DNA-Binding Proteins:
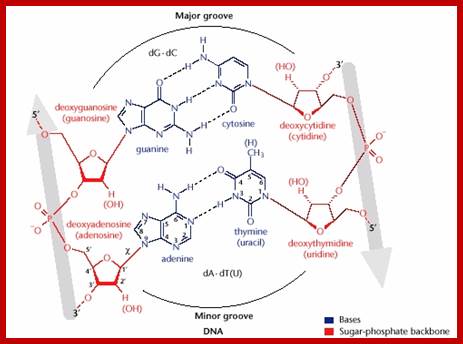
Helical DNA’s backbone is made of sugar-phosphate-sugar linkages, where phosphates in di-ester groups have negative charges. Base pairs A-T and G-C in a specific sequence provide interactive surfaces in the major groove and less commonly in the minor groove. Though the N and O atoms are involved in base pairing to each other by hydrogen bonds, the other groups NH and O groups do provide atomic surfaces for the side chains of amino acids in proteins.
· Proteins have charged amino acids, which can bind to specific bases or to charged phosphates or to both. But the binding to phosphate group in the back bone is not sequence specific but just interactive charges.
The Watson-Crick base pairing in a coiled structure provides charged surfaces all over the molecule, especially the one facing the major groove and the other the minor groove. The said surfaces in the base pairs provide provisions for interaction with charged ionic amino acids found in proteins.
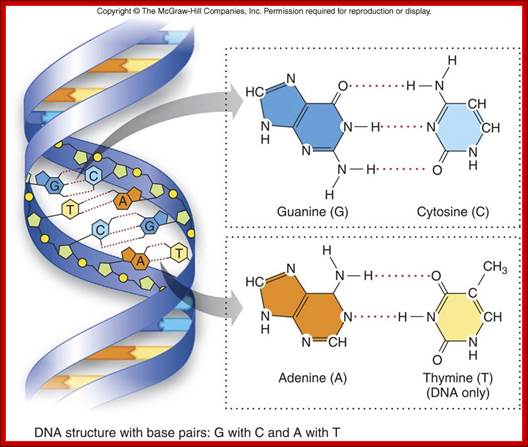
Basic Chemistry and the Macromolecules important to life; DNA structure with base pairs G with C and A with T; http://legacy.hopkinsville.kctcs.edu/

The figure above shows the pairings. The lone pairs of oxygen and nitrogen atoms act as H-bond acceptors for the protons of amine groups of the bases. Adenine and thymine share a pair of H-bonds and Guanine-Cytosine pairs share three bonds. These are the only base-pairs that ever occur in DNA: A-T and G-C; http://www.drcruzan.com/NucleicAcids.html

Nucleic Acids; Complementary base pairing with Hydrogen bonds, though not very strong but provides stable structure. legacy.hopkinsville.kctcs.edu
The positive and negative charge of a given atom depends upon the density of electrons found in that atom. This provides the forces that act with proteins and DNA to interact. Note in all these binding interaction there are no covalent bond formation only Hydrogen bonding, hydrophilic and hydrophobic forces act upon the interacting Protein side chains and DNA bases foun I the major groove and minor groove. The planetary model of the atom is most familiar. It was the first to make good predictions. But it does not work completely. http://thebigblogtheory.wordpress.com/
Each electron in an atom forms a "cloud" around the nucleus. Literally, it is in every possible position (some more than others). The nucleus itself takes the form of proton and neutron clouds, they're just much, much smaller. http://thebigblogtheory.wordpress.com/
Depending upon the number of electrons and protons the atom’s strength either as positive or negative is determined and they attract each other; positive to negative and vice-versa.In the modern picture of an atom, there are no orbits, just a cloud of probability of finding the electron. http://thebigblogtheory.wordpress.com/
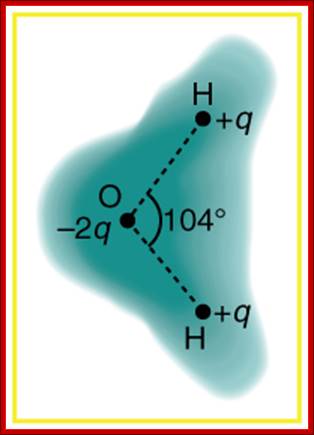
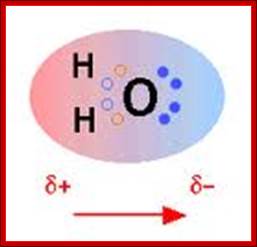
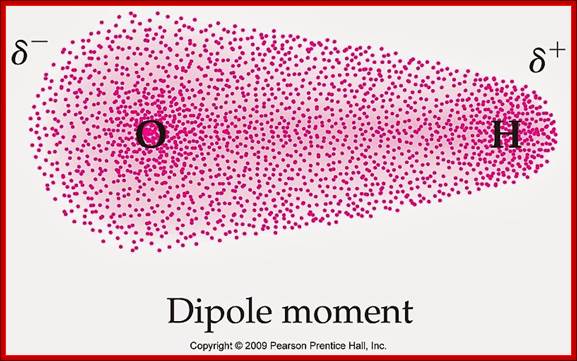
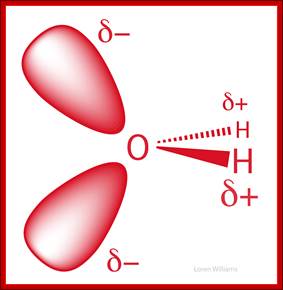
Schematic representation of the outer electron cloud of a neutral water molecule. The electrons spend more time near the oxygen than the hydrogens, giving a permanent charge separation as shown. Water is thus a polar molecule. It is more easily affected by electrostatic forces than molecules with uniform charge distributions; http://cnx.org/content

http://legacy.hopkinsville.kctcs.edu/

Structural features of DNA base pairing; https://www.studyblue.com
Watson-Crick Base Pairing ; ;http://www.boc.uu.se
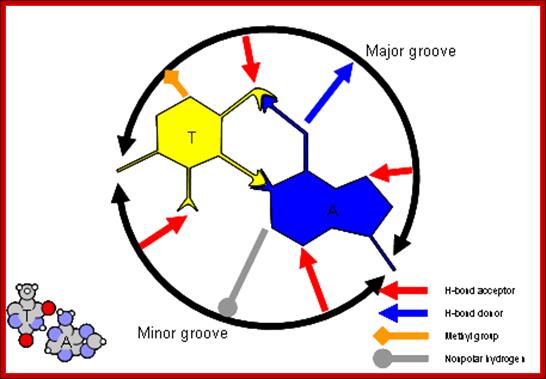
Figure: A-T base pairs provide H bond donors and acceptors; The helical DNA consists of a major groove and a minor groove; they contain a cloud of negative or positive charges, similarly the amino acid side chains also contain clouds of negative and positive charges, thus based on the nucleotide sequences and amino acid side chains, proteins bind in non- covalent fashion, among these Hydrogen bonds form important forces. http://www.boc.uu.se
Chains the major grove provides ionic charges for the binding of amino acid side chains in the protein in non-covalent fashion; however phosphates in sugar phosphate back bone provide negative charge to which positive charged side chains can bind. In this sequence specific bases in the major groove ( in most of the cases) bind to specific protein motifs, the nucleotide sequences provide such interactive domains and forces.
Proteins can interact specifically with DNA through electrostatic, H-bond, and hydrophobic interactions. AT and GC base pairs act as H bond donors and acceptors which are exposed in the major and minor grove of the ds DNA helix, allowing specific protein-DNA interactions.

