid pRil724 (217.6-kb). In this paper we provide information indicating that the agropine-type root-inducing (Ri) plasmid pRi1855 of Agrobacterium rhizogenes contains functional genes for auxin production (aux) in the right transferred DNA (T-DNA) region (T(R)-region).
Agrobacterium strains with an agropine-type Ri plasmid not only cause hairy root on certain plant species, but they also induce tumors on other plant species.
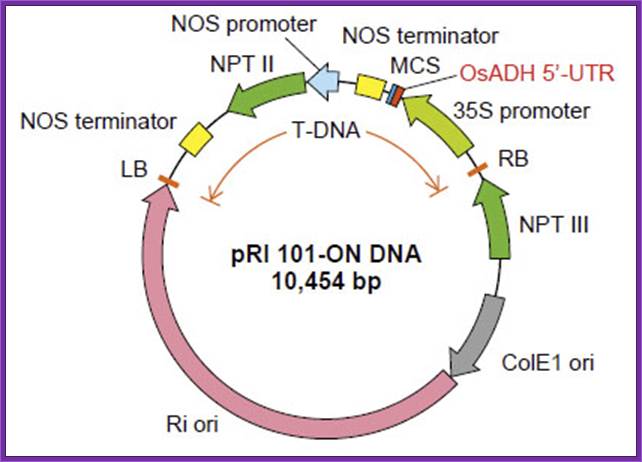
pRI
101 DNA series are binary vectors for expressing foreign gene in plant cells,
which has the 35S promoter of cauliflower mosaic virus (CaMV) and a 5'
non-coding region (5'-UTR) of alcohol dehydrogenase (ADH) gene. The 5'-UTR of
ADH functions as a translational enhancer in plants1). There are two
types of vectors, pRI 101-AN DNA having a 5'-UTR of Arabidopsis ADH (AtADH
5'-UTR) and pRI 101 ON DNA having a 5'-UTR of rice ADH (OsADH 5'-UTR). The pRI
101-AN is for dicotyledonous plants such as tobacco or Arabidopsis and the pRI
101-ON is for monocotyledonous plants such as rice.
The pRI 101 DNA are shuttle vectors, and replicate autonomously in E. coli
and Rhizobium (Agrobacterium). In E. coli, these vectors
are high copy number plasmid because these have replication origin same as that
of pUC type plasmid (ColE1 ori), and these are maintained stably in Rhizobium
(Agrobacterium) also with mutant type replication origin of Ri plasmid
(Ri-ori)2). The pRI 101 DNA are possible to integrate target gene in
plant chromosome stably because the cloning site of these products are located
at the position closer to Right Border (RB) of T-DNA than the selection marker
for plant, so the target gene is not deleted.
Description-pRI:
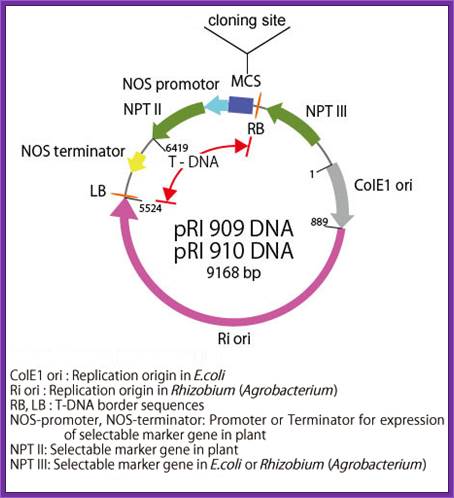
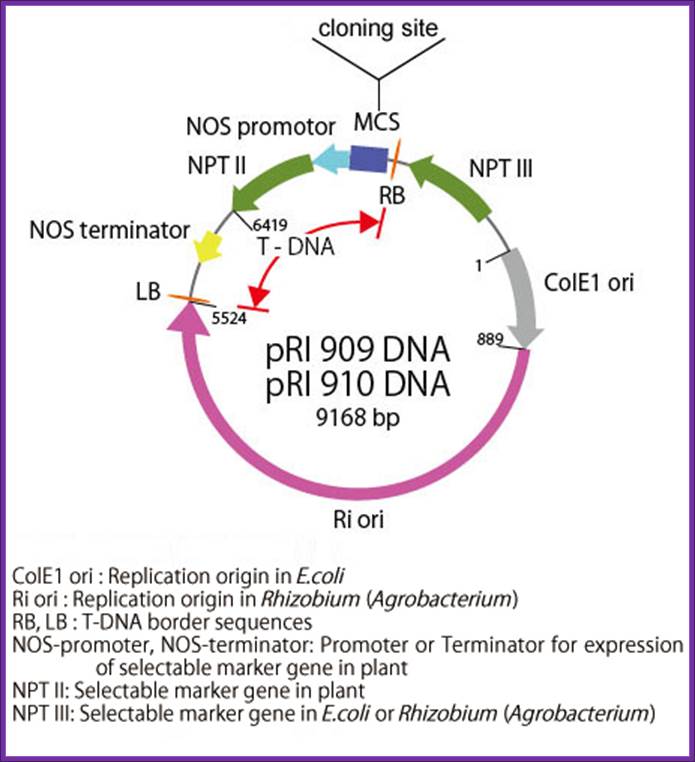
pRI 909 DNA is a binary vector
which has a region of T-DNA for plant transformation. It is originated from Ri
plasmid of Rhizobium (Agrobacterium) rhizogenes, lacking their
vir region. pRI 909 DNA is also a shuttle vector, and replicates autonomously
in E. coli and Rhizobium (Agrobacterium). In E. coli, this
vector is a high copy number plasmid because it has a replication origin same
as that of pUC type plasmid (ColE1 ori), and it is maintained stably in
Rhizobium (Agrobacterium) also with mutant type replication origin of Ri
plasmid (Ri-ori). This vector has a kanamycin resistant gene NPT III, as the
selection marker for E. coli and Rhizobium (Agrobacterium), a
mutant type kanamycin resistant gene NPT II, as the selection marker for plant,
and the same multi-cloning sites as pUC type plasmids are present.
