Prokaryotic Gene Structure:
Genes, based on their activity, can be grouped as house keeping genes and others are classed as induced to express or express in stage specific or tissue specific manner.
· House keeping genes express all the time under all normal conditions.
Most of the gene products of house keeping genes are involved in day-to-day metabolic activities responsible for the maintenance of the cell.
· But when cell confers with other signals such as, change in the temperature, change in th pH, and other environmental features such as exposure to toxic chemicals, or chemical inducers, light, non availability of nutrients, and any other factor that is not ambient to cells and not conducive to cells, specific genes respond to such changes or inductions and express to overcome such hostile or unfavorable situations.
Structural features of promoters of these genes, though basically have common features, but individually they vary slightly from one to the other. The RNA polymerase that is responsible for transcription of the gene is same with some variations in regulator sigma factors.
· Most of the housekeeping genes have following structural features in general.
The coding region starts with an initiator codon and the reading frame ends in a terminator codon.
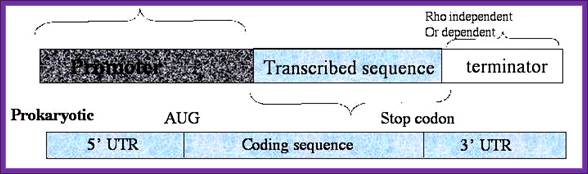
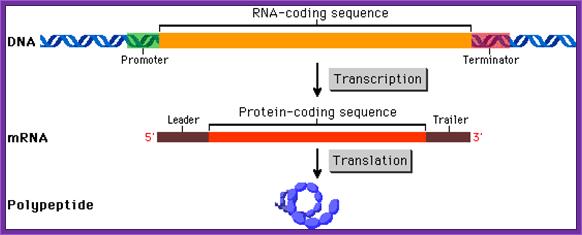
Prokaryotic coding region is collinear to its mRNA, which is collinear to polypeptide chain; The coding region is divided into cistrons separated y intercistronic spacers; each cistron codes for a polypeptide chain: Typical Prokaryotic Gene structural elements. http://www.phschool.com/;http://www.tvcc.edu/;
The coding region of structural genes is not split, but rRNA genes have spacers with in them.
· The upstream elements from the start of the coding region include promoter elements.
Nearly 50 to 100 ntds upstream of the start codon, is the first nucleotide at which transcription initiates, it means, it is at this site the first nucleotide is incorporated into the transcribed RNA.
· The site is called transcriptional initiation site or START.
Nearly 10 nucleotides upstream of the start, there is a sequence called TATAAT or Pribnow box.
· Any nucleotide present on the left of the start is denoted by (-) symbol and the region is called upstream element. The numbers are written as -10, -20, -35 etc.
The start site is the first ntds and symbolized by +1, any sequence to the right of the start is called down stream elements and numbered as +10, +35 and so on.

http://biocadmin.otago.ac.nz/
At –35 there is another consensus sequence TTGACA. These two sequences are the most important promoter elements, for if there is any change in their sequence and position, transcriptional initiation suffers.
· The meaning of a promoter essentially is a distinct sequence module recognized by transcription factors that recruits RNA polymerase (as a holozyme) and bind to the sequence tightly and initiate transcription by unwinding the helically coiled DNA into transcriptional bubble.
The said sequences not only facilitate the binding of TFs and enzyme and also provide sequence information for the site at which the enzyme to initiate transcription. If any one of the consensus sequences is deleted or changed drastically, the enzyme won’t bind, even if it binds, it initiates transcription at different positions.
PROMOTER
-200 -65 -60 -35 -10 +1
I----------I----//---I---------------I----------I------I----------I----------A
Enhancers/ Activator TTGACA TATAT
TGTGA--CTCACA
Consensus promoter sequence n E.coli:
-35-TCTTGACAT—11-15—TATAAT-5-8-InR +1A(G/T/C)
RNA polymerase binding region-
-35 --10 +1----->
TTGACA-------TATAAT----C A T-----
Thèse séquences are used for the binding of RNA polymerases guided by specific sigma factors
In addition to -10 and -35 sequences one finds up Stream sequences such as activator and ehancer elements. One also finds another sequence called Operator elements to which Repressors/Activators bind.
Gene promoters contain upstream and downstream elements from the start site sequences (InR)
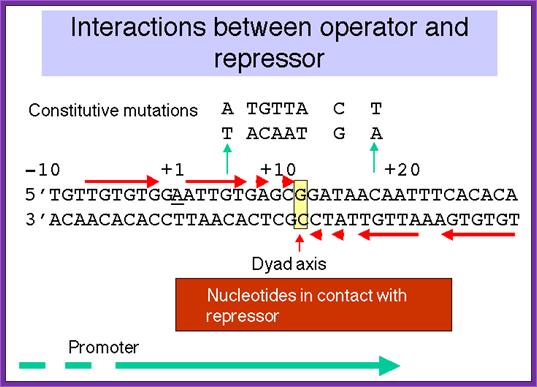
This is typical operator region of Lac operon, with a sequence of dyad axis, this facilitates the binding dimeric repressor proteins; the sequence extends from -10 to +10; this is identified by a dimeric Repressor and it binds and prevents RNA polymerase to act and transcribe. The operator sequences vary with other genes and other repressors. http://www.personal.psu.edu/
In lac operon an upstream sequence to TATAAT and -72- 52 sequence one finds a sequence for the binding of an activator protein complex. That regulates transcription of the gene or genes. Here operator sequence is shown in the figure below.