https://www.studyblue.com
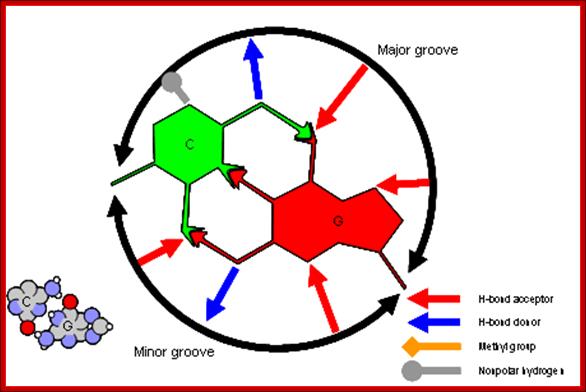
G-C Base pairing: Major groove provides interactive surface charges for interaction with amino acid side chains for non-covalent bonding. Only some proteins bind to minor groove. http://www.clipartbest.com/
· Proteins exhibit greater diversity and greater complex structural organizations than DNA sequences, yet DNA provide base specificities in the form of Nitrogen bases. The R-groups of amino acids, with basic residues such as Lysine, Arginine, Histidine, Aspargine and Glutamine can easily interact with adenine of the A: T base pair, and guanine of the G: C base pair, where NH2 and X=O groups of the base pairs can preferably form hydrogen bonds with amino acid residues of Glutamine, Aspargine, Arginine and Lysine.
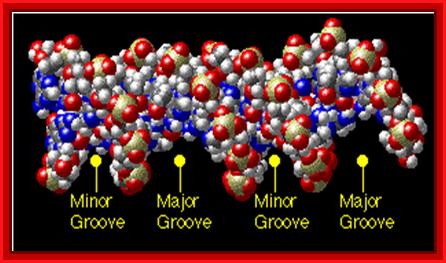
The major groove is 50% wider than the minor groove; http://www.biology.arizona.edu/
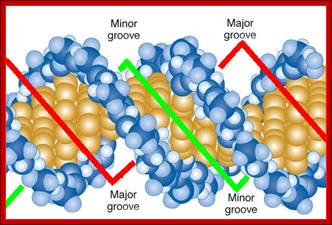
Major and minor grooves; www. Mun.ca
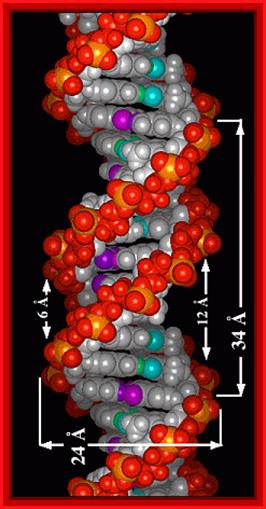
Look at the dimensions of one helical turn, the width of major groove and minor groove and the diameter of the DNA helix; http://elrinconde4eso.blogspot.com
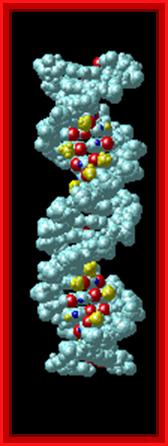

Proteins that interact with DNA often make contact with the edges of the base pairs that protrude into the major groove. The chemical groups on the edges of GC and AT base pairs that are available for interaction in the major (left picture) and minor grooves (right picture), color-coded for different types of interactions, are shown in the pair of illustrations. More information on the recognition elements in the major and minor grooves is found in the views "major groove recogniton" and "minor groove recognition".These pictures are from the views "Major groove recognition" and "Minor groove recognition".http://www.biology.arizona.edu/biochemistry/
Interactive forces that operate in the binding of proteins with sequence specific DNA nucleotides:
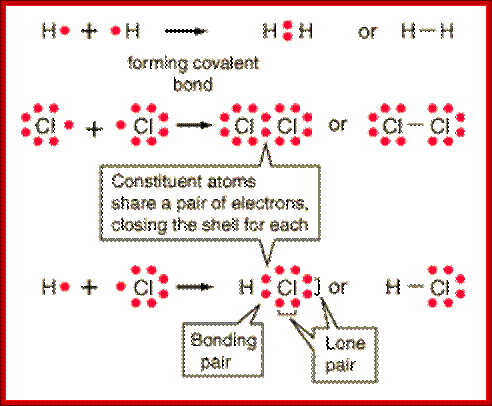
Covalent Bonds-In this type one or more pair of electrons are shared between two reacting atoms
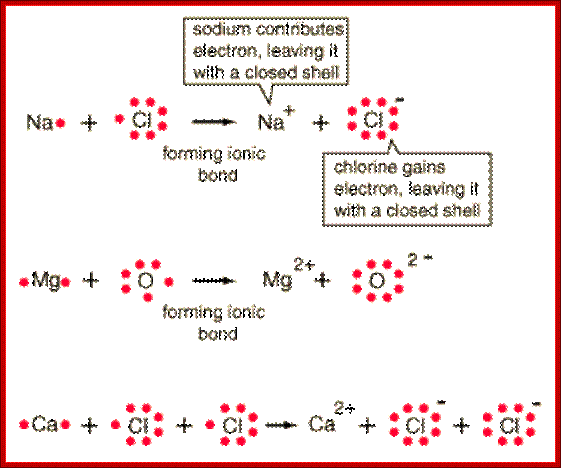
Ionic bonds-One or more electrons are transferred to the other, resulting in positive and negative ions which attract each other,
Metallic, hydrogen and Van der Waals bonds-Water molecules are attracted to each other were and held by electronic forces and such forces are called Va der Waal’s forces, another example is of dipole moment, where positive and negative centers and such polar molecules attract to each other in cohesion, ex. Water molecules; this also contributes to viscosity and surface tension.
http://hyperphysics.phy-astr.gsu.edu/
Chemical Bonding; http://hyperphysics.phy-astr.gsu.edu/

![]()

Diagram of repulsive and attractive electrostatic forces between two hydrogen atoms; https://www.quora.com
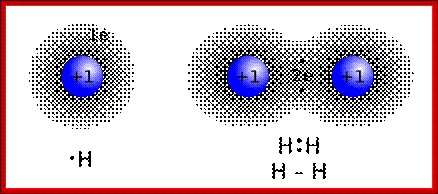
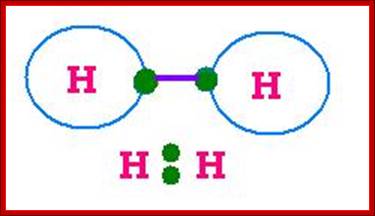
Lewis Electron Dot Structures; On the left is a single hydrogen atom with one electron. On the right is an H2 molecule showing the electron cloud overlap. The shared pair of electrons in the covalent bond can be shown in a Lewis structure by either a pair of dots or a dash.;Involvement of electronic clouds; https://www.ck12.org
World of Biochemistry, http://worldofbiochemistry.blogspot.in/

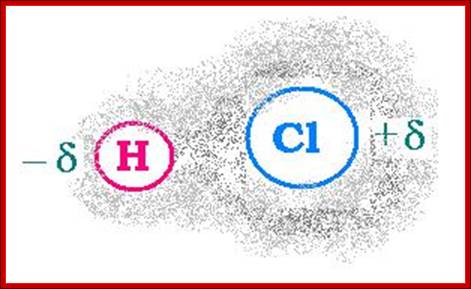
Electron cloud around Hydrogen and Chlorine; http://www.angelo.edu/
HCl; http://scientificsentence.net/
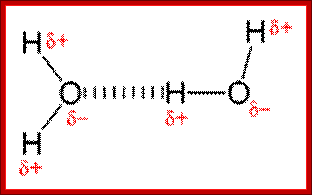
Hydrogen bonding-Noncovalent interactions; http://www.differencebetween.net/
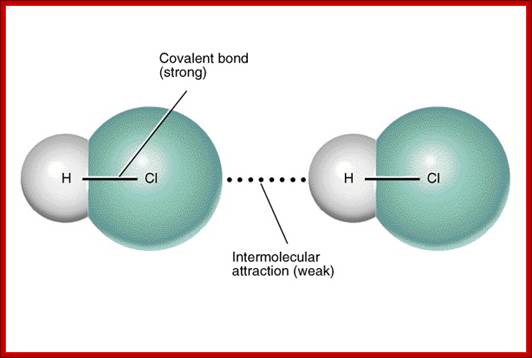
Intermolecular interactions-non covalent; http://www.differencebetween.net/
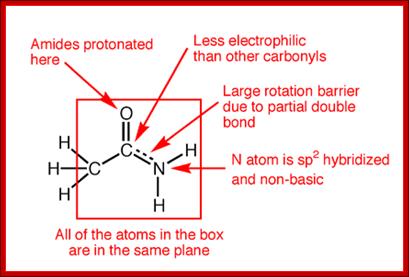
http://iverson.cm.utexas.edu/
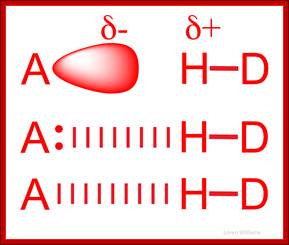
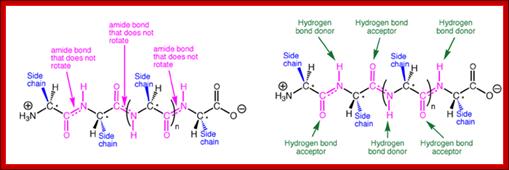
The amide -NH group is a good hydrogen bond donor, while the amide carbonyl is a good hydrogen bond acceptor, allowing primary and secondary amides to form strong hydrogen bonds;
A representative protein alpha helix and beta sheet are shown in the Figure with the trans amide bonds highlighted in magenta and backbone hydrogen bonds shown as green broken lines. The same backbone is shown above each structure for comparison, with side chains removed for clarity. The prominence of amide bonds in the protein backbone is apparent. These secondary structures are thus stabilized to a significant extent by the hydrogen bonding and conformational restriction of the protein backbone imposed by the amides.; http://iverson.cm.utexas.edu/