This vector is available for plant transformation in combination with Rhizobium
(Agrobacterium) by binary vector method. It is possible to integrate
target gene in plant chromosome stably using this vector because its cloning
site is located at the position closer to Right Border(RB) of T-DNA than the
selection marker (NPT II) for plant, so the target gene is not deleted. In pRI
909 and pRI 910, the sequence of MCS and its neighboring region is the opposite
direction.
pRI 201:
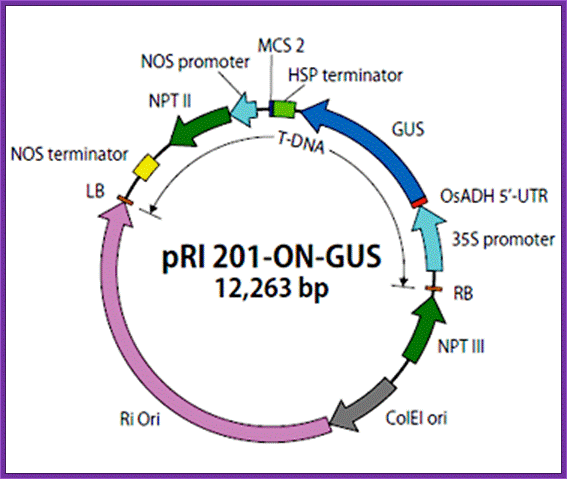
Description;
pRI 201 DNA
series are designed for expressing target genes in transformed plant cells.
This series of vectors retain the backbone of pRI 101 vectors1 (Cat.
#3262/3263), which includes an alcohol dehydrogenase (ADH) gene-derived 5'
untranslated region (5' -UTR) (translational enhancer region) downstream of the
35S promoter from cauliflower mosaic virus (CaMV). In addition, they have a
heat shock protein (HSP) gene-derived terminator in place of the nopaline
synthase (NOS) gene-derived terminator, allowing higher target gene expression
compared with the pRI 101 series2. Further, multigene transformation
with a single vector is possible by integrating an expression cassette
containing another gene (promoter + enhancer + gene of interest + terminator)
into the cloning site (MCS2) downstream of the HSP terminator.
The pRI 201 series has two types of vectors: pRI 201-AN DNA and pRI 201-ON DNA.
Carrying an Arabidopsis ADH-derived 5' -UTR (AtADH 5' -UTR), pRI 201-AN DNA is
suitable for dicotyledonous plants. With a rice ADH-derived x' -UTR (OsADH 5'
-UTR), pRI 201-ON DNA is compatible with monocotyledonous plants. Positive
control vectors (pRI 201-AN-GUS DNA and pRI 201-ON-GUS DNA) containing the
β-glucuronidase (GUS) gene are available outside the U.S. pRI 201-AN DNA
and pRI 201-ON DNA are binary vectors for plant transformation and have a
mutant-type replication origin (Ri ori) from the Rhizobium rhizogenes Ri
plasmid3. These vectors also have both a replication origin (ColE1
ori) derived from pUC plasmids, which allows maintenance at a high-copy-number
in E. coli , and a multicloning site located near the right border (RB)
of T-DNA relative to the plant selection marker (NPT II), which allows stable
integration of the target gene into a plant chromosome.
Agrobacterium strains used for Gene engineering:
Common disarmed strains-LBA 4404 and C58
The recombinant small replicon is transferred via bacterial conjugation or direct transfer to A. tumefaciens harboring a helper Ti plasmid,
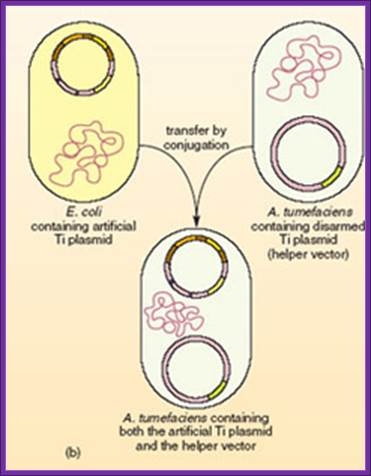
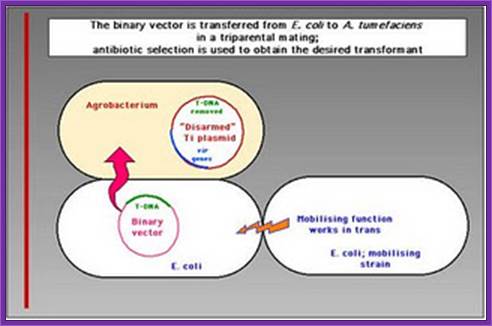
Transfer of E.coli binary vector into Agrobacterium is done by triparental mating, where the E.coli designed plasmid to transfer into Agrobacterium containing helper plasmid.
