Gene Expression II7- Response Elements
Factors and Cell Types:
Gene expression in eukaryotes is controlled by a variety of mechanisms that range from those that prevent transcription to those that prevent expression after the protein has been produced. The diagram below shows five kinds of general mechanisms that can be used.

Transcriptional-These mechanisms prevent transcription. Posttranscriptional-These mechanisms control or regulate mRNA after it has produced. Translational- These mechanisms prevent translation. They often involve protein factors needed for translation. Posttranslational - These mechanisms act after the protein has been produced
Gene Expression in Response to:
Biosystems live and perpetuate within an ambient environment, whether it is inter cellular or intracellular or it can be external.
· The kind of stimulus provided can be in the form of chemicals, change in the pH, change in the ionic concentration, change in the nutrients, change in the temperature, light, infectious attack from other organisms and many other situations like cell to cell contact and others; all of them act as stimulants which can have inducible effect and some have repressive effects.
· A particular inducer for one gene can be a repressor for the other gene. Here inducer means a factor that induces the expression of a gene.
· All these effects are transduced through certain factors ultimately to the gene.
· There is a time lapse between the time of stimulus application and the time at which response is manifested.
· Genes are endowed with genetic facility to respond, by having a variety of sequence modules, located at specific distance from the start site and beyond start site.
· Each module contains a specific sequence, located at specific position.
· There may be several modules, or combination of modules, positioned in combination.
· The kind of modules, location of them from the start and combination of them are gene specific and stimulus specific.
· Besides having response elements in their promoter regions for stimuli, cells do produce specific factors for specific genes and for specific response.
· The cellular factors may be present in cytosol, or in the nucleus or on the cell membranes.
· Whether the factors are in fixed position or free, they have many roles and many steps before they act on genes and express them specifically.
· The stimulus, whatever kind it may be, first it should contact or impinge upon the cell or enter into the cell, by whatever means possible.
· Then they bind to certain factors, or bring about changes in the factors to activate or inactivate them, and perform certain reactions that ultimately lead to the entry of them into the nucleus.
· In the nucleus they bind to respective DNA sequence elements. Once they bind they contact or interact with the basal transcriptional apparatus elements (there are several of them, which are tissue specific) and activate or modulate the RNAP enzyme to initiate transcription.
· However the efficiency and the rate of transcription depend upon the kind of interaction they do.
Response Elements, Factors and Cell Types:
In addition, eukaryotic gene contain sequences further upstream called response elements bind specific proteins (such as CREB or cyclic AMP response element binding protein) to further control gene transcription.
Few of the response elements and factor that binds is shown;
|
Module |
Factor; size; type |
Sequence |
|
Cell Type
|
GAL4 |
GAL4; 89;LZ/HLH |
>CGGAGGACT <GTCCTCCG |
|
Yeast, upstream activator |
|
GCN4 |
GCN4; 17kd; LZ |
ATGA C TCAT |
|
Activator cum enhancer, |
|
HSE |
HSTF; 93kd |
CNNGAANNTCCNNG (CNNGAANNTCCNG) |
|
Yeast, heat shock |
|
GRE |
GR receptor; 94kd; ZF |
TGGTACAAATGTTCT (>5’TGTTCT<AGAAC3’ (N) |
|
Glucocorticoid, General |
|
TRE |
TREF, 39kd |
TGA C TCA |
|
Tumor inducing |
|
SRE |
SRF; 52kd |
GGATGTCCATATTAGGACATCT (CCATATTAGG) |
|
Serum factor |
|
ERE |
EGRF; 60.8kd |
TGACCT N AGGCTCA |
|
Estrogen |
|
MRE (cadmium) |
MRF; |
CGNCCCGGNCNC (GGNCCCGGNCNC) |
|
Metal-cadmium |
|
PRE |
PRF; 104kd |
|
|
Progesterone |
|
T3RE |
TREF; 44.8kd |
TGACCT N4 TGACCT |
|
Thyroxin |
|
Vitamine D Retinoic acid RXR |
VDREF RAREF; 47.2kd RxRf |
AGGTA N3 TGACCT (ACGTCATGACCT (TGACCT N5 TGACCT) TGACCT>n1TGACCT>
|
|
Vit-D Retinoic acid 9cis Retinoic |
|
Phorbol |
TRE |
TGACTCA |
|
Phorbol ester |
|
Antioxidant |
ARE |
GTGACTCAGC |
|
Antioxident |
|
Hypoxia |
HRE |
CCACAGTGCATACGT GGGCTCCAACAGGTC CTCTCCCTCCCATGCA |
|
|
|
Peroxisome Proliferator Activated Receptor (PPAR) |
PPRE |
|
PPAR |
|
|
Steroid (general) (progesterone, androgen, mineralocorticoids, glucocorticoids |
|
AGAACAxxxACAAGA (inverted repeat) |
|
|
Heat shock gene expression:
· Both prokaryotes and eukaryotes contain specific kind of heat shock response genes, which produce heat shock proteins.
Cells of PK and EK including humans, if subjected to elevated temperatures and or severe environmental, conditions cells respond by changing their pattern of gene expression.
· Cells start producing few specific heat shock proteins, called HSPs; they are produced at higher level, while much other cellular gene expression may be down regulated.
· In Prokaryotes specific sigma factors become active and they in turn, switch on several heat shock and their related genes.
· Among eukaryotes, especially, Drosophila has been subjected to intensive studies.
· When 11th day larvae of this insect are subjected to elevated temperatures, with in minutes, specific changes, in the form of gene expressions, has been observed.
· At several loci, in the polytene chromosomes, chromosomal puffs develop. With time some puffs regress and some puffs develop.
· Chromosomal puffs are the sites at which chromatin has opened up to actively transcribing DNA loops, which is free from nucleosomes.
This is a par excellent example for large scale expression i.e. chromatin has to open up to provide DNA free from other chromatin proteins only to be bound by gene activating complexes.
The system is ensured of proper and preferential translation of the heat shock mRNAs.
· The HSPs assure protection to several cellular proteins.
Such gene loci are called heat shock loci. Seven such heat shock loci have been identified in Drosophila.
|
|
|
|
· They are named as HSP 28, 23, 1, 26, 22, 4 and 5. All of them are clustered in a 15-kbp region called 67B locus in the insect chromosomes.
· Most of the HSPs are chaperone proteins which assist deformed proteins into stable ones.
· There are two types of Heat shock proteins one chaperones and the other Cheronins.
Heat Shock Gene Cluster
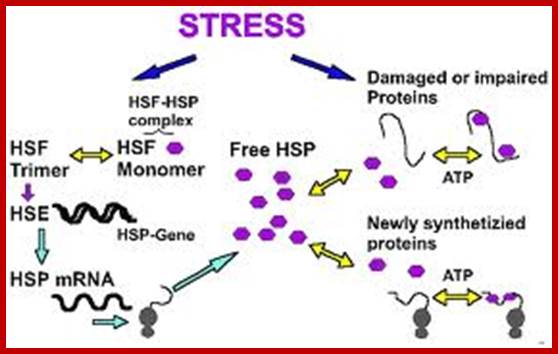
Stress-dependent regulation of HSP-gene expression by HSF (left), and functions of HSP as chaperone of denatured proteins or of nascent proteins (right), and the feed-back regulation of HSP-HSF-complex,: Yuefei Liu and Jürgen M. Steinack; http://www.bioscience.org/
Promoter Elements of HSP 70 Gene
Heat shock genes are expressed in response to heat shock; organisms one or the other time get exposed to heat shock, means high temperature, which has devastating effect in terms of proteins’ denaturation.
· Some of the heat shock genes are expressed in stage specific manner. HSP 26 and 28 are expressed during oogenesis, HSP22 and 26 and 1, 4 & 5 express at early and late pupa 3rd instar stage respectively.
· The Escherichia coli dnaK gene is homologous to the major heat shock-induced gene in Drosophila (Hsp7O).
· The two sequences are homologous; the dnaK gene could encode a 69,121-Da polypeptide, 48% identical to the hsp7O protein of Drosophila.
· In higher systems there are ten different heat shock response genes. All of them have similar sequence modules in the upstream of the start.