Acidic (Asp, Glu) and basic (Arg, Lys, His) amino acids take part in charge-charge interactions orionic bonds because they are charged. These tend to be stronger than the charge-charge interactions between amino acids that contain alcohols (Ser, Thr, Tyr) or amides (Asn, Gln) R groups which are polar due to the 'O' and 'N' atoms drawing electrons toward themselves. These also tend to be the amino acids that are better at Hydrogen bonding because they are polarised. Hydrogen bonds occur between the Hydrogen of an N-H or O-H group on one amino acid, and the lone pair on an Oxygen or Nitrogen atom of an adjacent amino acid.
Cysteine residues can form disulphide bridges either within a peptide, or between two polypeptide chains to join them together. These disulphide bonds are covalent bonds and unlike the other types of bond discussed do not form spontaneously in protein-ligand interactions, requiring a catalyst to join and break them.; http://www.fastbleep.com/
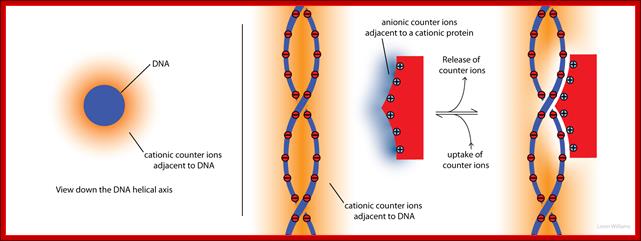
Molecular inyteractions_(left) shows an axial view of DNA, represented as a anionic cyclinder. Cationic counterions (orange shading) surround the cyclinder. The concentration of cations decreases with distance from the surface of the cyclinder. The deeper orange shading indicates more concentrated cations. The panel on the right illustrates how both anionic counterions (blue) associated with a cationic protein, and cationic counterions (orange) associated with anionic DNA, are released to bulk solution when the protein binds to DNA. This release of counterions drives the association (by contributing +TΔS). Molecular interactions (non-covalent); http://ww2.chemistry.gatech.edu/Loren Williams Home,, Georgia Tech
Most of the interactive forces that lead to binding to DNA and Proteins is due to their compatible electronic forces that overlap with one another to form non covalent bonds or say adherence.
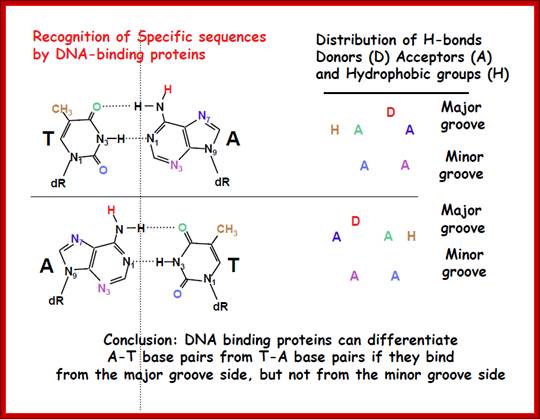
DNA binding proteins can differentiate AT base pairs from TA base pairs if they bind from the major groove side, but not from minor groove side; same with GC base pairs; http://webcache.googleusercontehttp://kc.njnu.edu.cn/ nt.com/
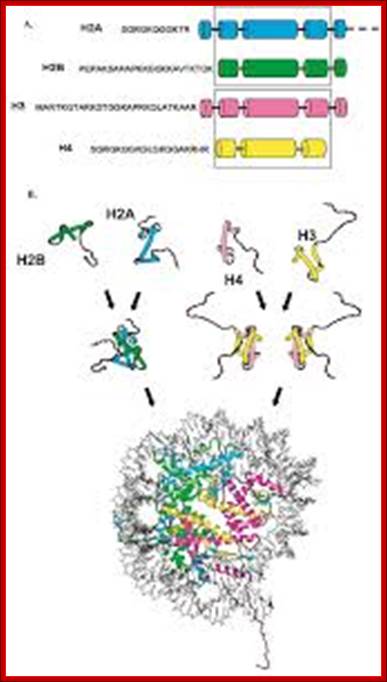
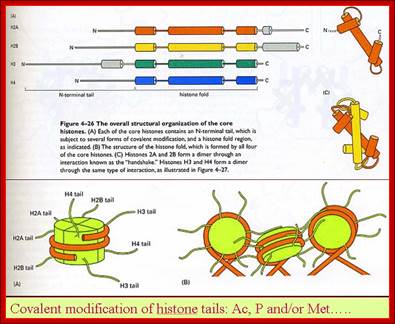
Histones and nucleosome formation: (A) schematic representation of the core histones. The boxes indicate the helixes of the histone fold domain, which is involved in the histone-histone interactions between H2A/H2B, and H3/H4. The amino-acid sequences correspond to the conserved sequence of the unstructured histone tail domain. (B) Individual core histones H2A (green), H2B (blue), H3 (yellow) and H4 (magenta) first heterodimerize to form the H2A/H2B and the H3/H4 complexes. The different complexes can either under different stringencies or with the help of histone chaperones associate together to form the nucleosome composed of a central tetramer of H3/H4 flanked by two heterodimers of H2A/H2B, and wrapped by 146 base pairs of DNA. By Angélique Galvani and Christophe Thiriet; http://www.intechopen.com/
Though hydrogen bonds are weak, but the binding of proteins to DNA over a length at different positions, in a sequence specific manner, can cause conformational change not only in the protein’s overall structure but also in DNA, either in the form of bending of DNA to 80 to 90 degrees or distortion leading to the melting of the DNA or compacting the DNA.
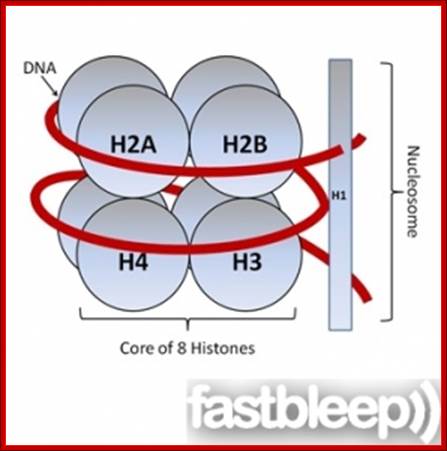
The histones form a complex called a nucleosome, and the DNA is wound around it, rather like beads on a string. These non-specific interactions rely on the formation of ionic bonds between the basic amino acid residues in the histones and the acidic sugar phosphate backbone of the DNA. It does not depend on the base pairs present in the DNA sequence. http://www.fastbleep.com/biology-notes
The key to DNA and protein interaction is specificity. For example binding of a tetramer lac-Z repressor is to 20 to 22 base pair sequence causes repression of gene expression. The genomic length is 4.6 x 10^6 base pairs with so many combination of sequences, yet the repressor binds tightly and binds only to the Lac operator and not to any other regions. This clearly exemplifies strict correlation between the sequence of amino acids in the protein motif and nucleotide sequences in the DNA should be absolute and perfect for the binding. Crystal structures of DNA bound to specific proteins delineate which group of protein binds to which group of bases.
· Bending and opening of the helix into a bubble are very important features for initiating transcription or replication. This is exactly the function of RNAP-TFs complexes, but the complex, if fails to succeed in efficient initiation, it is the other DNA binding proteins, by binding elsewhere provide that function to open and activate the RNAP-TF complex to transcribe efficiently or they can also inhibit transcription. Similarly the binding origin specific protein complexes responsible for Replication.
Proteins, which are specialized in binding to DNA in sequence specific manner, come in different sizes, structures and shapes, you name it; and you have it.
· Basically, some of the structural motifs found in DNA binding proteins have been well characterized. They are characterized as zinc finger proteins, leucine zipper proteins, helix turn helix and helix loop helix proteins and there are few more other modules. They have the said motifs perhaps with certain structural combination of motifs.
All the said types of proteins contain a region to interact with DNA in sequence specific manner (DNA binding domain), which is most important for identifying the site; then they have domains for protein-protein interaction by which they dimerizes or produce multimers. Some of the binding or interacting proteins have specific charged regions, whose contact and interaction with basal transcriptional apparatus can lead to activation of the proteins.
When dimers bind to palindromic sequences; one monomer binds a sequence on one strand and the other monomer binds to opposite strand with the same sequence running in opposite direction; such sequences are palindromic.
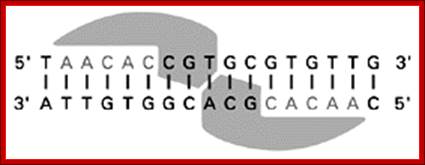
When dimeric proteins bind, one monomer binds a sequence in one strand and the other monomer binds to the opposite DNA strand in palindromic fashion; http://kc.njnu.edu.cn/
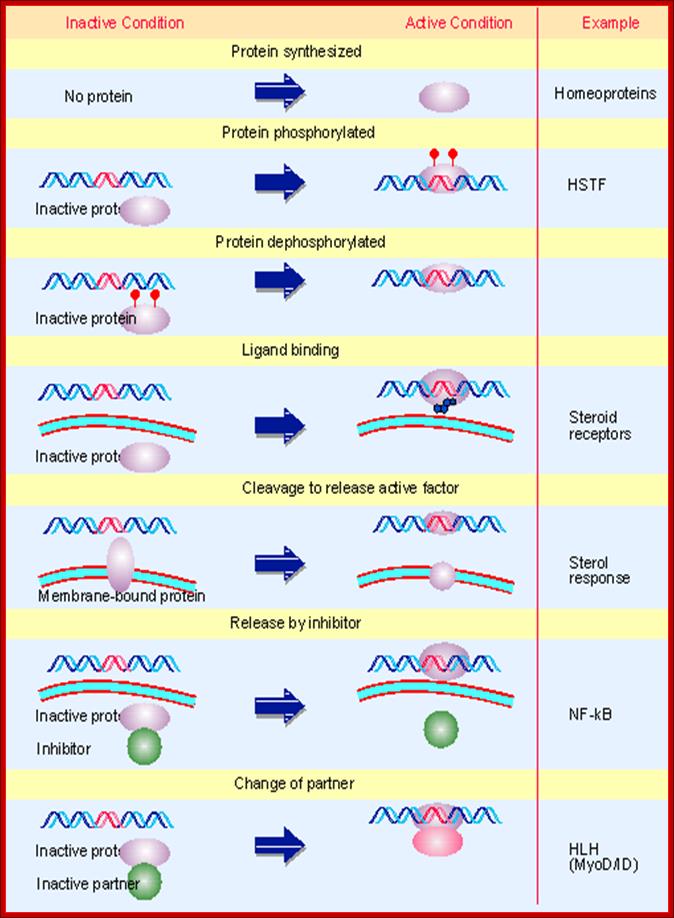
The activity of a regulatory transcription factor may be
controlled by synthesis of protein, covalent modification of protein, ligand
binding, or binding of inhibitors that sequester the protein or affect its
ability to bind to DNA.
Multiple figure. Here is a list of
cellular proteins, which bind to DNA in sequence specific manner
and perform certain important functions listed against the proteins. http://flylib.com/books/en
Helixes turn helix proteins:
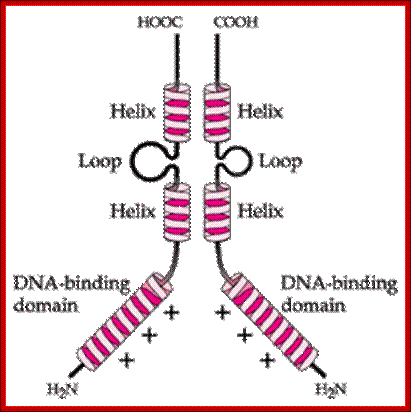
Helix turn helix proteins consist of two short helices of 7 to 9 amino acids long but separated or linked by non helical segment of 3 to 4 amino acids, which are actually responsible for turning the protein by nearly 180^o degrees in space (don’t take it literally); it is an average estimate; ex. Cro (61a.a) and cI proteins of Lambda (~78 and ~260 amino acids long), Lac-Z of E.coli’s lac-Z operon, catabolic receptor protein (CRP or CAP, 224a.a), and few other transcriptional factors.
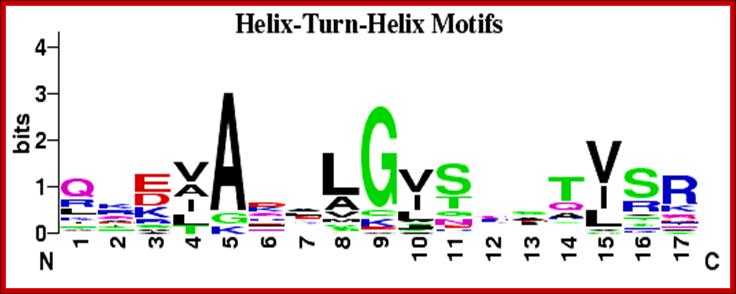
HtH proteins-DNA binding motifs fonjd in gibbs sampling system and below is CAP binding sequences;;http://weblogo.berkeley.edu/