Triparental mating:
From Wikipedia, the free encyclopedia
Jump to: navigation, search
Triparental mating is a form of Bacterial conjugation where a conjugative plasmid present in one bacterial strain assists the transfer of a mobilizable plasmid present in a second bacterial strain into a third bacterial strain. Plasmids are introduced into bacteria for such purposes as transformation, cloning, or transposon mutagenesis. Triparental matings can help overcome some of the barriers to efficient plasmid mobilization. For instance, if the conjugative plasmid and the mobilizable plasmid are members of the same incompatibility group they do not need to stably coexist in the second bacterial strain for the mobilizable plasmid to be transferred.
· A helper strain, Carrying a conjugative plasmid (such as the F-plasmid) that codes for genes required for conjugation and DNA transfer.
· A donor strain, Carrying a mobilizable plasmid that can utilize the transfer functions of the conjugative plasmid.
· A recipient strain, you wish to introduce the mobilizable plasmid into.
Five to seven days are required to determine if the plasmid was successfully introduced into the new bacterial strain and confirm that there is no carryover of the helper or donor strain.
The Agrobacterium strain used for transformation carries two plasmids. A nononcogenic disarmed tumor-inducing plasmid (Ti plasmid) containing the virulence (vir) genes, but lacking the T-DNA region. The T-DNA region is present on a second plasmid, the binary vector. The selection marker in the binary vector used in this study is the kanamycin resistance gene (kanr). DNA sequences to be transformed are cloned between the left and right border sequences of the T-DNA region and transferred to the host.
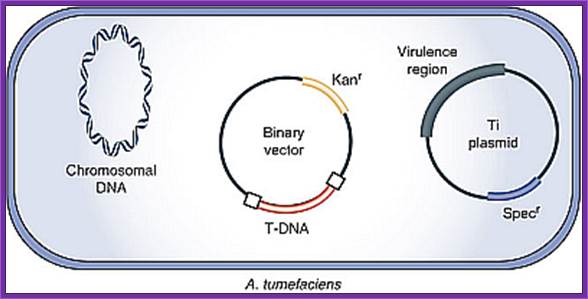
The Agrobacterium strain contains helper plasmid and the binary vector containing the desired gene. This bacteria is ready for transferring the desired T DNA construct to plant cells.
Tomato plant Transformation: Horticultural Science Departement, University of Florida ( the figure below).
Explant preparation
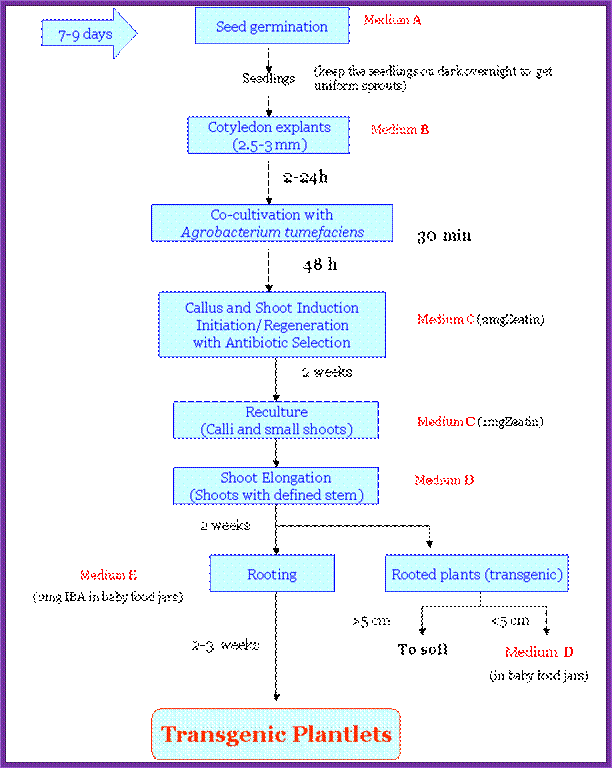
Just an outline of the mechanism by which the DNA is transferred. The mechanism has been explained in the text.