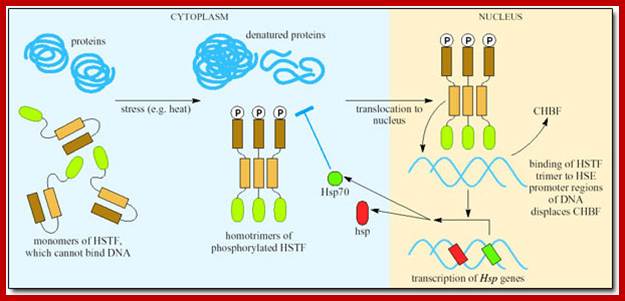
· Heat shock transcription factors are constitutively synthesized at low level and they remain inactive, because a protein called ubiquitin binds them.
· If the temperature of cytoplasm of in the cell is raised, the ubiquitin protein dissociates from the heat shock factors and the HSTFs become active and bind to upstream elements and activate transcriptional complex to initiate the synthesis of heat shock proteins.
· Heat shock proteins acts as chaperones for a variety of proteins otherwise the proteins become inactive.
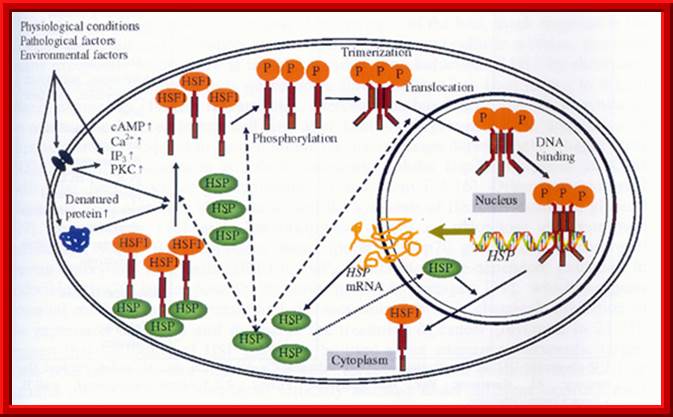
· cAMP, cyclic AMP; IP3, inositol triphosphate; PKC, Protein kinase C. (Reference: Tanguay and Wu, 2006) activate the expression of HSP genes.
· There are more than 26 members of the family from mouse to humans.
· Upon aging and longevity, the expression of Hsp chaperones has been reported in various organismes. Unbalanced chaperone requirement and/or chaperone capacity in aged organisms causes the accumulation of aggregated proteins, which often results in folding diseases, mostly of the nervous system, due to the limited proliferation of neurons. Therefore, over-expression of chaperones often delays the onset or diminishes the symptoms of the disease (Sőti and Csermely, 2002b), and increased chaperone induction can lead to increased longevity (Tatar et al., 1997; Morrow et al., 2006).
Heat Shock Gene Cluster:
<-----28-I<----23-I-1----->I-26----->I-22--->-<----4-I-<---5-I-----I--->
Promoter Elements of HSP 70 Gene

The human hsp70.1 promoter contains a proximal and distal HSE with five and six inverted nGAAn repeats. In addition, GC-, CCAAT-, and TATA-boxes exist for other transcriptional factor binding and constitutive hsp70.1 expression, and are involved in the maintenance of chromatin accessibility. (Reference: Hietakangas and Sistonen, 2006)
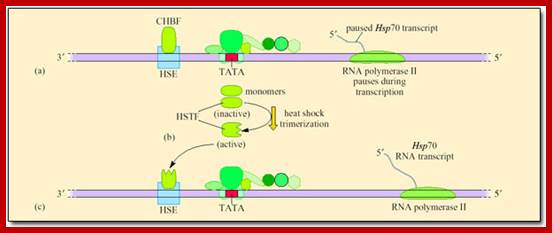
Control of mRNA synthesis during transcription of HSP gene; http://dspace.jorum.ac.uk
Control of mRNA synthesis during transcription of Hsp70. (a) RNA polymerase II pauses after synthesising about 25 nucleotides of the transcript as CHBF binds to HSE. (b) After a heat shock that produces part-denatured proteins heat-shock transcription factor (HSTF) is converted from an inactive into an active DNA-binding form. This occurs in response to the presence of part-denatured proteins. Monomers of HSTF link to form trimers that enter the nucleus. (c) Binding of activated (trimerized) HSTF to the heat-shock regulatory element (HSE) of the promoter of the Hsp70 gene, releases the paused RNA polymerase II, leading to rapid transcription of the Hsp70 gene
Pocley,G; Heat shock proteins in healtha nd disease;Animals at extreme temparatures; Expert Reviews in Molecular Medicine. Cambridge University Press; http://dspace.jorum.ac.uk/
Most work on Hsps has been carried out using cell lines and tissue cultures. Few studies have been carried out on vertebrates, and most of those have concentrated on fish. However, Zatsepina et al. (2000) studied the synthesis, properties and activation of Hsp70 in three species of desert lizard: Phrynocephalus interscapularis, a highly thermoresistant diurnal species, and Gymnodactylus caspiusand Crossobamon eversmanni, both nocturnal species. All three species were captured in a sand desert in Turkestan. Studies on a temperate species, Lacerta vivipara, provided comparisons with the desert species. All lizards were acclimated for 2 weeks at 25° C. Heat-shock treatment involved one hour exposure of lizards of each species to a specific T a at or greater than 39° C. Following heat shock the animals were killed and cell extracts from the whole body were prepared. Samples of the extracts were mixed with 32P-labelled HSE and incubated at 20° C for 20 minutes, during which time any HSTF or CHBF present would complex with the 32P-HSE. The free 32P-HSE was separated from the 32P-HSE-HSTF and 32P-HSE-CHBF complexes by gel electrophoresis. The gels were dried and exposed to X-ray film. Figure 43 shows the results of the analysis of binding of 32P-HSE to HSTF (complex III) and to CHBF (complexes I and II), in lizards kept at T a 25° C and lizards heat shocked at 42, 45 and 49° C.
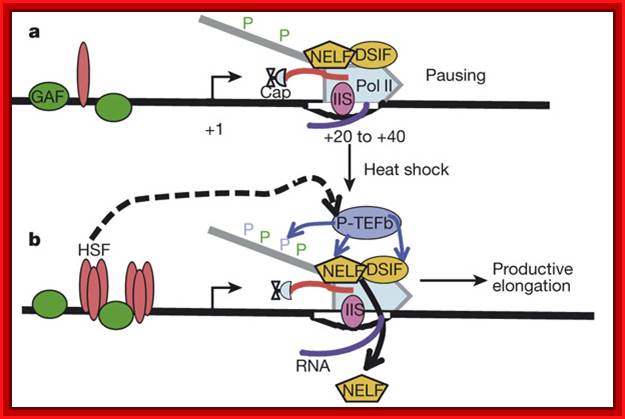
The Drosophila Hsp70 gene is regulated at the pause-escape step; Nicholas J. Fuda, M. Behfar Ardehali & John T. Lis; http://www.nature.com/
Drosophila Hsp70 was one of the first genes discovered to have promoter-proximal paused Pol II, and has been extensively studied. As a result, Hsp70 has served as the model for genes regulated at the step of early elongation. Its promoter resides in a nucleosome-free region extending to about 250 bases downstream of the TSS36, 62 (see figure, panel a). This open promoter is bound by GAGA factor (GAF, orange circles) and GTFs (blue rectangle)63. Studies have suggested that the GAGA elements are crucial for setting up the paused Pol II (red rocket)64, 65, 66, which is partially phosphorylated (red P). And in vitro evidence suggests that GAF bound to the promoter can recruit nucleosome remodelers to maintain this nucleosome-free state67. This open promoter allows Pol II to initiate and transcribe 20–40 bases downstream of the TSS, where it is held paused. This pausing is, at least partially, mediated by the SPT4-SPT5 complex (pink pentagon) and the NELF complex (purple circle). In vivo, NELF is present on uninduced Hsp70, and it is still present, but at lower levels, after heat shock68 (see figure, panel c). Furthermore, NELF depletion in vivo reduces the amount of engaged Pol II on uninduced Hsp70. Additionally, the downstream sequence may also be important for pausing. When the sequence within 30 bases downstream of the Hsp70TSS is switched with the sequence from another gene, the amount of pausing markedly decreases64. This may indicate that either the factors binding to downstream elements or the intrinsic pause-inducing characteristics of the transcribed sequence, or both, have a role in pausing. The paused Pol II is phosphorylated by the TFIIH subunit CDK7 on Ser 5 of the CTD repeats. This phosphorylation may be involved in pausing. A temperature-sensitive mutant of CDK7 decreases the amount of paused polymerase on Hsp70 at non-permissive temperatures; whether this affects pausing directly or at an earlier step remains to be resolved.