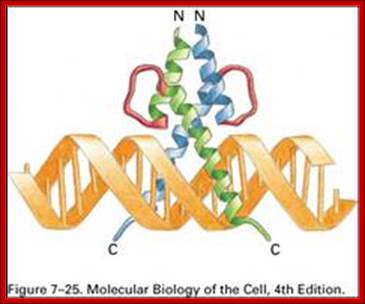
Interaction between the λ repressor dimer and DNA. Each λ repressor contains a helix-turn-helix motif. One of helices fits into the major groove of DNA. PDB ID = 1LMB. http://www.web-books.com/MolBio
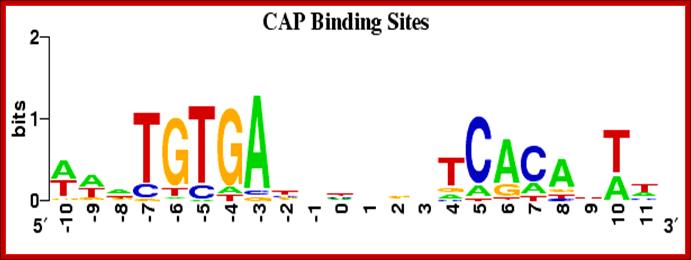
CAP binding HtH protein binding sequences; http://weblogo.berkeley.edu/
· TFs of homeo-domain of Hox- proteins, which are involved in the transcription of genes involved in body development of structures ex. like antp, eve, octa-1 octa-2, Hox and others.
Each of these homeo box proteins have three helical regions separated by a beta turn with 3 to 4 amino acids. Most of them have some similar domains, so they are called homeo-domains. Homeo-domain genes in humans and drosophila are clustered as complexes and they are involved in identity of organs; most of them act as transcription factors.
· One of the helix makes contact and binds to the major groove of the DNA in sequence specific manner (by hydrogen bonding), but the protein also binds to DNA nonspecifically to phosphate groups.
Each of the proteins also has another helix for protein- protein interaction, mostly stems from the helices containing hydrophobic amino acids on one surface of the protein.
· Many of the proteins also have nonstructural sites for the binding of substrates.
In the case of lactose repressor protein, by binding it does not allow RNAP to move forward thus it represses the expression of the gene.
· But the CAP protein when it is bound by cAMP binds to specific region of the promoter and activates the gene expression by means of contacting the RNAP-sigma complex. In this reaction one of the loops of the CAP protein interacts with one of the subunits of enzyme complex.
These proteins bind to the promoter region in sequence specific manner and interact with basal transcriptional apparatus by looping of the DNA by protein-protein interaction and the contact with basal transcriptional apparatus to activate the enzyme complex.
There are variations among HtH proteins such as- Di HTH, Tri Helical HTH, Tetra helical HtH and Winged HtH
Helix loop Helix proteins:
Proteins, with two helical segments, consisting of 12 to 15 amino acids that are separated by nonstructural, randomly organized segments, are generally named as helix loop helix proteins with the helix-loop-helix motif. Substantial number of these proteins are found to be Transcription factors or and regulative factors.
· These proteins have excellent features for dimerization, through hydrophobic amino acids or amphipathic helix-helix interactions in each of the helical segments.
Many such proteins are found in E12, E 47, Myo-D, My f-5 and myc proteins (Myc is the proto-Onco gene). In fact they are classified based on the type of tissue in which they express.
· Group A: ubiquitously expressed proteins include mammalian E12 and E47 and Fly-da.
· Group B: Expressed in tissue specific manner in mammalian tissues, Myo-D and Fly Ac-s.
· Myc class: It is a class by itself and its partner proteins are different and targets are different.
This protein shows Helix loop Helix domains plus zipper; http://www.utdallas.edu/
Helix-loop-Helix protein model (forrás):
http://8e.devbio.com/images/ch05/0504fig5.gif
The proteins, at C-terminal or at N- terminal, depending upon the kind of protein, have unpaired regions with more basic amino acids in specific positions. The basic residues with charges are required for the binding of proteins to DNA in sequence specific mode. Such proteins are called bHLH proteins (here b= basic amino acids).
· H-L-H proteins can dimerize with similar kind or of different kind proteins, to form either Homodimers or Heterodimers. Such proteins bind to DNA using one side of the protein and dimerize using the other side of the protein.
E12 and E47 bind to immunoglobulin enhancer regions. Myo-D (Myogenic) and myf-5 is involved in myogenesis. Myc gene is the counterpart of the Oncogene, involved in growth regulation; and the protein has such motifs.
· There are a group of gene products which have such motifs and they are involved in the development of Drosophila.
Some HLH proteins lack basic amino acids, e.g. IG proteins. Some such proteins, which lack basic amino acid ends, but have few Prolines in their place. These proteins play an important role in developmental process and they use what is called combinatorial dimrization.
Zinc finger proteins:
The zinc-finger proteins are globular proteins, but presents finger shaped motif to bind DNA, in sequence specific manner, and it is referred to as Zinc-Finger motif. There are innumerable examples of such proteins, to quote few, eg. Sp1, TFIII-A and ADR 1.
The name, Zinc finger protein, has derived from the kind of loop it generates when a covalent bond forms between a single zinc metal ion with 2 cytosine on one side and 2 Histidine on the other side either side of the polypeptide or it can be between 4 cys two each on either sides. Such a structure produces tetrahedral form. Each of the fingers is about 23 amino acids loop, if there is more than one finger the linker region between two loops is seven to eight amino acids.
· In most proteins with the zinc fingers, the N terminal region after cysteine has beta sheet and the right side of the loop has alpha helical structure.
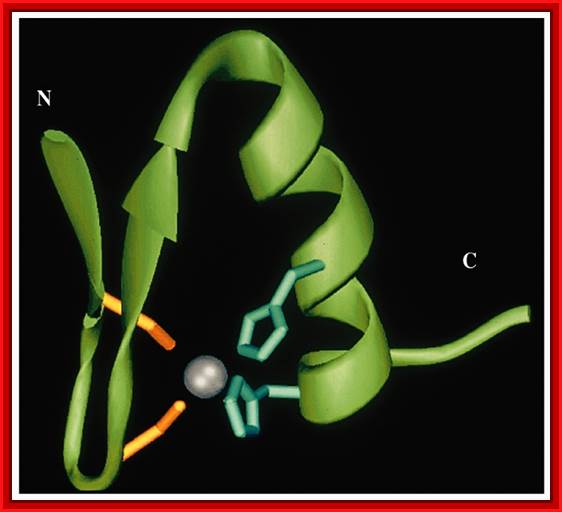
This diagram shows Helix and beta sheets brought together by Zinc ion bound to two Cysteine and two Histidine side chains. http://cbm.msoe.edu/
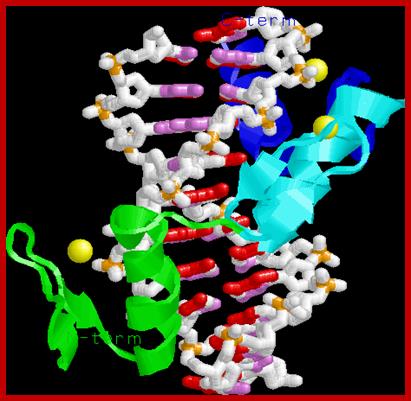
Note the three alpha helices pointing into the major groove, recognizing 3 bp each. The Zn binding domain is a structural element for protein folding, not directly involved in DNA binding; This is a nice ZiF protein diagram shows the components; http://www.biochem.umd.edu/
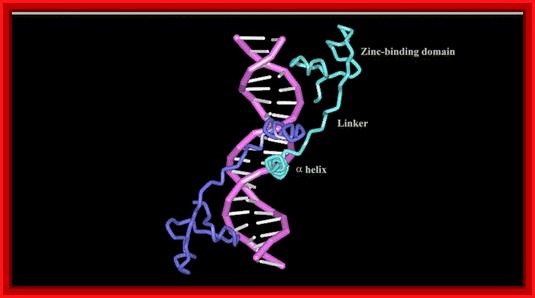
The X-ray structure of the GAL4 in the complex of 19bp DNA; www.nostatic.com/proteins/zincfinger/ZIncBinuclear.html
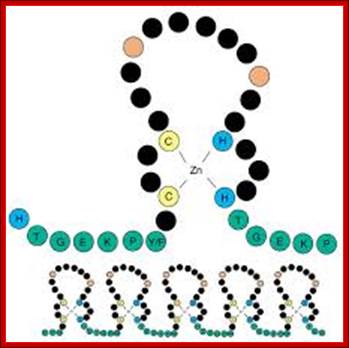
Schematic diagram of a C2H2 zinc-finger motif. The paired cysteines (C) and histidines (H) that bind the zinc ion are shown in yellow and blue, respectively. The linker sequence, shown in green with its consensus sequence in the single-letter amino acid code, frequently joins adjacent fingers. This is apparent in the lower panel, which shows the typical arrangement of fingers in a C2H2 ZNF protein. The two large hydrophobic residues, which are also structurally important, are shown in red. The black residues are not structurally important and include those responsible for contacting DNA during sequence-specific binding [16]. The precise number of 'black' residues between the cysteines, histidines and on the loop may vary; Knight and Shimeld Genome Biology 2001 2:research0016.1-research0016.8 doi:10.1186/gb-2001-2-5-research0016
|
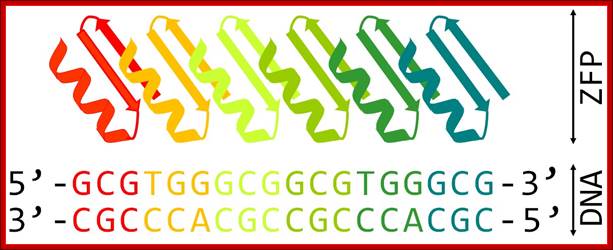
Schematic representation of six-finger ZFP attached to its DNA target. Each zinc finger motif composed of ββα fold binds specifically to 3 bp of DNA; http://openwetware.org/ |
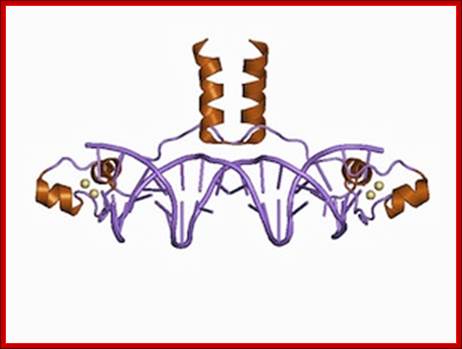
Ribbon model (cartoon) of Zn2Cys6 zinc finger dimer bound to DNA (Source: Wikimedia Commons.) Transcription factors motifs-Grant Jacobs; http://sciblogs.co.nz/