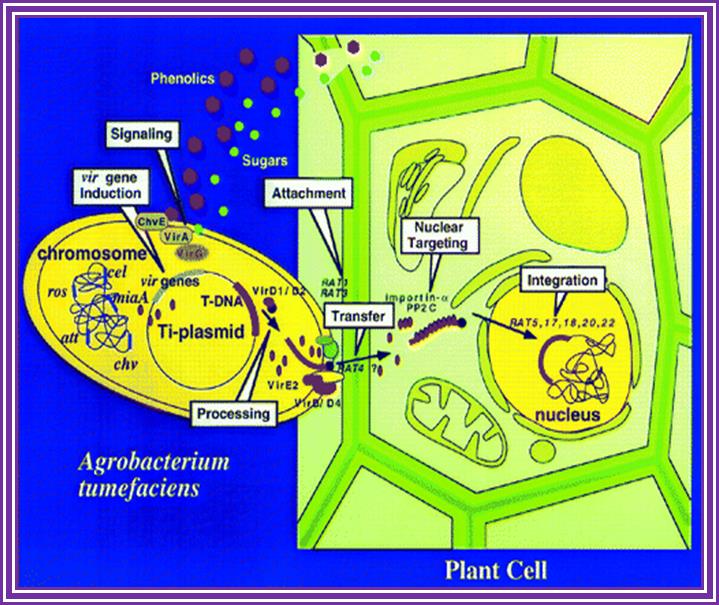
Phenolic signals make bacteria to bind to cells at at rthe wounded surface of plant explant. The signls are transmitted via VirA and VirG. This leads the transcription all Vir genes. This results in nicking the TDNA at right border and stripping off the ss DNA bound by VirD2 at the 5’end. This complex is transferred into the plant cells through transport complex formed at the interface between bacterium and lant cell walls. Once the TDNA is transferred the NA is bound by VirE2 and reeptors. Then it leads to the entry of te same into the nucleus, where TDNA gets integrated into host DNA. The diagrams below also show the same features. Infection mechanism of Agrobacterium. Credits: Gelvin, S. Annu. Rev. Plant Physiol. Plant Mol. Biol.http://microbewiki.kenyon.edu/;http://microbewiki.kenyon.edu/
An outline of DNA movment into plant cell. http://www.plantphysiol.org/

Hypothetical channel between Agrobacterium cell wall and Plant cell wall.
Once the recombinant T DNA inside the Nucleus gets integrated into chromatin DNA then, the Cre enzyme is activated and produces the segment of gene with proper promoter by recombination at LoxP sites. If the vector mediated TDNA is designed in such a way the marker gene is placed between the desired gene with LoxP direct repeats on either side of the marker gene. When a transgenic plant obtained can be hybridized with another transgenic plant that contain inducible cre gene. In the hybrid the cre gene enzyme excises the marker gene using LoxP sequences and ligates the adjacent segment restoring the full length coding region of the desired gene. Thus one can get transgenic plant free from antibiotic marker genes,
Protocol to produce transgenic plants- using recombinat Agrobacterium.
Agrobacterium mediated plant transformation;
· Use the desired recombinant construct with desired gene and selection marker gene.
· Transfer the construct into disabled (for the said characters) Agrobacterium.
· Grow them to sufficient density.
· Transfer the bacterial culture into plant culture medium with stimulant like aceto Syringone.
· The transfer the desired callus and allow the bacteria to infect overnight.
· Then wash the tissue with carboxylin or Carbamycin. The Carbamycin kills all agro bacterial cells.
· The transfer the tissue to regular differentiating medium contain in 100ug/ml of Hygromycin.
· Reculture the till you get rid of the agrobactrium contamination.
· Select the tissue that grows.
· From that one can generate shoots and check for the gene or gene produce by western blotting or PCR amplification.
Gene Gun method:
- Principle of gene gun is simple.
- It has an inlet for inert gas and a meter showing the pressure of the gas that allowed into the gun.

http://www.whatisthebiotechnology.com/
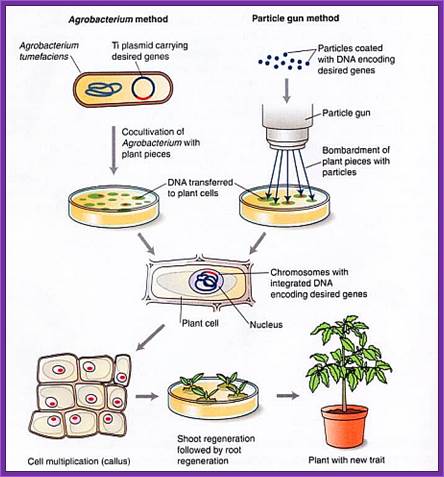
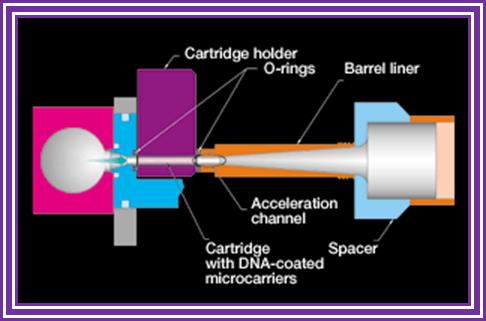
- At the base of the gun is a small slit into which one can insert a devise having a window, which is covered with a nitrocellulose membrane.
- Before inserting the devise into the window the DNA coated with the desired DNA laid and dried.
- Place the required tissue just below the gun.
- Allow the inert gas into the gun as the gas fills up the pressures builds up.
- At a particular pressure the membrane burst and the gold particle coated with DNA are shot into the cells with out harming the tissue.