Drosophila Hsp70 was one of the first genes discovered to have promoter-proximal paused Pol II, and has been extensively studied. As a result, Hsp70 has served as the model for genes regulated at the step of early elongation. Its promoter resides in a nucleosome-free region extending to about 250 bases downstream of the TSS36, 62 (see figure, panel a). This open promoter is bound by GAGA factor (GAF, orange circles) and GTFs (blue rectangle)63. Studies have suggested that the GAGA elements are crucial for setting up the paused Pol II (red rocket)64, 65, 66, which is partially phosphorylated (red P). And in vitro evidence suggests that GAF bound to the promoter can recruit nucleosome remodellers to maintain this nucleosome-free state67. This open promoter allows Pol II to initiate and transcribe 20–40 bases downstream of the TSS, where it is held paused. This pausing is, at least partially, mediated by the SPT4-SPT5 complex (pink pentagon) and the NELF complex (purple circle). In vivo, NELF is present on uninduced Hsp70, and it is still present, but at lower levels, after heat shock68 (see figure, panel c). Furthermore, NELF depletion in vivo reduces the amount of engaged Pol II on uninduced Hsp70 (ref. 69). Additionally, the downstream sequence may also be important for pausing. When the sequence within 30 bases downstream of the Hsp70 TSS is switched with the sequence from another gene, the amount of pausing markedly decreases64. This may indicate that either the factors binding to downstream elements or the intrinsic pause-inducing characteristics of the transcribed sequence, or both, have a role in pausing. The paused Pol II is phosphorylated by the TFIIH subunit CDK7 on Ser 5 of the CTD repeats. This phosphorylation may be involved in pausing. A temperature-sensitive mutant of CDK7 decreases the amount of paused polymerase on Hsp70 at non-permissive temperatures70; whether this affects pausing directly or at an earlier step remains to be resolved. Defining mechanisms that regulate RNA polymerase II transcription in vivo
Nicholas J. Fuda, M. Behfar Ardehali & John T. Lis ;http://www.nature.com/
Heat shock (see figure, panel b) causes the transcriptional activator HSF (yellow diamonds) to trimerize and stably bind upstream of Hsp70. Such a temperature shift also activates HSF, resulting in the recruitment of coactivators (green hexagon), a rapid general loss of nucleosome protection across the gene36 and release of the paused Pol II into productive elongation. Upon heat shock, P-TEFb (blue triangle) is recruited to the gene and phosphorylates (blue P) the CTD, SPT5 and NELF subunits; the NELF complex dissociates from the Pol II complex; and Pol II releases from the pause sites, allowing rapid recruitment of new Pol II to the gene (see figure, panel c). Although Pol II still resides in the canonical pause sites under these conditions, it is estimated that the pause is of much shorter duration, with Pol II escaping every 4 s rather than once every 10 min before heat-shock induction.
Several studies have demonstrated that P-TEFb is important for releasing the paused polymerase upon induction of Hsp70. In vitro assays show that P-TEFb relieves the inhibitory effects of SPT4-SPT5 and NELF. Depletion or inhibition of P-TEFb severely reduces Hsp70 RNA expression, and P-TEFb inhibition, either before or after heat shock, blocks Pol II escape from the 5' end of the gene. Additionally, TFIIS is important for Pol II escape from the pause sites through its maintenance of paused Pol II in an elongation-competent state. Depletion of TFIIS impedes the release of Pol II from the pause and reduces the rate of Hsp70 mRNA production.
Environ factors when impinge on cell surface they activate messengers such as cAMP, Ca2+, IP and PKC which activate HSTF. HSTF initially bound to HSP proteins, when activated transcriptions factors are released from HSP and gets phosphorylated. Phosphorylation leads to trimerization of transcriptional factors and then enter into the nucleus, where they bind to response elements and activate genes.
Minimal model of Pol II pausing at Hsp70 genes before and after heat shock:
For a more complete description of factors associated with heat shock genes see the more comprehensive reviews. a, Prior to heat-shock, paused Pol II, which is partially phosphorylated at Ser 5 residues (green P) of the carboxy-terminal domain, is in a complex with DSIF and NELF complexes and occupies a region between + 20–--+40 base pairs downstream of the start site of Hsp70. GAF is a sequence-specific binding factor, bound to upstream of the start, that is present before activation. b, HSF is the key activator protein that trimerizes and binds with high affinity to its DNA elements in response to heat shock. Both DSIF and TFIIS (IIS) are part of both the paused and the fully competent Pol II elongation complexes. P-TEFb is the kinase that is critical for the maturation of paused Pol II into a productive elongation product, and it phosphorylates DSIF, NELF and the Ser 2 residues (blue P) of the carboxy-terminal domain of Pol II and transcription ensues.
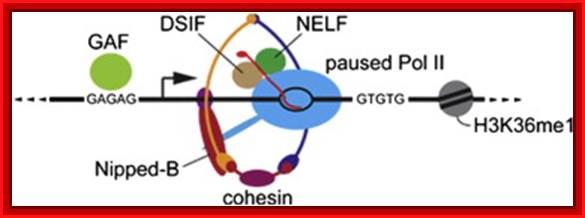
Cohesin selectively binds genes that have paused RNA polymerase ► Cohesin depletion alters gene expression without reducing polymerase pausing ► Cohesin and pausing factors regulate closely linked steps at repressed genes► Cohesin inhibits transition of paused polymerase to elongation at repressed genes; NELP-Negative elongation factor, DSIF-DRB sensitive inducing factor.
Plant Gene expression in response to Light: RBCs and RBCL:
· Plants have genetically adopted to respond to light and dark, day and night, short days and long days, cold and hot temperatures, humid and dry conditions and many other environmental vagaries, for all of them grow under natural conditions.
· Plants and light are intertwined in their interactions, one as the influencing factor and the other as the responder. Plants are influenced by light in their growth, flowering, photosynthesis, transpiration, dormancy, germination, opening and closing of the of the stomata, opening and closing of flowers, movement of leaves, shedding of leaves, pigment synthesis and many other metabolic processes.
· RuBisCo is composed of eight each of small subunits (SSU) coded for by the nuclear rbcLS multigene family and eight large subunits (LSU) code for by the rbcL gene in the plastome. For the production of holozymes both gene have to be expressed simultaneously. RuBisCo is a bifunctional enzyme (carbon fixation and oxygenase). The subunit is produced as precursor containing extension at the Carboxyl end, which is used for the transport across the chloroplast membrane into stroma.
· RuBisCo is the most abundant protein found in plant cells, than in any other cells; quantitatively it is about 50% of the total protein content of any cell. It is most abundant in leaves, less so in stems and other photosynthetic organs, and almost undetectable in roots.
· RuBisCo also exists in multiple forms in some system. It has an important biochemical role in Calvin-Benson-Basham reductive pentose pathway.
· Red light induces the expression of Ribulose Biphosphate Carboxylase/Oxygenase (RuBisCo) small subunit (RbcS) gene found in the nucleus, but it represses the expression of Phytochrome genes. RuBisCo’s large subunit (RbcL) gene is found and expressed in chloroplast.
· The Ribulose 1, 5-bisphosphate carboxylase small subunit (rbcS) in higher plants is encoded by a small multigene family. Members of the gene family contain 1-3 introns. The rbcS mRNA is differentially distributed in various plant organs. RuBisCo activase (RCA) activates (nuclear encoded protein) RuBisCo function.

