Eukaryotic Gene Structure:
The number of genes per genome in eukaryotes is large and they vary in their structure and function. There is division of labor among the RNAPs and their associated transcription factors that transcribe them. There are three basic types of RNAPs and they are associated with specific transcription factors and often found to be regulated by activators/Coactivator, enhancers and repressors/Corepressors.
Regulation of Gene expression in Eukaryotes; Principles of Cell Biology; http://www.mun.ca/biology
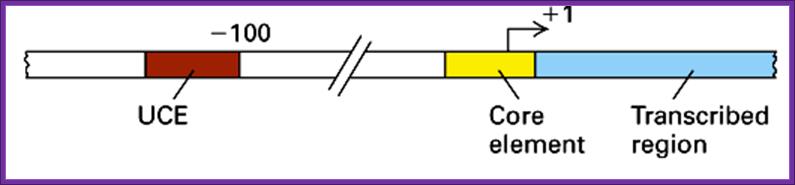
Promoter Structure for RNA Pol-I
Genes, whether they are prokaryotic or eukaryotic, have same structural features such as coding regions, promoter elements and terminal sequences.
· However the detailed organizations vary in terms sequence blocks and their positions. The major difference from prokaryotes is that the coding region is split into coding and noncoding regions.
Promoter regions and the ends of genes show different structural features, because eukaryotic genes, depending upon the kind of gene, they are transcribed by three different enzymes, where as in prokaryotic systems all types of genes are transcribed by only one type of RNA polymerase, of course with different sigma factors for different set of genes.
· It clearly means, eukaryotic gene structure, especially promoter regions, including their regulatory regions and their structure are different and more complicated.
So far except for few simple genes, understanding of others is nebulous; this is in spite of great strides made in gene cloning, sequencing and expressions of genes in different cell types.
· Genes transcribed by different enzymes have different structural features and different functions.
Promoter of rRNA genes:
--142 -- 110 +1 >
---------I----I--------I----I--------I-promoter I-- core—I---------ter-
GC rich --45-----I>
Core : [---CTCCGAGTCG (N) 5TGGGCCGCCGG—]
A. Promoter Structure: For RNA pol-I:
Genes for ribosomal RNA are exclusively transcribed by RNA polymerase-I. Total number of rRNA genes per haploid genome can be more than 200-300. Majority of them are continuously transcribed
· In eukaryotic system most active and highly productive genes, which are transcribed most of the time, are ribosomal RNA genes.
More than 90 % of the total RNA found in any eukaryotic cell is rRNA. rRNA genes 100-200 per chromosomes are transcribed in Toto and in massive scale with certain exceptions.
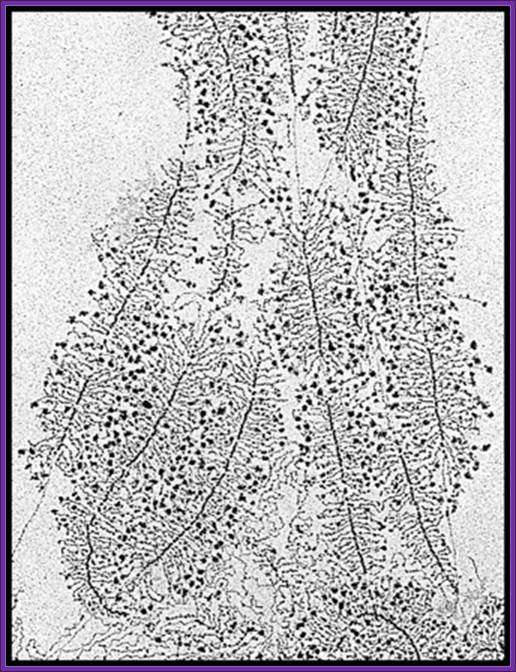
American Society for Microbiology. All Rights Reserved.): Electron micrograph of rRNA genes from a Saccharomyces cerevisiae strain containing only ~42 rRNA genes. Wild-type strains have ~140 rRNA genes. The reduced-copy-number strain grows as well as control strains by loading twice as many polymerases per active rRNA gene, indicating that RNA polymerase I transcription is not limited by the number of active genes in normal cells during exponential growth; http://mcb.asm.org/
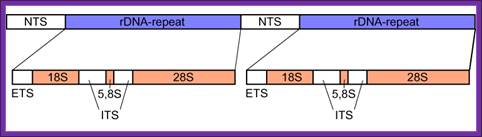
The gene segment of eukaryotic rDNA contains 18S, 5.8S, and 28S tracts and forms a tandem repetitive cluster; the 5S rDNA is coded separately. NTS, nontranscribed spacer, ETS, external transcribed spacer, ITS, internal transcribed spacers 1 and 2, numbered from 5' end; ;http://en.wikipedia.org/
Note- Most of the 5S ribosomal RNA genes are localized elsewhere and transcribed by RNA pol III.