- Take out the tissue and grow on culture media containing Hygromycin or any other antibiotic you are using.
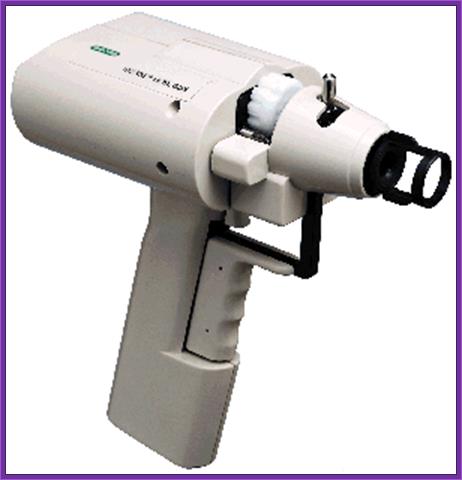
Hand held gene gun; http://www.bio.davidson.edu/

Electroporation method:
The required tissue such as small embryos and small pieces of callus or protoplast can be used for electroporation. However one has to determine the input voltage, resistance and the time required. Many times if dry embryo is soaked with water containing the required DNA, the DNA is efficiently taken into the embryonic cells. The expression or the presence of the gene can be checked either by western bolting or by PCR.
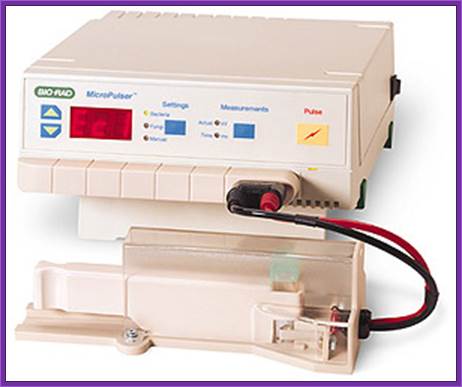
https://www.ices.a-star.edu.sg
Detecting the gene and its expression in the transgenic plants:
Any tissue grows in the culture media is definitely transgenic tissue for the tissue can grow only when it has the antibiotic resistant gene. As the media contains the antibiotic it is certain that the tissue that grows is a transgenic tissue.
In order to identify the gene of interest, whether or no it is inserted, isolate small amount of DNA from any tissue and amplify the gene or a part of the genetic DNA using specific set of primers.
To find out \whether or not the gene has expressed, one can perform western blotting using specific antibody or one use set of primers for amplifying specific mRNA. Even microarray assays can be performed.
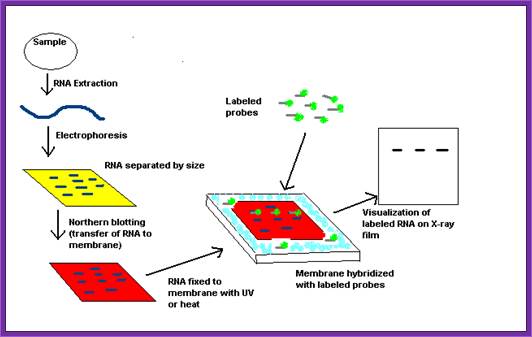
Flow diagram outlining the general procedure for RNA detection by northern blotting. http://en.wikipedia.org/

Fluorescent Plant

Insect resistant plant; http://www.usfq.edu.ec/

Two tomato plants artificially inoculated with bacterial speck disease. Plant on left has been genetically engineered with a gene for resistance to the disease, and plant on right is a susceptible, non-engineered variety. Source: Dr. Steve Tanksley, Cornell University.Transgenic tomato plant; http://cls.casa.colostate.edu/

Other projects aim to enrich tomatoes with substances offering health benefits. All of these products, however, are still many steps away from receiving authorisation.; http://www.pharmatutor.org/
Tests underway for new HIV drug farmed from GM tobacco plants; ;Tobacco GM plant; http://cordis.europa.eu/
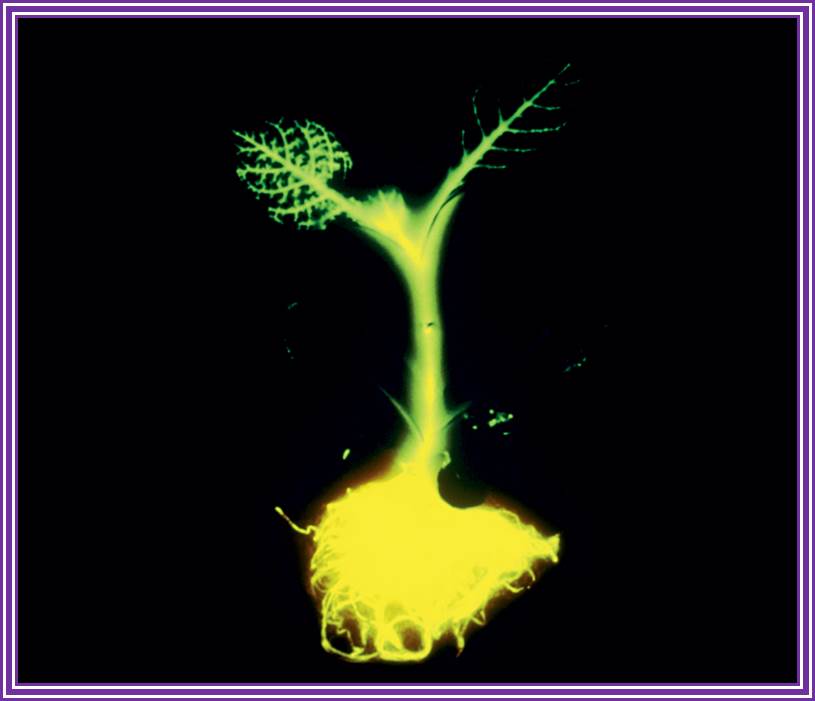
Transgnic plant with Fluorescent gene; Jim Haseloff; http://link.springer.com/
Nutrient medium for plant tissue culture:
Murshige / Skoog medium:
Basically this entire medium contains basic components such as macronutrients, micronutrients and vitamins and carbohydrate sources such as sugars. Addition of hormone such as Auxin and Cytokinin vary depending upon what one requires. For the development of shoot and plant regeneration one uses higher concentration of cytokinins and on the other hand if one wants only callus formation one uses more of Auxin in relation to cytokinins. Manipulation of hormonal concentration is important in obtaining regeneration. For regeneration of explants that one has to determine the combination and concentration of hormones. For example for tomato plant regeneration one can use MS medium with 0.5mg/L of auxin and 2mg/l of cytokinin. Each plant and the tissue used, require some changes in the media and it has to be determined by trial and error methods.