· In the upstream of RuBisCo-S (RbcS) gene promoter there are several sequence modules, which act as light response elements, called LREs. The TATA box is located at –30 and light response elements at –166 and –149. There are three boxes in this region, called Box I, Box-II and Box-III.
· Plant genes for RuBisCo-s also have upstream elements at –410 and –160 called ARE (Age Response Elements) which respond to the age. LRE is recognized by GT-1 TF.
· In immature leaves RuBisCo-S genes express at very high levels.
· I----- 2112----// - 410---- -166--II---- -160 -140-------30-----I+>
ARE Box1 LRE Box2 LRE Box3
Light induced signals lead to activation of light response signal proteins which enter into the nucleus and bind to their respective response elements and recruit the transcriptional complex and induce gene expression.
RuBisCoL is coded for by plastid genome and transcribed by plastid multisubunit RNAP. The RbcL gene is a part of operon consisting RbcL, ATPase subunit atpB and QB polypeptide osbA genes. This is an operon.
-----P------------+1-----RbcL---I----atp------I----qb----I—t/t
-35 -10
TTGCGC TACAATC
Prrn promoter −35 (TTGACG) and −10 (TATATT)
Promoter elements; Transcribed by plastid RNAP (PEP)

Exogenous and endogenous factors such as light, hormones, photosynthesis, plastid type and chloroplast development differentially modulate transcription of plastid genes. Cues with repressing action on plastid transcription are depicted with lines terminated with a bar while arrows represent promoting signals (see text for references; effects of respiration on chloroplast transcription: T. Potapova, Y. Zubo, Y. Konstantinov, T. Börner, unpublished data).
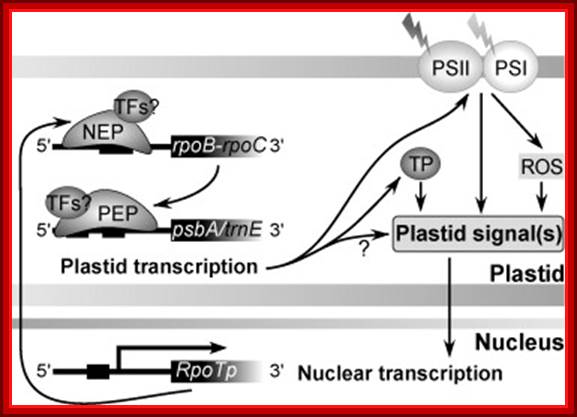
Fig. Model on the role of nuclear-encoded phage-type RNA polymerases in regulating plastid gene expression (updated and modified after Liere and Börner, 2007b). The nuclear RpoTp gene encodes in part the NEP transcription activity. NEP transcribes and therefore may regulate the expression of the plastid rpoB operon encoding subunits of the plastid-encoded RNA-polymerase (PEP). PEP, however, transcribes genes encoding components of the photosynthetic complexes (PSI, PSII) that modulate nuclear transcription by generating various ‘plastid signals’ (e.g. ROS, reactive oxygen species). PEP also transcribes trnE encoding a tRNA (trnAGlu). The tRNA is required for the synthesis of δ-aminolevulinic acid (ALA), the precursor of the tetrapyrrole (TP) biosynthesis (chlorophyll and heme), which too is thought to provide ‘plastid signals’. Thus, the regulatory network of the nuclear and plastid transcription machineries may be a key element for adjusting the expression of genes located within different compartments of the plant cell in response to exogenous and endogenous factors.
Expression of Genes in Response to Steroids and Corticoids:
Cells respond to a variety of hormones. The hormones can be lipid soluble or some may be water-soluble.
· Cells are also endowed with specific receptors and they respond to specific hormone in a specific way.
Some have general effects like growth hormone, cortisols, but some hormones are specific to certain tissues and their effect and response is hormone specific and tissue specific. For example Glucocorticoid activates 100 or more genes, some of them respond very early and some respond late and long lasting. Cortisols effect carbohydrate metabolism and increase blood sugar level and also have anti-inflammatory effects metabolism.
· Mineralo-corticoids maintain water and salt balance.
· Ecdysone acts on insect larvae and induces pupa formation, estrogens and androgens act as sex hormones in the development of sex organ development
· Estradiol activates Ovalbumin gene in chicks.
· Vitamin-D helps in bone development and calcium metabolism.
· Retinoic acid (9-cis Retinoic acid) acts as a morphogen and it is also required for the function of eyes.
· Thyroid hormones control basal metabolism and reaction kinetics.
· Steroid hormones are synthesized in response to a variety of neuro endocrine activities and effect major activities such as growth, tissue development and body homeostasis.
· Adrenal glands synthesize nearly 30 or more steroids, majority of them are Glucocorticoid and Mineralo-corticoids.
· Thyroid hormones, from thyroid tissue, in the form of iodinated Tyrosine such as T3 and T4 control the rate of basal metabolism.
· Pituitary glands produce four hormones; specific cell types in the same tissue produce different hormones, ex. islets of langerhans produce pro insulin; other cell types produce somatostatin and other two related hormones.
· Hormones are classified to four based on their target tissue and action.
Paracrine: Signal released act on nearby cells.
Synaptic: Neurotransmitters released are transmitted via synaptic cleft or by synaptic mechanism.
Endocrines: Most of the substances produced are hormones.
Autocrine: The released signals act on their own cells and stimulate their own cells to higher level.
Peptide and non-peptide signaling molecules perform a variety of functions through, phosphorylation and dephosphorylation. There are at least 1000 kinases and equal number of phosphotases. All of them are specific-to-specific biomolecules including proteins and other biomolecules.
· Phosphorylation of proteins can be at threonine (5-10%), serine (90-95%) or tyrosine (10%) and it is sequence and motif specific. Activation and inactivation of the proteins is mainly due to kinases and phosphotases activities.
The said enzymes are strongly influenced by the signal molecules. Most of the early reactions take place either at cell membrane level or in the cytoplasm before the factors execute their function and move into the nucleus and activate specific genes.
· Most of the lipid soluble hormones or signaling molecules have intracellular receptors, for ex. Steroid receptors belong to a super family of receptors, which are intracellular, yet each is distinctly different. However, though different from one another, they show some characteristic features in their protein structures.
All of them have sequence specific DNA binding domains that to bind to response elements, and interact with activators, and co activator domain. The dimers can be of the same kind to form homodimers or of different types to produce hetero dimers. Each of these dimers bind to two half sites, one to one, found in response elements of the genes. The C- terminal region of these proteins they have domain for dimerization. The N-terminal region of all these receptors varies considerably in size and character; these regions contain activation domain.
All together interacts with PIC and activates the RNAP. It is not unusual to find these response element binding factors actually recruit and assemble PICs. .
The sequence elements of gene are more or less similar pattern, yet distinct.
· The receptors, many of them just don’t bind to response elements as soon as they are bound by the ligand. After ligand binding, they may undergo further modifications or associate with other factors, before they bind to DNA. Some of the said factors are already associated with response elements; perhaps act as repressors; but the binding of signals can activate them.
DNA Binding Domains of Few Inducers:
|
|
NH3 end-Variable domain |
Activator domain |
DNA binding domain |
Dimrization domain |
Ligand binding domain---COO^- |
|
Cortisols
|
3^+NH----- |
----------------- |
--------------- |
------------------- |
-----COO^- |
|
Estrogen |
|
+NH3----- |
--------------- |
------------------- |
------COO- |
|
Progesterone |
3^+HN----- |
----------------- |
---------------- |
-------------------- |
---COO- |
|
Thyroxin |
|
+NH3-------- |
--------------- |
-------------------- |
-----COO |
|
Retinoic
|
|
+NH3------- |
--------------- |
-------------------- |
------COO |
|
Vit.D |
|
+NH3--------- |
----------------- |
-------------------- |
-------COO |
Mechanism:
Most of the steroid hormones have intracellular receptors, which can be located within the cytoplasm or in the nucleus.
· Glucocorticoid receptors are located in cytoplasm and sex hormone receptors are found in the nucleus.
In the case of cytoplasmic receptors, the steroids diffuse into the cell through cell membrane because of their lipid solubility.