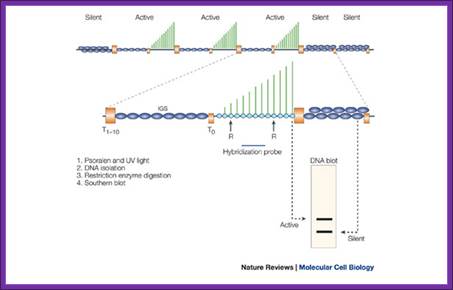
Psoralen is a three-ringed furocoumarin that is produced in the plant genus Psoralea. The compound can intercalate between the two strands of duplex DNA and, following exposure to long-wavelength ultraviolet (UV) light (for example, 366 nm), it reacts with thymidines to form covalent interstrand crosslinks. If the DNA is then purified, cut with a restriction endonuclease (denoted by 'R' in the figure) and subjected to electrophoresis, psoralen-crosslinked DNA migrates more slowly than unmodified DNA. Subsequent DNA blotting and hybridization with a probe specific for a particular gene fragment allows a quantitative estimate of the proportion of psoralen-accessible and -inaccessible genes. This technique has been used extensively in yeast, and to a lesser degree in other species, to estimate the proportion of transcriptionally active to inactive rRNA genes. Nascent precursor ribosomal RNA (green lines) has been shown to be associated with the subset of psoralen-accessible genes, indicating that the slow-migrating band corresponds to active genes. The orange boxes mark terminator elements upstream (T0) and downstream (T1–10) of the transcribed region. The dark-blue ellipses indicate the nucleosomal chromatin structure of the intergenic spacer (IGS) and the more compact, heterochromatic conformation of silent rRNA genes. Psoralen crosslinking; Epigenetic silencing of RNA polymerase I transcription; http://www.nature.com/
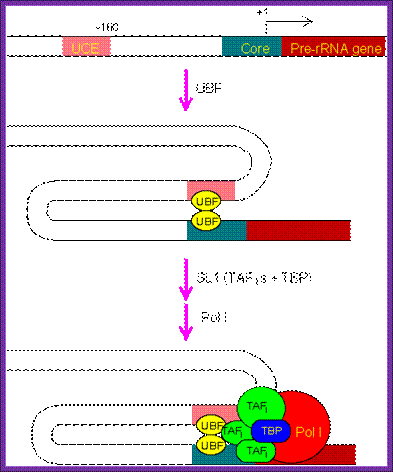
The PIC assembly for RNA polymerase I. The regulatory region of pre-rRNA gene contains a core element and an upstream control element (UCE). Binding of two upstream binding factors (UBF) to both elements may induce DNA looping, and subsequently recruiting TATA-binding protein (TBP) and TBP-associated factors (TAFI). Finally, RNA polymerase I joins the complex and completes the assembly process; http://www.web-books.com/MoBio
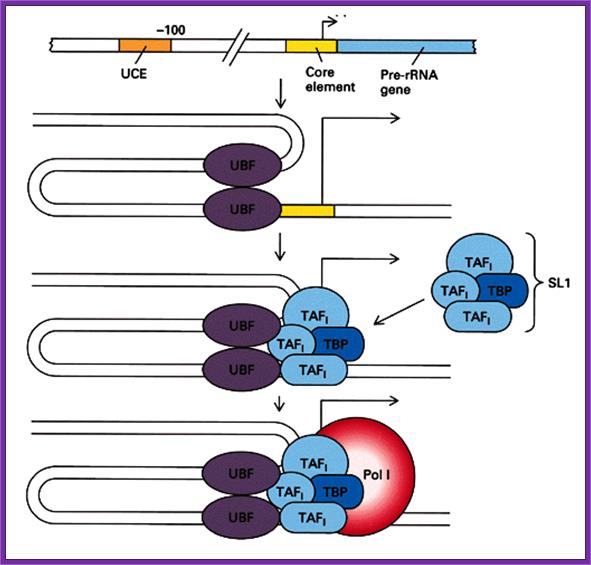
This illustrative diagram represents the assembly of Transcriptional complex on specific RNA Pol-I promoter elements.
· Its synthesis is triggered, when cells are activated for cell proliferation, in such situations tremendous increase of rRNA takes place, ex. rRNA synthesis during oogenesis is a par excellent example.
The rRNA genes themselves, at least in some cases get amplified by rolling circle mode of replication. The demand for rRNA is very high in developing oocyte for when eggs are formed; rRNA is not synthesized for at least 6-7 cell cycles after fertilization, which means there should be high concentration of stored rRNA in the Oocyte.
· In almost all eukaryotic systems, rRNA genes are organized as clusters of tandemly repeated genes in secondary constriction region of chromosomes.
The number of genes found in the region range from 200 to 600 per genome. These clusters are distributed among their chromosomes. For example Homo sapiens, have secondary constrictions for rRNA genes in five pairs of homologous chromosomes- 13, 14, 15, 21 and 22.