Basic salts:
Name mg/L
NH4NO3 600.
KNO3 1900.
KH2PO4 170.
KCl 300
MgSO4 300
CaCl2 600
MnSO4 010,
ZnSO4 002,
Na2MOO4 0.25,
CuSO4 0.025,
H3BO3 003,
KI 0.75,
Sequestrene 028,
Sugar sources:
Sucrose 250,
Xylose optional
Mannose, “ “
Sorbitol, “ “
Mannitol “ “
Organic acids:
Na Pyruvate 20,
Malate 40,
Citrate 40,
Fumerate 40,
Cassaminoacids 250
Coconut water 20,
Vitamins:
Thiamine HCl 10,
Riboflavin 0.2,
Ascorbic acid 002,
Folate
Pyridoxine HCl,
Inositol 100,
Biotin,
Vit-A,
Vit-D,
VitB12,
In addition one has to add hormone to the desired concentration.




Protoplast culture:
Use Cellulase 2gm (1%),
Pectinase 1%,
Pectolyase 0.1%
Macerase 0.1%
Sucrose 13.7%.
Surfaces sterilize the tissue. Use the very young growing leaves or meristems and chop them into small pieces. Place the tissue into the liquid mentioned above containing the culture medium. Allow the tissue over night. Hold the tissue in a forceps and shake so that all cells freed come into solution. Filter in cheesecloth. Gently centrifuge at 500 rpm for 5 minutes. Discard the upper liquid continue for another three or four times so as to get rid of the enzyme. Then suspend the cells in regular nutrients media. Now count the cell number using Hemocytometer. The plate the same on nutrient agar plate or use them for liquid culture.
Good protoplasts show no cell wall and all cell are spherical. To finds there is any remnant of cell wall one can use fluorescent whitener, which actually binds to cellulose fibers. If there is any remnant of cell wall one can observer the fluorescence at the surface of the cell. Protoplasts should be free from cellulose. To find out how many cells are living one can stain with Trypan blue. Those cells take the stain are dead and those don’t are living.
Transplastomics:
Introducing desired recombinant DNA into plastome (genome of the plastid) is deemed as Trans-Plastomics.
A transplastomic plant is a genetically modified plant in which the new genes have not been inserted in the nuclear DNA but in the DNA of the chloroplasts. The major advantage of this technology is that in many plant species plastid DNA is not transmitted through pollen, which prevents gene flow from the genetically modified plant to other plants (Wikipedia).
Plastid Biotechnology: Food, Fuel, and Medicine for the 21st Century:
1. Pal Maliga* and
2. Ralph Bock
The number of genes encoded by the plastid and mitochondrial genomes is much smaller, approximately 120 and approximately 60, respectively. The number of processes that may be modified by engineering the organellar genomes is much higher than the number of genes would suggest. That is because approximately 10% of the nuclear gene products are targeted to plastids and approximately 10% to mitochondria (Leister, 2003), making these nucleus-encoded processes amenable to manipulation by plastome engineering.
Engineering of the plastid genomes (plastomes; plastid DNA or ptDNA) was first accomplished in Chlamydomonas reinhardtii, a unicellular green alga (Boynton et al., 1988), followed by plastid transformation in tobacco (Nicotiana tabacum), a flowering plant species (Svab et al., 1990). Since 1988, plastid transformation has been expanded to a diverse group of species (see below). However, commercial applications are lagging behind, and currently no crops are grown commercially utilizing this technology.