· The cytoplasmic receptors are anchored in the cytoplasm with an inhibitor such as HSP. The binding of the hormone to cytoplasmic receptor releases the inhibitor from the receptor and forms complex S-Rc (S=steroid, R=receptor, C=cytoplasm and n=nucleus)
The receptors are also found in nucleus but maintain equilibrium status between cytoplasmic and nuclear.
· The S-RCs enters the nucleus, they are now called S-RNs, which seek response elements in the promoter region of the gene and binds to DNA and activates RNAP in the PIC. However activation by the S-Rn complex requires another additional protein called co-activator.
Among the Co- activators are CBP (CREB-binding protein) and or P300. The CREBp is a cAMP Response Element Binding protein, which is a common protein, responds to cAMP mediated actions. The co-activator mediates between the ligand bound receptor, which is already anchored on to a DNA sequence and with one of the TFs of the transcriptional apparatus. The CBP has acetylating property, by which it can Acetylate histone and make them to dissociate from the DNA, and facilitate the assembly of the transcriptional apparatus properly for initiation.
G-protein mediated activation:
Many neurotransmitters and few hormones bind to cell surface receptors, which are, not all, G protein linked receptors.
· When a ligand binds to such G linked receptors, on the cytosolic side, G–protein activate Adenyl Cyclase. The enzyme acts on ATP and converts it to cAMP, which is an important second messenger. cAMP in turn binds to cAMP dependent protein kinase and activates the catalytic subunit of CAPK.
The catalytic subunit moves into the nucleus where it phosphorylates ser 133 of the cyclic AMP response element binding protein called CREBp. The phosphorylated CREBp then binds cAMP response element and contacts the PIC and activates the RNAP for inducting transcription.
· Higher levels of calcium ions also activate the CREBP. When there is an increase in Ca2+ ions they bind to inactive Calmodulin. Binding of Ca2+ activates Calmodulin, which in turn activates CaM dependent CaM-kinase, which phosphorylates serine 133 of CREBP, which then binds to CRE elements and activates transcription.
· Thyroid hormone receptors, when not bound to thyroxin, remain bound to TRE or Thyroxin Response Elements and repress gene activation or expression. But when thyroxin, lipid soluble compound, is present, it enters the nucleus and binds to thyroxin receptor. Binding of the thyroxin to the receptor, transforms it into an activator.
Fig: Organization of CBP/p300-binding proteins. Association of CBP/p300 with transcriptional activators (top) and basal transcription factors and HATs (bottom) is shown. The zinc fingers (Zn), CREB-binding domain (KIX), Bromodoamin (Bromo), HAT domain, and glutamine rich domains are indicated,
Corticosteroid effects on cell signaling and gene expression:
Corticosteroids are the most effective anti-inflammatory therapy for asthma. Inflammation in asthma is characterized by the increased expression of multiple inflammatory genes regulated by pro-inflammatory transcription factors, such as nuclear factor-κB (NFkB) and activator protein-1, which bind to and activate co activator molecules that acetylate core histones and switch on gene transcription. Corticosteroids suppress the multiple inflammatory genes that are activated in asthmatic airways, mainly by reversing histone acetylation of activated inflammatory genes through binding of glucocorticoid receptors to co activators and recruitment of histone deacetylase-2 to the activated transcription complex.
Activated glucocorticoid receptors also bind to recognition sites in the promoters of certain genes in order to activate their transcription, resulting in secretion of anti-inflammatory proteins, such as mitogen-activated protein kinase phosphatase-1, which inhibits mitogen-activated protein kinase signaling pathways. Glucocorticoid receptors may also interact with other recognition sites to inhibit transcription, for example of several genes linked to their side-effects.
In some patients with steroid-resistant asthma, there are abnormalities in glucocorticoid receptor signaling pathways. In chronic obstructive pulmonary disease patients and asthmatic patients who smoke, histone deacetylase 2 is markedly impaired as a result of oxidative/nutritive stress, and so inflammation is resistant to the anti-inflammatory effects of corticosteroids.
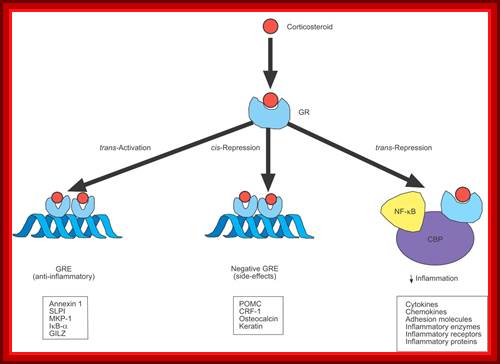
Corticosteroids may regulate gene expression in several ways. Corticosteroids enter the cell to bind to glucocorticoid receptors (GRs) in the cytoplasm that translocate to the nucleus. GR homodimers bind to glucocorticoid response elements (GREs) in the promoter region of steroid-sensitive genes, which may encode anti-inflammatory proteins. Less commonly, GR homodimers interact with negative GREs to suppress genes, particularly those linked to side-effects of corticosteroids. Nuclear GRs also interact with co activator molecules, such as cAMP-response-element-binding-protein-binding protein (CBP), which is activated by pro-inflammatory transcription factors, such as nuclear factor (NF)-κB, thus switching off the inflammatory genes that are activated by these transcription factors. SLPI: secretory leukoprotease inhibitor; MKP: mitogen-activated protein kinase phosphatase; IκB-α: inhibitor of NF-κB; GILZ: glucocorticoid-induced leucine zipper protein; POMC: pro-opiomelanocortin; CRF: corticotrophin releasing factor. ↓: decrease
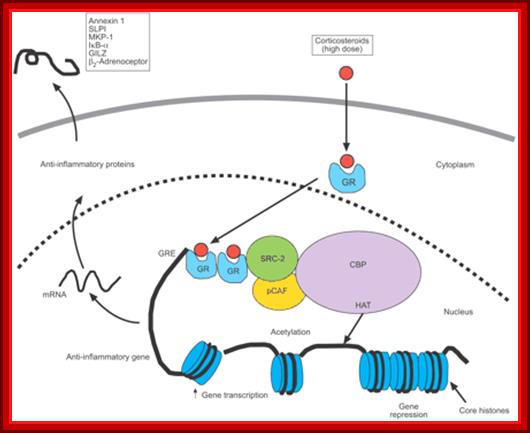
Corticosteroid activation of anti-inflammatory gene expression: Corticosteroids bind to cytoplasmic glucocorticoid receptors (GRs) that translocate to the nucleus, where they bind to glucocorticoid response elements (GREs) in the promoter region of steroid-sensitive genes and also directly or indirectly to coactivator molecules such as cAMP-response-element-binding-protein-binding protein (CBP), p300/CBP-associated factor (pCAF) or steroid receptor coactivator (SRC)-2, which have intrinsic histone acetyltransferase (HAT) activity, causing acetylation of lysines on histone H4, which leads to activation of genes encoding anti-inflammatory proteins, such as secretory leukoprotease inhibitor (SLPI), mitogen-activated protein kinase phosphatase (MKP)-1, inhibitor of nuclear factor-κB (IκB-α) and glucocorticoid-induced leucine zipper protein (GILZ).

Corticosteroid suppression of activated inflammatory genes: Inflammatory genes are activated by inflammatory stimuli, such as interleukin (IL)-1β or tumour necrosis factor (TNF)-α, resulting in activation of inhibitor of I-κB kinase (IKK)2, which activates the transcription nuclear factor (NF)-κB. A dimer of p50 and p65 NF-κB translocates to the nucleus and binds to specific κB recognition sites and also to coactivators, such as cAMP-response-element-binding-protein-binding protein (CBP) or p300/CBP-associated factor (pCAF), which have intrinsic histone acetyltransferase (HAT) activity. This results in acetylation of core histone H4, resulting in increased expression of genes encoding multiple inflammatory proteins. Glucocorticoid receptors (GRs), after activation by corticosteroids, translocate to the nucleus and bind to coactivators in order to inhibit HAT activity directly and recruiting histone deacetylase (HDAC)2, which reverses histone acetylation, leading to suppression of these activated
C-Fos gene expression:
Many growth factors such as EGF (epidermal growth factor) and platelet derived growth factor (PDGF); stimulate quiescent cells, under cell culture conditions, to enter into cell cycle from G-0 to G1 to S and G2 and back to G1. Stimulus, from growth factors, induces more than 100 genes.
