Mechanism of Transcription:
In Eukaryotes, there are three RNAPs and each have distinct promoter elements and functions, and their promoter elements have distinct elements.


Core promoters in transcription: old problem, new insights; Anand l.Roy and Dinah S.Singer;
http://www.cell.com
Assembly of RNAP II-Transcriptional Apparatus:
Whether the class II promoter contains TATA boxes or InR sequences and DPE or not, the transcriptional apparatus consisting of RNAP-II and transcription factors TF-II class of factors, assemble at promoter region in a sequence. A promoter need not be restricted to the above mentioned structures; it can also consist of few more upstream elements. But the core complex of promoter elements does contain all the three sequences or just DPE. Assembly of the basal transcriptional factor and RNAP II as Pre Initiation Complex (PIC) is often activated and assisted by activators; co activators, mediator complex proteins and enhancer bound factors upstream. Most important feature is that they can assemble only when chromatin loosens and opens up at specific promoter sites, so as to make the space available for the assembly of TFs at promoter sequences. Even chromatin Promoter is also the region at which regulators assemble either to repress or to express genes. Between certain genes with enhancers may contain Insulator regions which prevent the expression of the neighboring genes.
The basic promoter regulatory elements are from the left BRE-TATA---InR---MTE--DPE. The assembly of transcriptional complex need not in sequence, they may assemble in complexes.

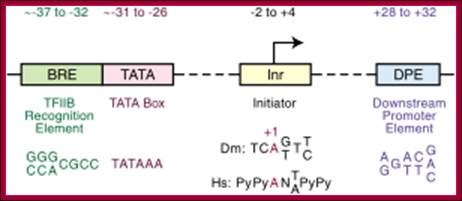
Downstream promoter elements

Some core promoter elements for transcription by RNA polymerase II; https://www.researchgate.net
-Overview of the four core promoter elements B recognition element (BRE), TATA box, initiator motif (Inr), and downstream promoter element (DPE), showing their respective consensus sequences and their distance from the transcription start site
- Some core promoter elements for transcription by RNA polymerase II. These elements include the BREu and BREd (TFIIB recognition element, upstream and downstream), TATA box, Inr (Initiator), MTE (motif ten element), DPE (downstream core promoter element), Bridge, TCT (polypyrimidine initiator), XCPE1 (X core promoter element 1), and DCE (downstream core element). The locations of the motifs are drawn roughly to scale. The BREu, TATA, Inr, MTE, DPE, and TCT motifs have been found in both Drosophila and humans. These motifs are typically found in focused core promoters, although there are probably Inr-like elements in dispersed promoters. There are no universal core promoter elements that are found in all promoters. Moreover, it is likely that many other core promoter motifs remain to be discovered. The functional properties of a core promoter are determined by the presence or absence of specific core promoter motifs. For example, some enhancers will activate transcription from DPE-dependent core promoters but not from TATA-dependent core promoters.