Zn
finger binding specificity
*Cannot predict this from sequence alone!
*Despite conservation of structure it is not easy to predict which amino acids
in the recognition helix contact DNA and determine sequence specificity.
-e.g.; http://web.sls.hw.ac.uk/teaching/Derek_J
The number of fingers varies from protein to protein, ex. Sp1 protein has 3 zinc fingers, with 2 cys and 2 his. Each of them have beta on the left side of the finger and an alpha helix on the right.
· Each of the fingers bind to each turn at major groove, they bind by consensus sequences.
Many Drosophila transcriptional factors have these zinc finger motifs in their protein domains.
· In the case of Xenopus laevis, the transcriptional factor called X-fin has only one zinc finger protein, but the protein is folded into a globular structure as in other Z-finger proteins. However this protein has two successive beta sheets and one alpha helix. Two cys and two link the two beta sheets and a single alpha helix his by single zinc ion. In this the one cys from each of the beta sheets are involved in bond formation.
Zif 268 from mouse has three zinc fingers and each of them binds to a major groove and binds to 3 bp; in this binding process, Arginine and Guanine are involved in hydrogen bonding.
· It is interesting to note some of the Zinc-finger motif containing proteins can bind specifically to stem loop structure of certain RNA. Ex. TF-III-A which has zinc finger motifs bind to internal promoter region and also binds to 5s RNA.
A translational Initiator factor eIF2 (has zinc finger motifs). Mutations in this gene influence the recognition of initiator codons.
· Retroviral capsid protein binds to genomic RNA using its (Capsids) zinc finger motifs.
Steroid receptor has 4 cys bound to a central Zn, i.e. 2 cys and 2 cys pattern. They bind to short palindromic sequences.
· Gluco-corticoid and estrogen receptors have 2 zinc fingers each. The fingers of Gluco corticoid receptor form dimers.
Depending upon the protein, the number of fingers in a protein range from one to ten.
· The tetrahedral structure can develop either by binding of 2Cys and 2 His or 4Cys.
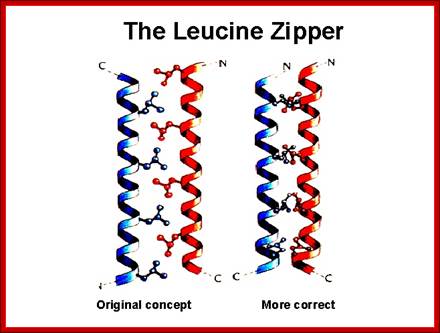
Leucine Zipper proteins:
These proteins have a stretch of amino acids rich in hydrophobic leucine and they are on one side of the right-handed helix.
· The repeat of Leucine is for every 3.5 residues per turn and this pattern repeats for every seven amino acid residues.
To illustrate this with an example, take a. b. c. d. e. f. g. h. as a sequence of amino acids as one segment of the helix, where a and d are hydrophobic, then one finds hydrophobic amino acids with hydrophobicity on the same side at every 3.5 amino acids, which is actually one turn of the helix.
· If two such chains having the same type of helices and hydrophobicity, they can easily interact with one another by protein-protein interaction and form coiled coils.
Two right handed helices, when coil to each other they form left handed coiled coils, but helically coiled region at the base bind to certain DNA sequences such as CAAT box of the rat liver and also to the core of SV 40 enhancer.
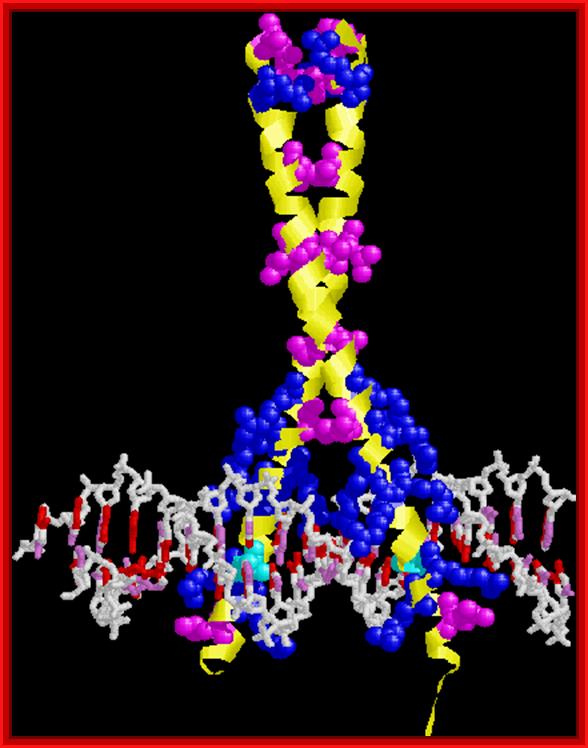
GCN4. Note leucine zipper, basic region, recognition of adjacent major grooves with "chopstick" motif. http://www.biochem.umd.edu/biochem