· One of the most important genes that respond very early to the stimulus is C-Fos gene, which in turn stimulates the expression of several other genes required for cell cycle.
The regulatory region of C-fos gene has serum response elements (SRE). Also it has several other sequence modules, which respond differently for different signals.
· Serum contains several growth factors; among them serum growth factor is one. The SGF activates some specific gene expression.
When the ligand binds to cell surface receptors, the cytosolic region of the receptor gets activated by phosphorylation (auto-phosphorylation). This activates RTK-Ras signaling pathway.
· RTK-ras activates MAP- kinase (mitogen activating protein kinase). The activated MAP-kinase now enters into the nucleus and phosphorylates a protein called Ternary Complex Factor (TCF) at serine found at C- terminal part of the protein. MAP-kinase also activates another kinase called pp90. This protein in turn activates two SRF (serum response element binding factor) by phosphorylating ser at 103 position. Unphosphorylated TCFs and SRFs can bind to the response element but cannot activate transcription.
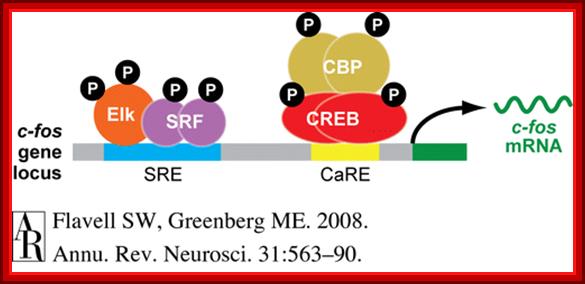
Serum response elements and CRE bound by their respective proteins such as SRF and CREBp and activate their respective gene expression.
Phosphorylated SRFs and TCFs form a complex in the nucleus and bind to SRE and activate C-Fos gene. The SRF-TCF factors are similar to yeast’s MCM1 and Ste-12 proteins.
Gene Expression in Response to Viral Infection:
Animal viruses infect, through cell surface receptors and infected cells respond by synthesizing glycoproteins called Interferons and the same are secreted. They are called as IFNs for they interfere with the viral multiplication and act as defense systems against second viral attack.
· Depending upon the cell types, they produce three different kinds of Interferons named; IFN-alpha by Leukocytes, IFN-beta by Fibroblasts and IFN-gamma by Lymphocytes. Interferons are classified as Type I, Type II and Type III. Each of these classes consists of many subsets of IFNs not less than a dozen or so.
Interferons are a kind of cytokines. Animal cells have specific receptors for each of the mentioned glycoproteins. When released, IFNs bind to receptors, then cells are induced to develop resistance to viruses; an antiviral state. They activate certain genes and their products act as defense systems against further viral attack. They also influence other cell types in the neighbor.
· One of the products that is very effective in defense system is double stranded RNA and another is dsRNA dependent protein kinase.
· IFN alpha, within 20 minutes of binding to cell surface receptor, induces the expression of some subset of genes by 20 fold. The genes, respond to IFN-alpha, contain Interferon Stimulation Response Elements (ISRE) in their regulatory regions. Four proteins have been found to bind to ISRE and their genes have been cloned.
Investigations on IFNs receptors have revealed that there are few sets of protein kinases and transcriptional factors called STATS, meaning Signal Transducers and Activators of Transcription. They form a kind of link between cell surface receptors to gene expression.
STAT1-alpha is 91 KD proteins, STAT1-beta is 84kd protein, and they are identical with exception that STAT1-alpha is 30 amino acids longer at C-terminal. The same gene codes both, but by alternate splicing they generate two different polypeptides.
· The third is STAT-2 is 113kd protein. This protein shows 59% homology with STAT 1-alpha and STAT 1-beta.
All these proteins have SH2 domains for protein-protein interaction to produce homodimers or heterodimers among themselves
· The fourth protein, called P48 is actually the DNA binding protein and it is unrelated to the first three stats and it has no SH2 domains.
Only those Cells, which are stimulated by IFN-alpha, contain the above-mentioned three STATs, which are found in cytoplasm. Following stimulation they move into the nucleus.
· On the contrary, P48 whether the cell is stimulated or not, it is found both in cytosol and nucleus. The P48 can bind to DNA to specific ISRE elements even without signals and its binding is independent of STATS.
The receptors for IFNs, all the other three, belong to a super family of receptors. They don’t have intrinsic protein kinase activity, but the cytosolic domain of the receptor on binding to the ligand they get activated. In this state they associate with one or more cytosolic protein kinases and activate them. But receptors without ligand remain inactive.
· The activated IFN’s receptor is believed to interact with TYK-2 and JAk-1 (tyrosine kinase-2 and Janus kinase-1) and activate them. TYK-2 or JAK-1 phosphorylates STATS at Tyrosine residues.
Phosphorylated STATs interact with one another and generate heterodimers (one with phosphorylated protein dimerizes with one with another containing SH2 group). The dimerized STATS move in to the nucleus and interact with P48, which is already bound to ISRE; this leads to activation of the transcriptional apparatus.
· Each type of IFNs induce unique subset of genes by using IFNs-STATS signaling pathway. This specificity stems from the fact that each receptor is different and their associated proteins are different and the STATS are different and their ISRE elements may be different but may be located in different positions.
Specificity stems from cell lines, e.g. Mutants for TYK-2 are insensitive to IFN-alpha, but are sensitive to IFN-gamma, which suggests the TYK-2 is a component of IFN-alpha specific pathway.
· Cell line mutants for JAK-1 are sensitive to both IFN-alpha and IFN-gamma, thus they play a role in both pathways. However IFN-gamma receptors have shown to associate with second type of kinases called JAK-2.
Antigen stimulated T-helper cells, release or secrete IFN-gamma kind of Interferons. Most of the other cells contain receptors for IFN-gamma.
When IFN-gamma binds to its receptors, cells get activated and activate a set of genes resulting in cells to be in antiviral state. Each of the genes that respond to IFN-gamma stimulation contains, in their regulator regions, the GAF Response Elements (GAF-RE). GAF means gamma-associated factors.
· Transcriptional factors, required for IFN-gamma mediated activation, have been identified and cloned. Its Mol.wt is 91 KD and it is called STAT-1 (which is a signal transducer and activator of transcription).
IFN-gamma transduces the signal by kinase activity, which results in the phosphorylation of STAT-1 at one of the tyrosine moiety. The phosphorylated STAT-1 dimerizes and moves into the nucleus and a bind to GAF, which is already bound to GAF-RE and the gene, is activated for transcription.
· Cells, mutants for STAT-1 alpha, cannot be phosphorylated because of the absence of tyrosine residues and STAT-1 Alpha does not move into the nucleus, in spite of the stimulation by IFN-gamma.
Interestingly, stimulation of cells by IFN-gamma, in the absence of IFN-alpha, leads to phosphorylation and dimrization of STAT-1 and they move into the nucleus and bind to GAF and stimulate only GAF bound to GAF response elements containing genes, it is an exclusive gene expression.
· Signaling pathway of cytokine receptor families is more or less similar to interferon JAK-STAT or TYK-STAT pathways.
In an generalized version, it now known, that binding of the ligand to a receptor causes dimrization of the receptor at cytosolic side, which interact with kinases of JAK family and activates them, which in turn specifically phosphorylate a set of transcriptional factors, which then move and bind directly to response elements or bind to a factor which is already bound the response element and activate the transcriptional apparatus.
The role of HMGI(Y)
in the assembly and function of the IFN- enhanceosome: Junming Yie1, Menie
Merika1, Nikhil
Munshi1, Guoying
Chen1 and Dimitris
Thanos1;Department
of Biochemistry and Molecular Biophysics, Columbia University, 630 West.
enhanceosome: Junming Yie1, Menie
Merika1, Nikhil
Munshi1, Guoying
Chen1 and Dimitris
Thanos1;Department
of Biochemistry and Molecular Biophysics, Columbia University, 630 West.