Promoters have Puase sequences, Stalled, Active DPE
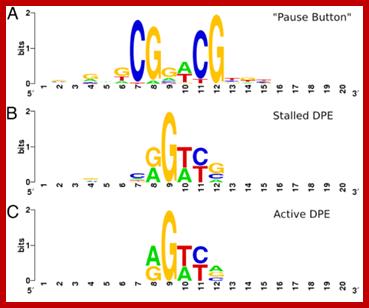
PB vs. DPE. (A) LOGO for the “Pause Button” (PB) motif, computed from instances in stalled promoters. (B) LOGO for matches to the DPE consensus (RGWYV) in stalled promoters. (C) LOGO for matches to the DPE consensus in active promoters. Stalled promoters show a DPE base composition that more closely matches the PB LOGO. Positions on the x axis of LOGOs do not represent position relative to the TSS, but show a 20-bp window around the core of each motif, centered such that positions of similar base composition match for each motif. ; http://www.pnas.org

http://slideplayer.com

The RNA polymerase II core promoter: a key component in the regulation of gene expression; Some modes of regulation via the core promoter. These models are discussed in the text. TATA1 vs. TATA2regulation usually involves a canonical TATA sequence (TATAAA) relative to a weak, noncanonical TATA sequence. http://genesdev.cshlp.org
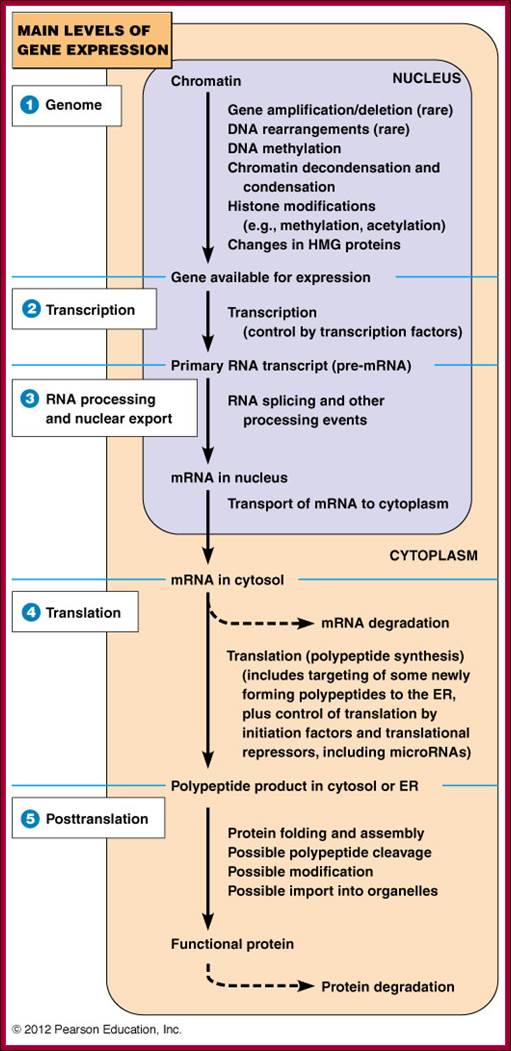
Genomic –Gene, Transcription, Processing, Translation and Post translation events; http://www.mun.ca/
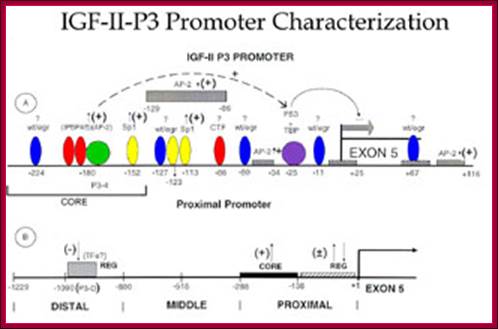
Transcriptional factors and cis elements known to bind the proximal and distal segments of IGF-II P3 promoter as an example; The ~ ntds position of the cis elements along the promoter is indicated.+ = up-regulation of promoter activity; - = down-regulation of promoter activity. A = cis element in the proximal promoter region of the IGF-II P-3 promoter. B = cis element recently identified by our laboratory in the distal segment of the P3 promoter. The approximate position of the transcriptional factors that have been shown to bind to the indicated cis elements in the proximal and distal segments of the P3 promoter are indicated.Wt/egr = wilms tumor antigen 1/early growth response element; IPBP4/5 = transcriptional factors that bind the novel cis element IPBP4/5 as described in References 58,59; AP-2 = activating protein2; Sp1 = Sp1 transcriptional factors, binding GC rich areas; CTF = transcriptional factors binding a novel cis element as described in Reference 59; TBP = TATA box binding proteins; P53 = tumor suppressor protein; TFs = transcriptional factors; P3-D = a novel cis element described by us in J. Biol. Chem. 276: 6937-6944, 2001; CORE = represents the core elements required for optimal activation of the P3 promoter; REG = regulatory elements; EXON 5 = the EXON 5 of IGF-II human promoter.; http://bmb.utmb.edu/

A full Gene- Upstream regulatory elements, Inr, coding region with introns and exons and Poly-A tail region; http://www.mun.ca/
http://www.en.wikipedia.org
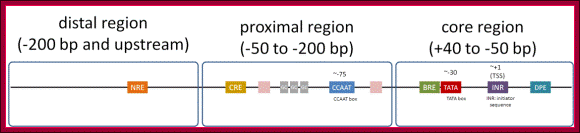
Eukaryotic promoter structure: Example, human insulin gene promoter. The general structure is common to most eukaryotes, though the detailed sequence arrangements are highly variable.