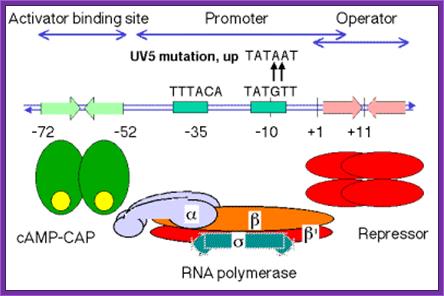
Regulatory region of lac operon, including CAP-CRP binding site; http://www.personal.psu.edu/
The prokaryotic RNA-pol is a Holozyme, when it binds properly in a sequence context; it covers a length from –60 to +20 or little more. It is this segment of the gene that is called Promoter. Whether it is a house keeping gene or special gene, either from prokaryote or eukaryote, the meaning and the function of the promoter is same.
· So promoters act as defining set of sequence defined structural elements, which positions the transcriptional apparatus to initiate transcriptional process. Whether transcription is successfully initiated or not the criteria, but positioning and potentiality for initiating, is an important criteria, then only such sequence modules are called promoters.
For the RNA pol to recognize different genes, the promoter elements have recognition sequences (signature sequences), which are recognized by specific sigma factors that associate with RNA-Pol.
· In prokaryotes there are other sequences in the upstream of the promoter, beyond –35 sequences. Such sequences may present at –65 to –60 or they may be present at –200 or they may be present at—1000 bp upstream or they may present in downstream regions. They are called activator and enhancer sequences. Here enhancer means it increases the efficiency of transcription by 100 to 200%.
The –65 to –60 sequences, position certain factors, whose binding leads to the activation of the polymerase, which was hitherto remained inactive even though it is bound to correct promoter elements. This process is termed as activation and the element as activator elements.
· The other sequence at –200 or ---1000, is called enhancer, for it enhances the rate of transcription by 100 to 200 fold. This is achieved through certain proteins bind to enhancer elements, and then contacts RNA Holozyme by protein-protein interactions, by way of DNA bending or looping, and enhances the efficiency of enzyme.
Some proteins, after binding to their DNA sequences, they interact with transcriptional apparatus and activate the enzyme.
· The kind of sequences, however, and the position of the sequences vary from one gene to another.
It is important to realize, that the proteins that bind have specific motifs called DNA binding motifs, and also possess protein-protein interacting domains.
· The DNA also provides a structural motif in the form of sequence.
Understanding of DNA sequence context and 3-D structural organization of DNA binding protein is of great importance to appreciate the regulatory processes.
· Genes that are regulated in response to the needs, basically have the promoter components. In addition, they have operators, activators and enhancers in different positions, which require specific regulator proteins for operation.
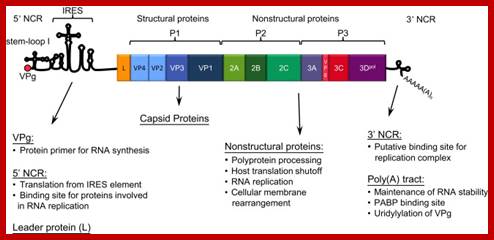
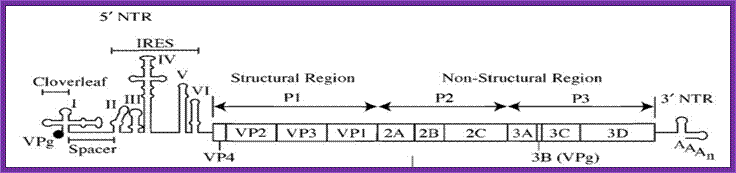
Picorna viral RNA genome with IREs elements; http://www.sciencedirect.com/
The diagram is of picorna virus genome. This genome replicates and produces viruses in bacterial cell, so this can be considered as prokaryotic genome. This can also be called prokaryotic promoter for this genome is of RNA and it has 5’ sequences for internal ribosomal entry. For its replication and translation the plus strand is used. Those RNA genomes infect eukaryotic cells and multiply, and then such RNA genome can be considered as eukaryotic genomes
The viruses containing DNA genomes that infect bacterial cells and multiply can also be considered as Prokaryotic type ex. T4 phage, Lambda, M13 phage and other DNA phages.