The number of plastids per cell and the number of ptDNA copies per plastid varies for example, an Arabidopsis (Arabidopsis thaliana) leaf mesophyll cell contains about 120 chloroplasts, and these harbor approximately 1,000 to 1,700 copies of the approximately 154-kb plastid genome (Zoschke et al., 2007). Tobacco leaf cells contain a comparable 100 chloroplasts containing approximately 10,000 copies of the approximately 156-kb ptDNA (Shaver et al., 2006). The ptDNA in chloroplasts is localized to membranes in clusters of approximately 10 ptDNA copies, referred to as nucleoids.
When T-DNA (Transforming DNA) is introduced into plastids only few of the plastids get the foreign DNA. Meristematic cells contain 10 to 14 proplastids, each of which carries only one or two nucleoids. Reduction of plastid number from 100 to 10 to 14 greatly accelerates plastid sorting during cell division, during which plastids carrying the T-ptDNA are dividing at a faster rate. Plastids carrying only the wild-type ptDNA are ultimately lost by dilution during cell division. Depicted in is a selection for an aurea construct that confers a pale pigment phenotype to plants (Lutz and Maliga, 2008; Tungsuchat-Huang et al., 2011).
The transforming DNA is introduced on the surface of microscopic (0.4–1.0 μm) gold or tungsten particles or by polyethylene glycol treatment. Biolistic DNA delivery is used when the targets are plastids in intact tissue; polyethylene glycol treatment is used for DNA introduction into protoplasts (Dix and Kavanagh, 1995). The most commonly used selective marker gene is aadA, encoding spectinomycin resistance (Svab and Maliga, 1993). Kanamycin (Carrer et al., 1993), chloramphenicol (Li et al., 2011), and the amino acid analogs 4-methylindole and 7-methyl-dl-Trp (Barone et al., 2009) have also been successfully employed as selective agents.
Depicted in Figure 1 is a selection for an aurea construct that confers a pale pigment phenotype to plants (Lutz and Maliga, 2008; Tungsuchat-Huang et al., 2011).
Sorting of T-ptDNA at the organelle and cellular levels yields homoplastomic plants. A, Replication and sorting of T-ptDNA at the organelle level yields homoplastomic organelles. Sorting is facilitated by the conversion of chloroplasts (CHL) to proplastids (PP), which contain only one to two nucleoids instead of 10. Wild-type ptDNA and T-ptDNA (blue circles and red circles, respectively) are anchored to membranes by proteins (black dots) in nucleoids (N). Sorting of ptDNA and T-ptDNA in heteroplastomic nucleoids (#1) yields nucleoids with only T-ptDNA (#1a) and wild-type ptDNA (#1b). For details, see Maliga (2004). B, Division and sorting of plastids yields genetically stable transplastomic plants. Sorting is accelerated by reduction from approximately 100 chloroplasts in leaf cells to approximately 10 to 14 proplastids in meristematic cells. In the cells, the nucleus (Nu) is also marked. C, The plastid genotype of long-term stem cells in the three layers (L1, L2, L3) of the shoot apex determines the plastid genotype in leaves. PZ and CZ are the peripheral and central zones, respectively. On the left is a shoot apex with T-ptDNA in all three layers and a homoplastomic plant carrying only T-ptDNA encoding the aurea spectinomycin resistance (aadA) gene. The variegated plant in the middle has cells with wild-type ptDNA and T-ptDNA in its shoot apex. The regenerated plant on the right has only wild-type ptDNA. C is modified from Lutz and Maliga (2008); the plants were described by Tungsuchar-Haung etal 2011.
Shoots emerging from a bombarded leaf are chimeric, consisting of transplastomic and wild-type sectors. The transplastomic sectors, often very small, can be visualized by the expression of aurea transgenes that confer a golden-yellow phenotype to leaves (Fig. 1C, bottom middle). The pigment phenotype is due to posttranscriptional interference with the plastid clpP1 (ATP-dependent protease proteolytic subunit) gene expression by the aurea aadAau (aminoglycoside 3′′-adenylyltransferase, aurea) transgene. Wild-type sectors in the shoot are present because transplastomic sectors protect wild-type cells against antibiotics in culture.
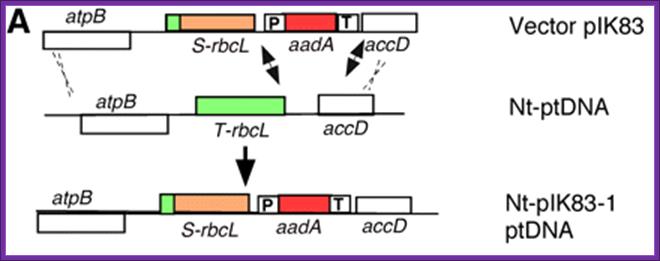
Vector containing DNA is now incorporated into the plastid DNA removing its counterpart.
The transforming DNA is introduced on the surface of microscopic (0.4–1.0 μm) gold or tungsten particles or by polyethylene glycol treatment. Biolistic DNA delivery is used when the targets are plastids in intact tissue; polyethylene glycol treatment is used for DNA introduction into protoplasts (Dix and Kavanagh, 1995). The most commonly used selective marker gene is aadA, encoding spectinomycin resistance (Svab and Maliga, 1993). Kanamycin (Carrer et al., 1993), chloramphenicol (Li et al., 2011), and the amino acid analogs 4-methylindole and 7-methyl-dl-Trp (Barone et al., 2009) have also been successfully employed as selective agents.