Leucine zipper protein on opposing amphiphilic helices; https://www.researchgate.net
Control of Genetic Systems in Perokaryotes; http://www.uic.edu/
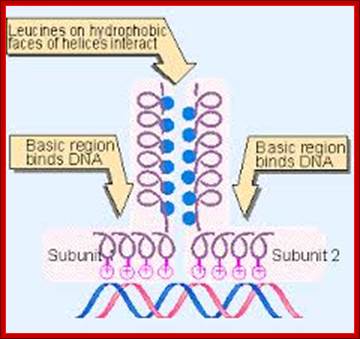
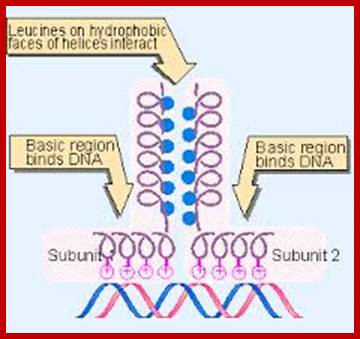
The leucine zipper is a stretch of amino acids rich in leucine residues that provide a dimerization motif. Dimer formation itself has emerged as a common principle in the action of proteins that recognize specific DNA sequences, and in the case of the leucine zipper, its relationship to DNA binding is especially clear, because we can see how dimerization juxtaposes the DNA-binding regions of each subunit. The reaction is depicted diagrammatically in Figure; Regulation of Transcription; http://genes.atspace.org/
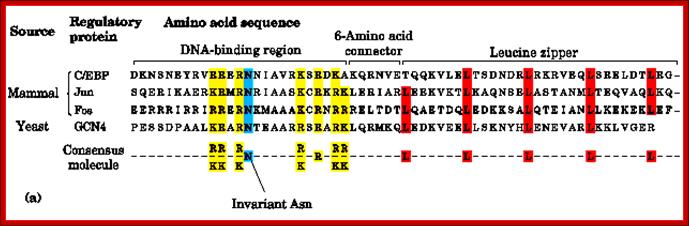
Most of the LZ proteins have amino acid sequences with specific functional motifs such as DNA binding, connector, Leucine zipper and an invariant Asn next to DNA binding domain; http://www.bioinfo.org.cn
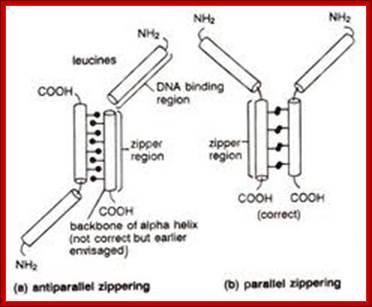
This diagram shows the view from the top of the coiled coils showing the opposing hydrophobic residues providing the force for interaction and binding to each other. http://genes.atspace.org
Bioencyclopedia;
http://www.eplantscience.com/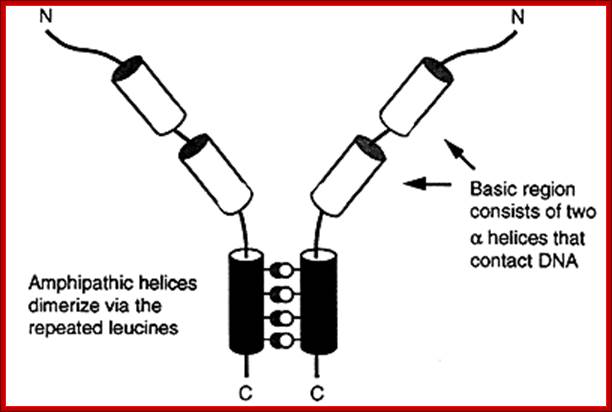
Regulation of Gene Expression; http://www.gravitywaves.com/chemistry