Transcriptional
activation of the virus inducible enhancer of the human interferon- (IFN-
(IFN- ) gene in response to virus infection requires the assembly of an
enhanceosome, consisting of the transcriptional activators NF-
) gene in response to virus infection requires the assembly of an
enhanceosome, consisting of the transcriptional activators NF- B, ATF-2/c-Jun, IRFs and the architectural protein of the mammalian
high mobility group I(Y) [HMG I(Y)]. Here, we demonstrate that the first step
in enhanceosome assembly, i.e. HMG I(Y)-dependent recruitment of NF-
B, ATF-2/c-Jun, IRFs and the architectural protein of the mammalian
high mobility group I(Y) [HMG I(Y)]. Here, we demonstrate that the first step
in enhanceosome assembly, i.e. HMG I(Y)-dependent recruitment of NF- B and ATF-2/c-Jun to the enhancer, is facilitated by discrete regions
of HMG I and is mediated by allosteric changes induced in the DNA by HMG I(Y)
and not by protein–protein interactions between HMG I(Y) and these proteins.
However, we show that completion of the enhanceosome assembly process requires
protein–protein interactions between HMG I(Y) and the activators. Finally, we
demonstrate that once assembled, the IFN-
B and ATF-2/c-Jun to the enhancer, is facilitated by discrete regions
of HMG I and is mediated by allosteric changes induced in the DNA by HMG I(Y)
and not by protein–protein interactions between HMG I(Y) and these proteins.
However, we show that completion of the enhanceosome assembly process requires
protein–protein interactions between HMG I(Y) and the activators. Finally, we
demonstrate that once assembled, the IFN- enhanceosome is an unusually stable nucleoprotein structure that can
activate transcription at high levels by promoting multiple rounds of
reinitiation of transcription.
enhanceosome is an unusually stable nucleoprotein structure that can
activate transcription at high levels by promoting multiple rounds of
reinitiation of transcription.
The IFN-β enhanceosome complex: Assembly of the IFN-β enhanceosome creates a stereospecific interaction surface for recruitment of CBP/p300 and the basal transcription machinery to allow multiple rounds of transcription. GTFs indicate general transcription factors. p160 refers to SRC/TIF pCIP family of coactivators View larger version:
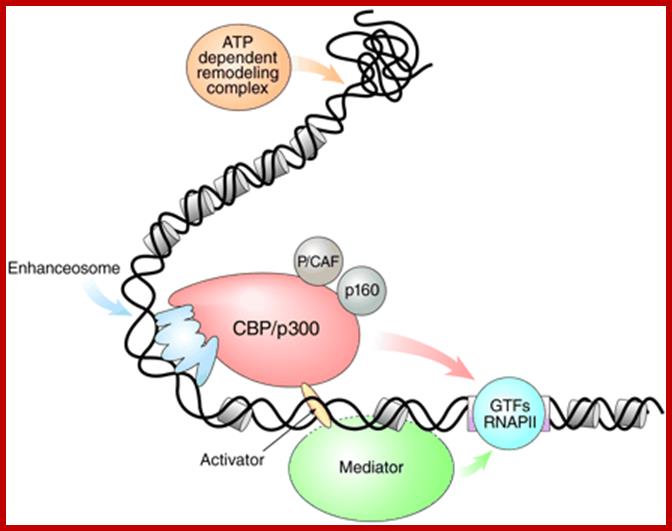
The multistep model of transcription: First, ATP-dependent remodeling complexes alter the structure of chromatin. Second, co activator HATs facilitate the formation of enhanceosomes and permit the actions of mediator complexes. CBP/p300 may facilitate the recruitment of the mediator complex to active sites of transcription. Mediator, in turn, regulates transcription through interactions with components of the basal transcription machinery. The dashed line indicates an unknown association with DNA. GTFs, general transcription factors.
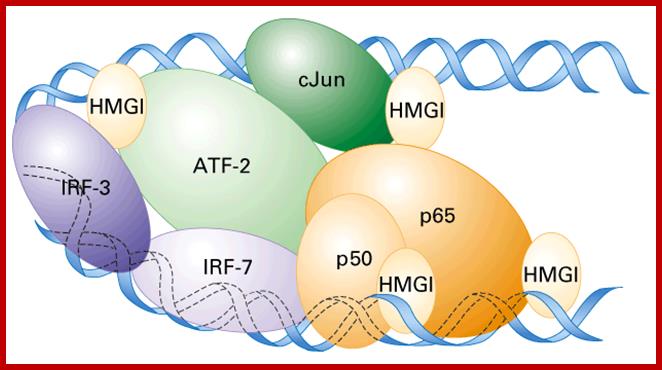
Enhanceosome- Interferon beta gene is activated by the binding of various factors where the factors such as HMG1 by binding to DNA bends the DNA so as other factors can contact with each other and activate RNAPII. Activation of IFN beta gene requires factors such as IRF (interferon response factors,) activating transcription factor (ATF), cJun and p50 and p65 are additional factor; all these come to interact with one another by bending of the DNA in the form of a loop by HMG1 protein.
Due to the
unfavorable intrinsic DNA curvature of the promoter, the IFN-![]() activators recognize their binding sites with low affinity. However,
binding of HMG I(Y) to the promoter unbends the DNA, thus lowering the free
energy required for activator DNA binding. This allosteric effect on DNA results
in a significant enhancement of activators' promoter binding affinity in the
absence of protein–protein interactions with HMG I(Y). Enhanceosome assembly is
completed by protein–protein interactions (arrows) between all the components,
thus leading to a remarkably stable nucleoprotein structure, the enhanceosome.
activators recognize their binding sites with low affinity. However,
binding of HMG I(Y) to the promoter unbends the DNA, thus lowering the free
energy required for activator DNA binding. This allosteric effect on DNA results
in a significant enhancement of activators' promoter binding affinity in the
absence of protein–protein interactions with HMG I(Y). Enhanceosome assembly is
completed by protein–protein interactions (arrows) between all the components,
thus leading to a remarkably stable nucleoprotein structure, the enhanceosome.
This pathway appears to operate in other hormone stimulated gene expression, ex. EGF binding to the receptor activates the kinase domain i.e Receptor tyrosine kinase ( RTK), which then stimulate cytosolic STATs, which move into the nucleus and bind to C-fos-SRE and activate the genes, called serum inducible genes.
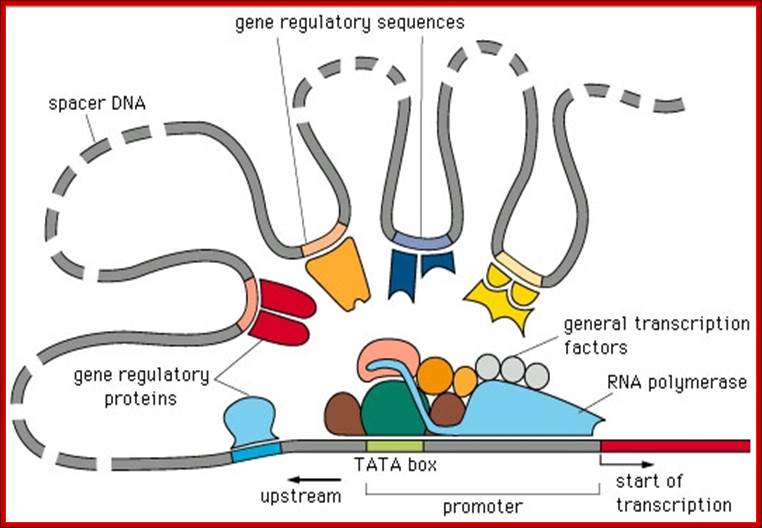
Transcriptional regulation in eukaryotes.; enhanceasome; Christoph Weigel; http://pbil.univ-lyon1.fr/
Perpetuation of Status-Activated or Repressed genes;
Specificity of the gene, which to be activated or which to be repressed, in what tissue and at what time, is already inbuilt in their regulatory elements; where gene specific proteins are already bound to their respective regulatory sequences. Identification of a gene which to be expressed or to be repressed is determined not by its coding sequences but by its imprinted regulatory sequence elements and their bound factors. These imprinted elements are differentially identified by regulatory proteins which not only identify which gene to be expressed or repressed in tissue specific, stage and time specific manner. Depending upon the kind and creed of a gene, the regulatory proteins during successive cycles of cell divisions the bound proteins are also perpetuated. While the DNA is replicated nucleosomal proteins are disassembled and reassembled for any number of generations, similarly the regulatory proteins are also perpetuated for they are made available during interphase. Depending upon the stage, time and tissue certain factors are made available either to activate the required genes or silence genes that are not required to the appropriate functions. Some of the pioneering proteins, which identify specific gene or genes, remain bound to specific loci in chromatin.