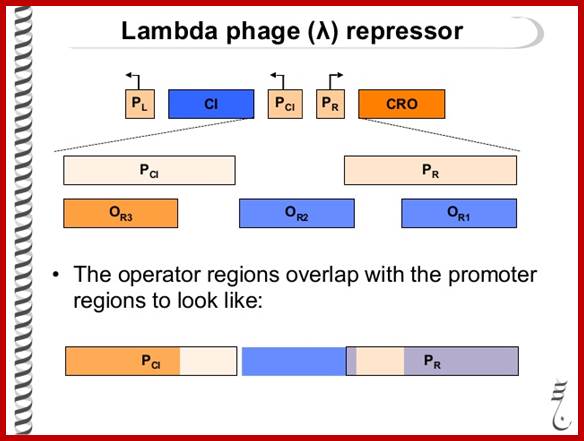
Lambda- promoter with its operator structures; this figure shows Bidirectional mode of Promoters. https://www.slideshare.net
|
PRM |
<-TTAGAT---ATAGAT <-+1-(-10) TAATAT---(-35)-ACAGTT |
|
PRE |
TTGC GTTTGT TTGC <-- 13 --> AAGTAT |
|
PI |
TTGC GTGTAA TTGC <-- 13 --> TGTACT |
|
PaQ |
TTGC GAGCAC TTGC <-- 13 --> TAGTAT |
|
Conserved CII binding sites are shown in red; -35 sequences are in green; -10 sequences are in blue. |
|
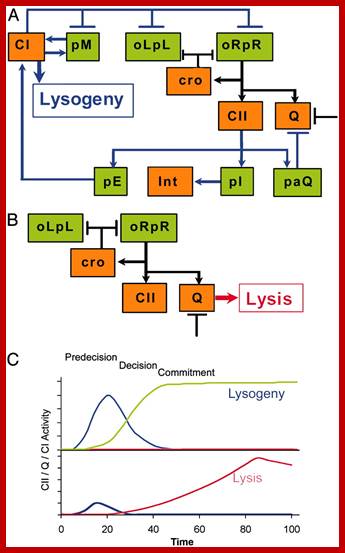
Quantitative kinetic analysis of the bacteriophage λ genetic network; http://www.pnas.org
Terminal Region of the Gene:
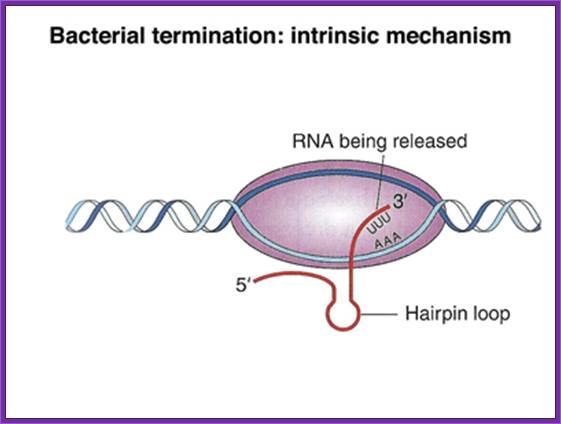
In bacteria transcription termination takes place in two modes, one stem-loop Poly (U) mode called Rho independent mode and the other is called Rho dependent mode, it is also called intrinsic termination.
Rho dependent mode:
It uses a factor called Rho (hexamer) and a specific sequence in the terminal region of the gene.
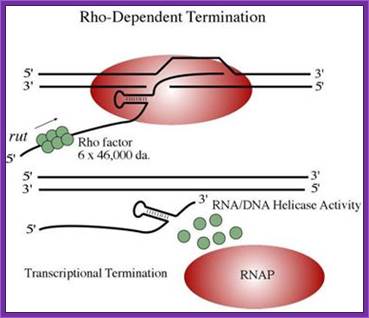
www.jb.asm.org
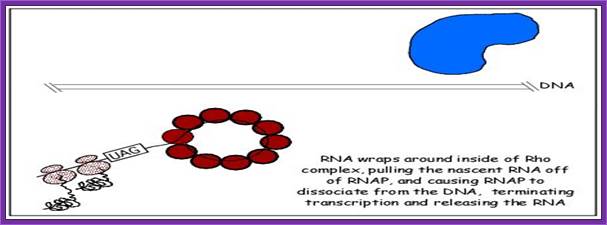
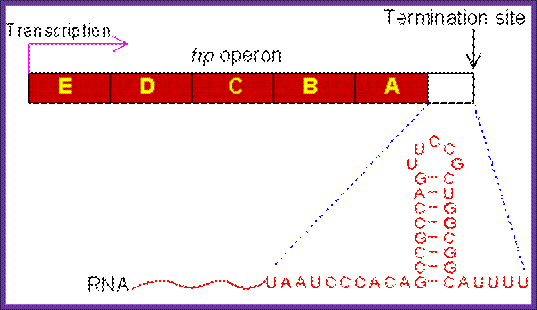
Rho Independent mode: At the end of coding region that is beyond the terminator codon or codons, there are certain sequences positioned which provide a sequence for the transcript to generate a secondary structure that facilitates the termination of transcription.
· One of the structural motifs that the sequence provides is the formation of stem with GC rich sequence and open loop and terminates in 2 to 4 U sequences.
· Transcription termination takes place in two modes one Rho independent manner and the second Rho dependent manner.
www.discoveryandinnovation.com
In some of the transcriptional terminator sequences there are specific sequences rich in Cs; they are little longer and far away from the TER codon.