Leucine rich repeats; It is structural motif; http://en.wikipedia.org/
Rat liver TF called C/EBP, an enhancer binding protein, binds to a response element CAAT sequences. In this a 28-a.a region has Leucine at every 7th position. This causes the dimrization of the proteins into coiled coils called zipper proteins.
Yeast’s GCN4, a transcriptional factor, and many proto- Oncogenes encoded proteins have such structural domains. It is not true for all such transcriptional factors. The GCN4 b. a Zipper protein consists of 281 residues. Its first 30 residues contain 3.6 or 3.5 heptad repeats. They coil into an 8 amino acids turn alpha helix.
Two such proteins can dimerize. Its N-terminal region remains open region, consisting of 16 amino acid residues, which are basic in nature. They engage in sequence specific DNA binding.
· But its C-terminal region forms a coiled-coiled helix as a Leucine-zipper pattern.
Such Leucine zipper proteins can sponsor both homo and hetero dimer formations.
· The C-EBP, an enhancer factor, binds to CAAT box of the rat liver and also SV 40 core enhancer sequence, and the protein has four such segment repeats, where each Leucine is found at 7th residue, that amounts to eight turns and 3.5a.a per turn.
An enhancer binding protein called AP 1 was found to bind to the enhancer region of the SV40, they are ‘jun’ and ‘fos’ factors that form heterodimers called AP-1 factor. They contain 5 such Leucine repeats.
Below some DNA binding proteins in their 3-D glory are shown for a sheer pleasure see them in their intimate binding positions.
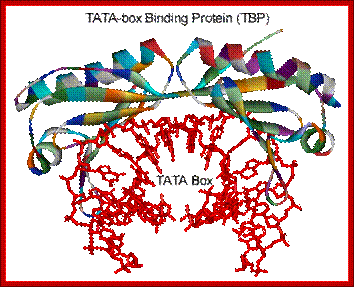
Transcription Mechanism in Eukaryotes; Structure of the human TBP core domain complexed with DNA as determined by x-ray crystallography. The TBP binding bends the DNA into curved structure and facilitates the opening of the double stranded into single strands;, Binding of the TBP bebds the away from it as if it is crushing. The DNA includes the TATA element. PDB ID = 1CDW. http://www.web-books.com/
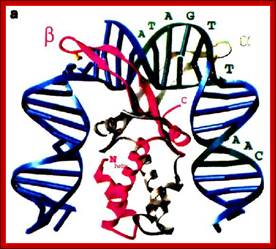
This another protein model showing how it is bound to DNA; the binding bends DNA; The binding of Protein bends the DNA around it http://genome.cbs.dtu.dk/
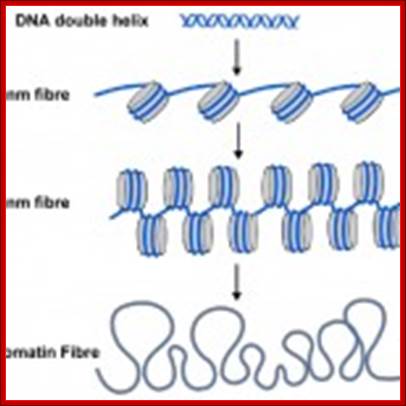
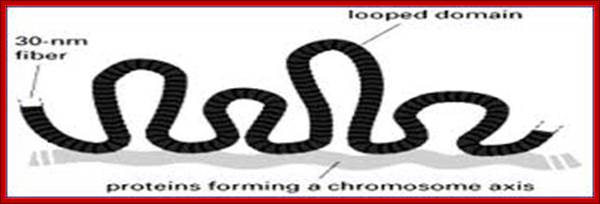
The familiar DNA double helix (blue) is wrapped around nucleosomes (grey cylinders) in cells. The string of nucleosomes can be coiled into a thicker filament, called the 30 nm fibre and this can be further coiled into a still thicker chromatin fibre. When genes are switched on their nucleosomes are more uncoiled like the 10nm fibre.;Scaffold proteins binding to SARS region makes the DNA looping; University of Newcastle-Upon-Tyne, UK; http://bscb.org/learning-resources.