· Most complicated organization of rRNA genes is, one of the rRNA components called 5s RNA. This rRNA gene is not located with major rRNA gene segments but found elsewhere.
The 5s rRNA genes are distributed all over the genomic chromosomes, some are found near the tips of certain chromosome just behind telomeres, and others are located elsewhere.
· In real terms, though the rRNA genes are clustered as tandem repeats, each of the ribosomal genes show their own structural features. And they separated by intergenic spaces IGS, which are not transcribed, but contain core promoter elements. The spacer regions can be as large as 3.3kb to 11.2kbp in species of Drosophila. This length varies among the animal and plant species long.
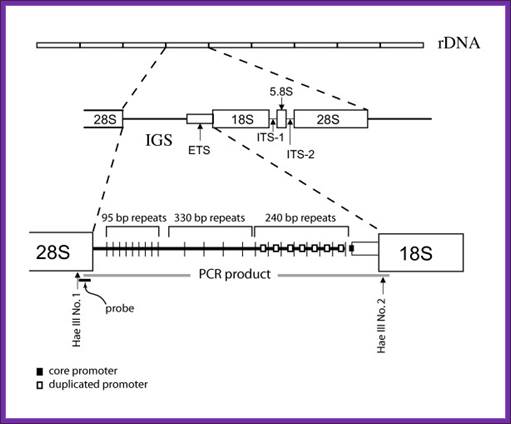
Drosophila-Diagram shows position of PCR product and primers, Hae III restriction sites at the ends of the intergenic spacer (IGS) region, and hybridization probe used in this study [modified from 9, 14].Mateos and Markow BMC Evolutionary Biology 2005 5:46
· And the spacer regions themselves are organized into blocks, each of which have their own sequence elements that promote transcriptional initiation, perhaps they act as enhancers. Basically they all have what is an essential structural elemental of a promoter.
The consensus promoter elemental feature of all rRNA is presence of core structure encompassing about 40 bp from the start site.
· A consensus sequence that has been established from several rRNA genes is the presence of a sequence from start site of transcription.
It is important to note the rRNA genes don’t have TATA boxes in their promoters. But they contain an 11 base pair elements surrounding the start site called INR sequences; (py) 2-C- [A]-(py) 5. This is to some extent true for protein coding genes. At start site the sequence invariably starts with A and some times it is G.
· It has, what is termed as core promoter region between (-) 10 and (-) 45 and an upstream control elements (UCE)at (-) 110, it is the region to which upstream element binding factors bind.
Between –110 and –142 regions a GC rich sequence is present.
· At the start region it has --3(py) - A – 3-4 (py).
· The core region attracts selectivity factor SL-I, 3 TAFs (TBP associated factors) and TBP (TATA binding factors). Positioning of the TBP is assisted and determined by the SL-I and then TAFs bring TBP.
· It is now known that two histone like proteins are also associated with this complex.
This assembly ultimately brings RNA pol-I to the site. But the activation depends on upstream control element binding factors UBF 1; they bind not only to the core but also to UCE.
· UBFI binding results in protein-protein interaction in such a way two units of UBFs join with one another with a DNA loop, and activate the RNA pol-I complex.
Transcription Terminator regions and Termination:
Transcription termination takes far beyond the coding region of rRNA, and the region contains a (T)n sequences recognized by a set of protein-factors called ancillary factors (TTERF)/TTFs, which make the RNAP I to dissociate from the rRNA and the DNA template.
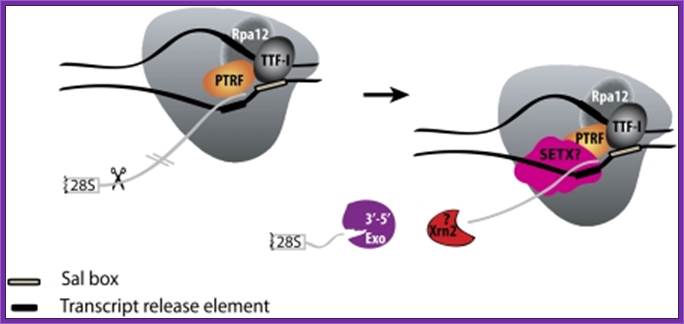
Mammalian RNAPI termination. Transcription terminates at the transcript release element composed of a stretch of Ts, upstream of the TTF-I-mediated pause site. The release factor PTRF, which interacts with RNAPI and TTF-I, recognizes the transcript release element and most likely binds the stretch of Us on the nascent RNA. The RNAPI-specific subunit Rpa12 also plays a role in yeast RNAPI termination that is possibly conserved in mammals. After cleavage of the precursor downstream from the 28S gene, Xrn2 degrades the downstream RNA, “catches up” with RNAPI, and participates in RNAPI release, possibly in conjunction with SETX. An unidentified 3′–5′ exonuclease might be involved in processing of the rRNA precursor. http://genesdev.cshlp.org/