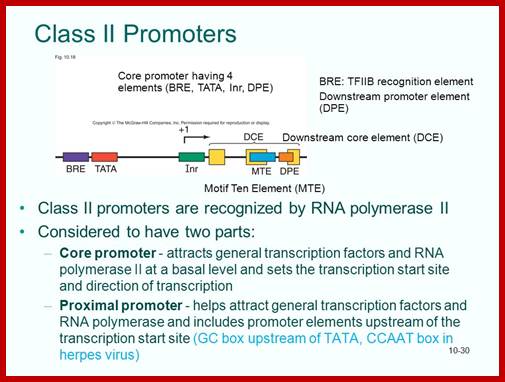
http://slideplayer.com
Just to remind the components of RNAP II the following diagram is given.
.
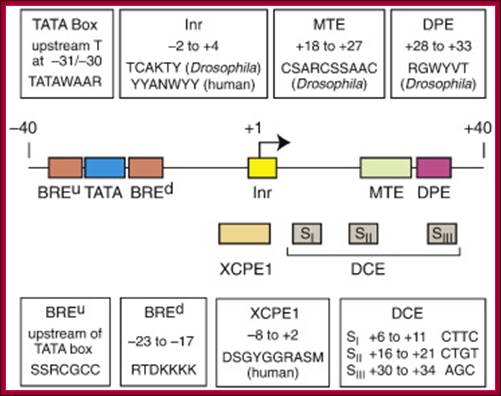
Details of upstream , Inr +1MTE and DPE+40; http://slideplayer.com
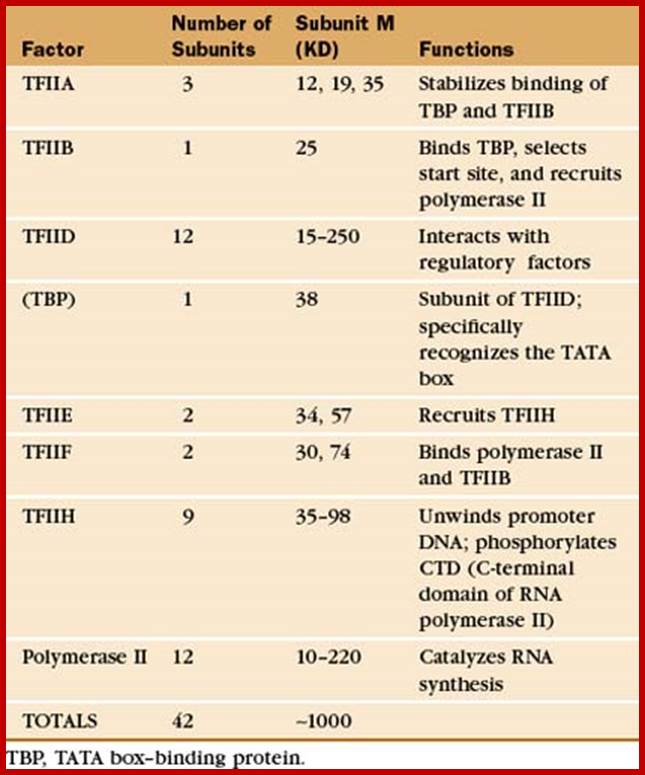
RNA Polymerase subunits:
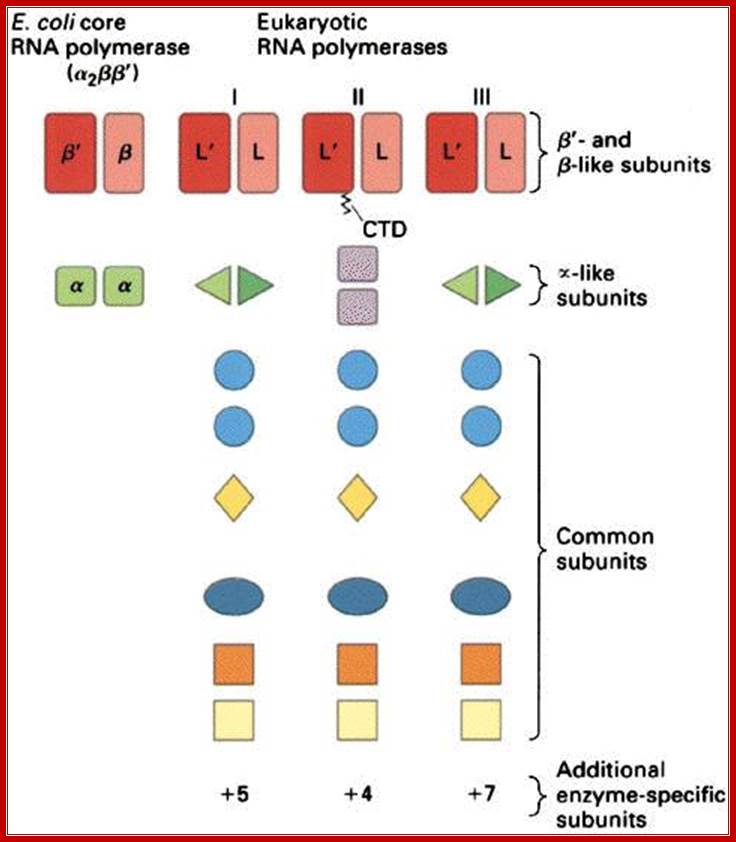
E.coli and Ek polymerase subunits; www.cbs.dtu.dk
• The core components of RNAPII are RPB1,RPB2 and RPB3-4,
• All others are associated components,
• The largest RpB of RNAP II has c-terminal tail,
• The tail consists of certain amino acid repeats-YSPTSPS.
• The number of repeats 22 (yeast) and 52 (humans).
• They play very important roles, in capping, splicing and polyadenylation.
Assembly of Transcriptional Apparatus:
Transcriptional apparatus consists of a number of components, but they act as General Transcriptional factors (GTF), assemble at core region of the promoter as Basal Transcriptional Apparatus (BTA) or Pre Initiation Complex (PIC). Whether they assemble as pre-assembled BTA complex or they assemble one after another in a sequence, at the time of transcription, is yet to be resolved unambiguously. But many opine they assemble as a complex. Often, the assembly of PIC in activated form facilitated by upstream factors. The assembly of this PIC by itself does not assure initiation of transcription nor its efficiency; they are controlled by other factors.
The assembly of GTF as PIC starts with TF II-D which contains TATA identifying and binding factor TBP (30KD), all beta ribbons. A monomer saddle shaped, bind to TATA box. Binding to TATA box is facilitated by 11 or more TAFs, whose number and characters varies from tissue to tissue. Though most of them are common, but one finds specific TAFs