How DNA determines the chromatin fiber geometry;;We postulate that the major driving force for fiber formation is the dense packing of the underlying DNA-protein spools, the nucleosomes, allowing for fibers with four possible diameters. G. Lanzani and H. Schiessel; http://iopscience.iop.org/
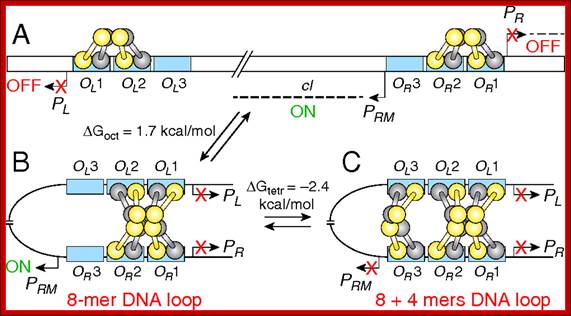
(A) Promoters (PR, PL, and PRM); operators OL (OL1, OL2, OL3) and OR(OR1, OR2, OR3) are in blue rectangles; CI dimers (one monomer is shown in yellow, the other is in gray). The bent arrows show the transcription start points of promoters. The dashed line indicates transcripts from PR, PL, and PRM. The cI gene is transcribed from PRM. (B) DNA looping and octamer formation (8-mer) by CI tetramer binding to OL1 ∼ OL2 interacts with that at OR1 ∼ OR2. (C) Octamer and tetramer (12-mer) of CI binding to OL and OR. Red X means promoter is turned off. ΔGoct and ΔGtetr values for octamer and tetramer loops stability are from Zurla et al. (25), measured on linear DNA under different conditions. See refs. 7, 18, 20, and 59 for models on CI regulation..Binding of lambda cI repressor proteins in sequence specific manner and protein-protein interaction leads to looping of the DNA; http://www.pnas.org/
Gene Expression; Ecitable; http://www.nature.com/
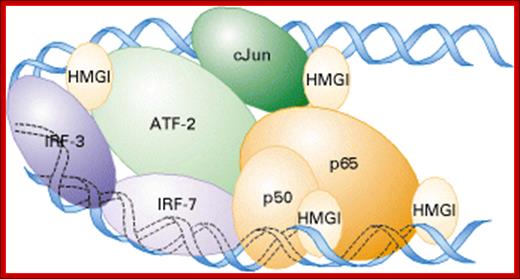
Model of the enhancesome that forms on the b-interferon enhancer. Heterodimeric cJun/ATF-2, IRF-3, IRF-7, and NF-kB (a heterodimer of p50 and p65) bind to the four control elements in the ≈70-bp enhancer. Cooperative binding of these transcription factors is facilitated by HMGI, which binds to the minor groove of DNA;Regulation of Transcription Initiation, Molecular cell biology; http://www.1cro.com/
Recent data base entries (2785 or more) with respect to transcription factors and other DNA binding or otherwise are grouped into many super class proteins.
Single stranded DNA and RNA Binding Proteins:
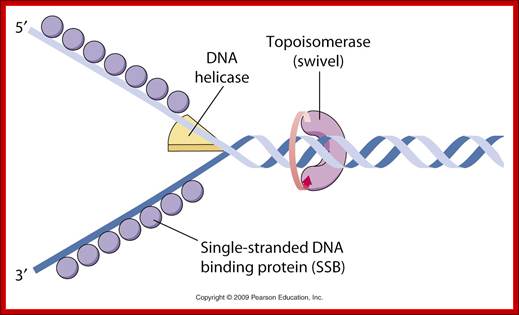
http://carolc13.wordpress.com/
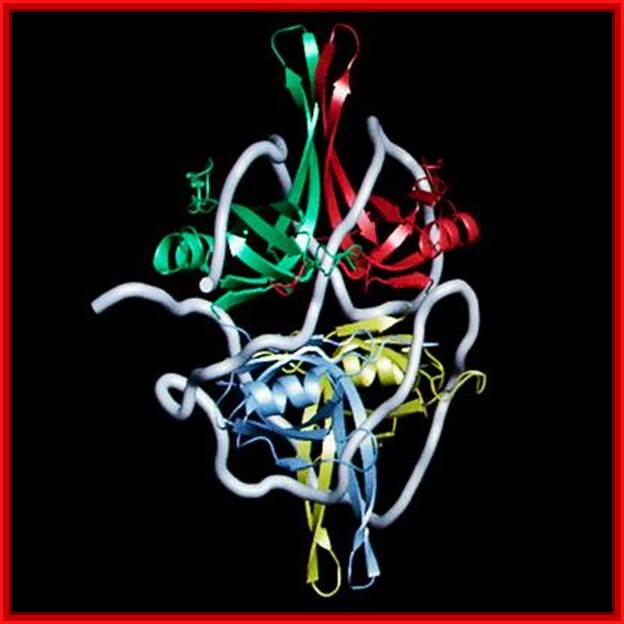
Protein bound to helix ;L. Brian Stauffer, ;http://esciencenews.com/
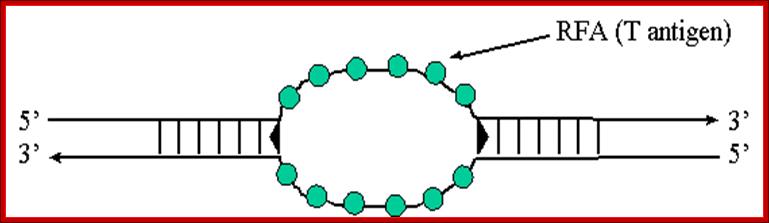
RFA proteins; http://www.ufrgs.br/
RNA binding: Most of the mRNAs are associated with proteins during processing and translation, almost all nc RNAs are also associated with specific proteins without which they cannot function.
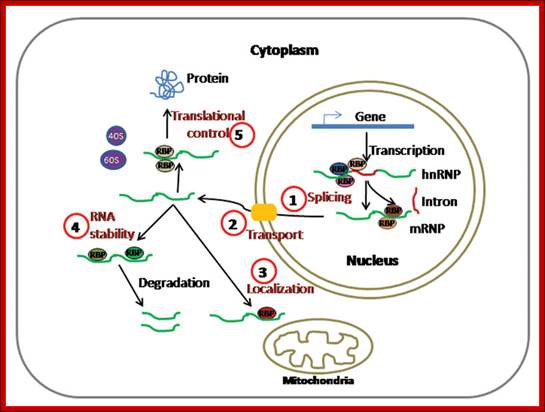
Dynamics of
Post-Transcriptional Regulatory Network Directed
by RNA-Binding Proteins; Sarath Chandra Janga and Nitish Mittal,http://www.landesbioscience.com/
Some of the TFs have been classified into super classes:
Superclass: Basic domains:
Leucine zipper (bZIP),
Helix loop helix (bHLH),
Helix-loop Helix/leucine zipper (bHLH-Zip),
NF-1,
RF-X
bHSH,
Super class: Zinc-coordinating DNA binding:
Cys4 ZIF-nuclear receptor,
Cys2His2zif domain,
Cys6 cysteine-zinc cluster,
Zinc fingers of alternating composition.
Super class: Helix-turn-helix:
Homeodomain,
Paired box,
Fork head/winged helix,
Heat shock factors,
Tryptohan clusters,
TEA domain,
Super class: Beta-scaffold factors (with minor groove contact).
RHR (Rel homology),
STAT,
P53,
MADs box,
Beta barrel alpha helix-transcription,
TATA binding proteins,
HMG,
Heteromeric CCAAT factors,
Grainy head proteins,
Cold shock domain factors,
Runt,
Super class; other TFs,
Copper first proteins,
HMGI(Y),
Pocket domain,
E1A like factors,
AP2/EREBP related factors.
DNA binding proteins:
The number and characters of DNA binding proteins that bind directly in sequence specific manner and proteins that bind to DNA bound proteins are many in numbers that can run to several thousands.
Eucaryotes and Prokaryotes :
Helix Turn Helix proteins:
Homeodomian Protein (Drosophila Hox genes),
RAP1 protein (repressor n activator protein of telomeres),
TFIIB family, include winged HTH members,
IFN regulator proteins,
Transcription factors (a family of proteins),
UBX-homeodomain,
POU domain,
Fork head/winged,
HSPs
Helix loop helix proteins:
Class A factors, MyoD, Archaeate- cute, Tal/Twist/Hen.
Helix loop helix –Leucine zipper proteins:
SREBP, USF1 and USF2,
c-Myc.
Zinc finger Proteins: cys4- Zn finger, cys2-His2 zinc factors
Beta-beta-Alpha zinc finger proteins,
Nuclear receptor proteins (a family),
Loop-sheet-helix type proteins,
GAL4 type proteins,
GATA-factors,
Steroid hormone receptors,
Thyroid hormone receptors,
TFIIIA
Leucine Zippers:
Basic zippers,
Zipper type (helix loop-helix),
AP-1, CREB, C/CREB,
Plant G-box proteins
Beta scaffold factors: RHR Rel homology region),
NFkB,
Ankyrin,
NFAT, STAT, p53, MADS box proteins,( SRF),
CCAAT factors
TBP, HMG box proteins- SOX and SRY,,
Other alpha helix:
Cre recombinase,
E2 protein of Papilloma virus),
Histone family of proteins,
HMG family of proteins,
MADs box family proteins.
Beta-sheet:
TATA box binding protein (TPB),
Beta-hairpin/ribbon type:
Arc receptors,
Met repressor proteins,
Tus replication Ter proteins,
IHF (integration host factor),
Transcription factor t-domain protein,
Others:
Rel homology region proteins,
STAT.
DNA binding enzymes:
C-methyl transferase,
Endonuclease V proteins,
DNA mismatch endonucleases,
DNA pol I,
DNA pol7,
Most of the restriction endonucleases,
Dnase I,
DNA pol beta,
Uracil glycosylase,
Homing endonucleases,
Topoisomerases.
Transcription factors:
That bind directly or indirectly to DNA, classified according to their functions: 1. Constitutively active-they are present in all cells and all times. E.g Sp1, NF1, CAAT binding proteins,
2. Conditionally active- they are required for activation; they can be- Developmental specific (cell specific),- GATA, HNF, MyoD, Myf 5 , Hox, winged Helix.
Signal dependent- extracellular ligand dependent, or intracellular ligand dependent (SREBP, p53,), cell membrane receptor dependent; resident nuclear factors (CREB, AP1), or latent cytoplasmic factors, when activated they move into the nucleus- STAT,R-MAD,NF-kB, Notch, NFAT etc.
Super class: basic domains:
Based on their structural (domains) and functional features they have been classified different classes and families.
Leucine zipper factors; Helix loop helix factors; Helix loop helix/leucine zipper factors, NF family, RF-X family and bHSH family.
Super class: Zinc binding;
Cys 4 ZFs, cys4 zinc finger, diverse zinc finger, cys2-His2 zinc finger, cys6cystein zinc clusters, Zinc fingers alternating composition and their related members.
Superclass: Helix turn helix- Homeodomain, paired box, Fork head/winged helix, Hsps, Trp clusters and TEA members.
Superclass: Beta-scaffold factors- RHR, STAT, p53, MADS box, beta-barrel alpha helix, TATA (TBP), HMG box, heteromeric, grainy head, Cold shock domain Runt and others.
0 superclass TFs:
HMG like, Copper fist, Pocket domain, E1A, AP2, EREBP, ARF,ABI and RAV.
· There are few more proteins that bind to DNA-
· Single-strand binding protein
· ssdNA binding Proteins ,
· RNA binding proteins,