In the case of Wilms tumor, the EGR gene is always found repressed by a specific repressor WT. When this gene mutates, the EGR gene becomes active and the cells develop into tumors. Similarly Retino Blastoma (RB) proteins, suppressors of tumors, bind to E2F transcriptional factors all the time, to be released from binding only when the cell enters cell division.
In the case of yeast cells, GAL4 protein is perpetuated for many generations as repressor of GAL1< GAL 10 and GAL 7 genes, by binding to its regulatory sequences. Though this GAL4 protein is also an activator bound to its upstream activator sequences (UAS), remains inactive for its active site remains masked by GAL80. But it gets activated when Galactose substrate is provided. Galactose binds to GAL 80 through GAL3, and the GAL80 by conformational change releases itself from the GAL4, then the GAL4 undergoes conformational changes in such a way its activator domain is formed to interact with promoter elements. In the Galactose genes, the upstream is made up of UAS sites containing four clusters of GAL4 binding elements to which the factors bind. When once they are released from repressor they all together act as clusters in recruiting TFs and activate transcription from a distance. When Galactose is removed the GAL4 with GAL 80 remains in its place as a repressor.
Even in prokaryotic organisms the perpetuation of repressor suppressing a specific gene or genes is propagated for hundreds of generation without any let up. Lambda phage repressor cI (it is a repressor) by self-regulation of its own gene expression perpetuates repression for any number of generations; it is a par excellent example. The repression of lac operon, Tryptophan operon and Arabinose operon genes are few of many examples.
The repression in Lac operon is released only when glucose level goes down and the alternate carbohydrate source lactose is provided, so it is the presence of lactose and the absence of Glucose that induces the expression of Lac operon, but in the presence of Glucose lactose gene suppression perpetuated. In these examples specificity of genes that to be expressed and that to be repressed is delineated by genetics of gene mechanism. Removal of Lactose or Arabinose induces their gene activation because of the absence of repressor factors; this state is perpetuated till the conditions change. In eukaryote the DNA of genes are associated with histones and nonhistones all the time. Chromatin provides the sites for such binding.
Some of the regulator proteins bound to chromatin in specific loci render the genes in that region inactive and this state is perpetuated for any number of generations; for ex. GAL4 repressor bound to GAL1, Gal10 and GAL7 promoter elements. Similarly specific proteins bound to specific loci maintain the activation state of the genes, ex. When Galactose is added the GAL4 protein becomes active and activates the said genes and activation is perpetuated till the Galactose is removed. The most perplexing and intriguing problem in eukaryotes is how each of the genes or gene blocks is identified. There are many such examples- NF-cB-dependent induction of the NF-KB p50 subunit gene promoter underlies self-perpetuation of human immunodeficiency virus transcription in monocyte cells by Carlos V.paya*tt, Rosa M.tens et al.
Note, in recent years molecular biologists have found the assembled RNAP complex at the 5’end of genes. The activation or repression of the RNA complex requires specific factors.
Arabidopsis embryo/cell fate/self-perpetuation/ shoot meristem/stem cells:
Bernard Moussian, Heiko Schoof, Achim Haecker, Gerd Ju¨ rgens and Thomas Laux1
e-mail: thomas.laux@uni-tuebingen.de
Postembryonic development in higher plants is marked by repetitive organ formation via a self-perpetuating stem cell system, the shoot meristem. Organs are initiated at the shoot meristem periphery, while a central zone harbors the stem cells. Here we show by genetic and molecular analyses that the ZWILLE (ZLL) gene is specifically required to establish the central–peripheral organization of the embryo apex and that this step is critical for shoot meristem self-perpetuation. zll mutants correctly initiate expression of the shoot meristem-specific gene SHOOT MERISTEMLESS in early embryos, but fail to regulate its spatial expression pattern at later embryo stages and initiate differentiated structures in place of stem cells. We isolated the ZLL gene by map-based cloning. It encodes a novel protein, and related sequences are highly conserved in multicellular plants and animals but are absent from bacteria and yeast. We propose that ZLL relays positional information required to maintain stem cells of the developing shoot meristem in an undifferentiated state during the transition from embryonic development to repetitive postembryonic organ formation.
Bookmarking the Genome: Maintenance of Epigenetic Information*
1. Sayyed K. Zaidi‡, Daniel W. Young‡,1, Martin Montecino§, Andre J. van Wijnen‡, Janet L. Stein‡, Jane B. Lian‡ and Gary S. Stein↵ To whom correspondence should be addressed. E-mail: gary.stein@umassmed.edu.
Abstract:
Mitotic inheritance of gene function is obligatory to sustain biological control. Emerging evidence suggests that epigenetic mechanisms are linked to transmission of cell fate, lineage commitment, and maintenance of cellular phenotype in progeny cells. Mechanisms of epigenetic memory include gene silencing by DNA methylation, transcriptional regulation by histone modifications, regulation of gene expression by noncoding small RNA molecules, and retention of regulatory machinery on target gene loci for activation and repression. We will focus on the regulatory implications of epigenetic memory for physiological control and for the onset and progression of disease.
Mammalian Epigenetic Inheritance - Explained; A technical executive summary:
The maintenance of a repressed or activated status of a gene is often necessary for cellular differentiation. This observation should not be very surprising: A person’s liver cells, skin cells and kidney cells look different and behave quite differently, yet the all contain the same genetic information. With very few exceptions the differences between specialized cells are epigenetic (e.g. “post-genomic” or “post-translational”), not genetic. The remarkable thing about specialized cells is that not only can they acquire specialized traits and functions through development; they can also pass on these phenotypic manifestations to their own daughter cells. Although their DNA sequences remain unchanged during development, differentiated cells nevertheless acquire information that they pass on to their succeeding generations. The transmission of this sort of information is known as epigenetic inheritance systems (EIS). Chromatin Marking Systems:
The Types of Epigenetic Inheritance Systems:
1. Self-sustaining loops
2. Architectural changes
3. Chromatin marking systems
Chromatin marking systems:
1. DNA methylation and demethylation.
2. Histone methylation and demethylation.
3. Histone acetylation and de-acetylation.
4. Phosphorylation and dephosphorylation,
5. Ubiquitination, Deubiquitinating and Sumoyalaytion,
6. RNA interference (Secondary).
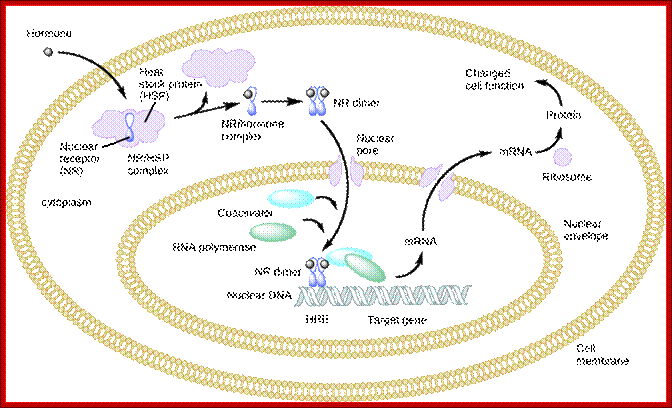
An intracellular nuclear receptor (NR) is located in the cytoplasm bound to a heat shock protein (HSP). Upon hormone binding, the receptor dissociates from the heat shock protein and translocates to the nucleus. In the nucleus, the hormone-receptor complex binds to a DNA sequence called a hormone response element (HRE), which triggers gene transcription and translation. The corresponding protein product can then mediate changes in cell function; . Derived copy of biology,Text Book by Safianu Rabiu. http://cnx.org/content/