Although there are reports of partial success in many species, reproducible protocols for plastid transformation have been described only in tobacco (Svab and Maliga, 1993), tomato (Solanum lycopersicum; Ruf et al., 2001), petunia (Petunia hybrida; Zubko et al., 2004), potato (Solanum tuberosum; Valkov et al., 2011), soybean (Gycine max; Dufourmantel et al., 2004), lettuce (Lactuca sativa; Kanamoto et al., 2006), and cabbage (Brassica oleracea; Liu et al., 2007). Monocots as a group appear to be the most recalcitrant species.

Purple Tomato Genetically Engineered to Fight Cancer; The scientists created these Super Tomatoes by transferring the gene containing the "intense purple" pigment from snapdragons to regular red tomatoes. British scientists have been working on developing a dark purple tomato that contains high amounts of the chemical anthocyanin, which is normally found in berries such as blueberries, cranberries and blackberries. As we reported earlier this week, anthocyanin has strong anti-inflammatory effects, and tests have shown that the purple tomatoes slow down cancer in mice. They also have double the shelf life of regular tomatoes. http://www.laweekly.com/
Apel and Blok: Metabolic engineering of the carotenoid pathway in transplastomic plants. A, Schematic overview of the carotenoid biosynthetic pathway. Lycopene, the major storage carotenoid in tomato, is indicated in red, and β-carotene (provitamin A) is indicated in orange. Enzymes that have been used for plastid genome engineering are given in italics. Parts of the pathway not occurring in higher plants are shown in blue. Multiple arrows denote conversions involving multiple enzymatic steps. The reversible reactions of the xanthophyll cycle are indicated by double arrows. B, Phenotypes of fruits from transplastomic tomato plants expressing a lycopene β-cyclase transgene from daffodil. Fruits from a wild-type plant (top two panels) and a transplastomic line (bottom two panels) were harvested at different ripening stages and photographed from the side and from the bottom. The orange color of the ripe transplastomic tomatoes comes from the efficient conversion of the red storage carotenoid lycopene into the orange provitamin A (β-carotene). The provitamin A levels reached 1 mg g−1 dry weight, while wild-type fruits had less than 200 ng provitamin A g−1.
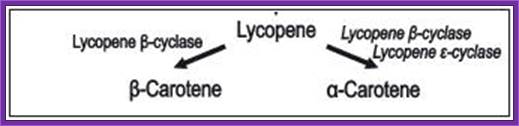
Conversion of lycopene to either β-carotene or ά-carotene.
Modified from The Plant Journal 49, 276-288.
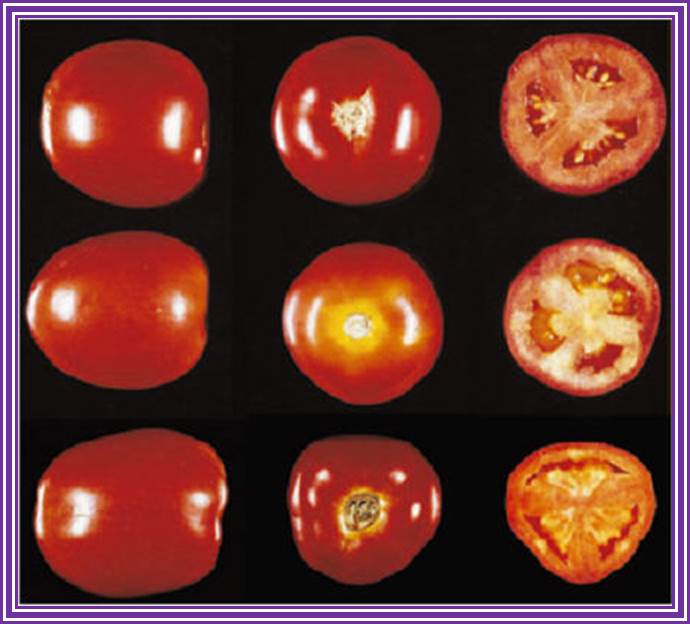
Figure: Phenotype of ripe tomato fruits harvested from
transplastomic tomato plants expressing the lycopene-cyclase gene from Erwinia.
Top row: Wild type; Middle row: Transplastomic tomatoes expressing lycopene
b-cyclase; Bottom row: Fruits from transplastomic plants treated with 2.5 mM
CPTA. Source: The Plant Journal 49,
276-288. Tawanda Zidenga http://www.isb.vt.edu/
Although there are reports of partial success in many species, reproducible protocols for plastid transformation have been described only in tobacco (Svab and Maliga, 1993), tomato (Solanum lycopersicum; Ruf et al., 2001), petunia (Petunia hybrida; Zubko et al., 2004), potato (Solanum tuberosum; Valkov et al., 2011), soybean (Gycine max; Dufourmantel et al., 2004), lettuce (Lactuca sativa; Kanamoto et al., 2006), and cabbage (Brassica oleracea; Liu et al., 2007). Monocots as a group appear to be the most recalcitrant species.