in specific tissues. Some of the TAFs (TAF42 and TAF62) have homology to histone proteins (H3 and H4) few of them are associated with histone modifying enzymes such as SAGA like. Certain TAFs themselves bind to DNA elements at promoter and InR sequences. Binding of TBP is like a positioning factor for the whole transcriptional complex. It protects DNA from –37 to –25. The protein binds to TATA bases pairs by hydrogen bonding. TPB not only binds to DNA, its C-terminal intercalates into minor groove of the DNA, thus distorts and expands the minor groove by 9Ĺ and also bends the DNA by 80˚ degrees and unwinds one third of the turn; and creates positive writhe. However the N-terminal of TBP remains exposed for interaction with other components. The TAF 230 assembles at the concave surface of the TBP; the said TAF mimics the minor groove.
The TBP binding acts as the focal center for the assembly other GTFs in sequence. The next factor that binds is TF II-A; it relieves TBP from TAF230 and positions to the left or upstream of TF II-D (a complex of protein subunits), then TF II-B binds. TF II-B covers upstream of TATA sequence. The TF II-B is considered as an important factor for it contacts DNA on either sides of TBP. It intercalates into major groove at the upstream and minor groove at downstream of the TATA box. This facilitates in identifying the start as well as determines the directionality of the promoter. This is considered as rate limiting factor. It also contacts with RNAP when it binds.
In the next line of assembly is TF II-E (protects +30) followed by TF II-F which actually stabilizes TBP–IIB complex. The TF II-F actually associated with RNAP-II, brings on the polymerase and positions itself in the transcription complex. The TF II-E recruits TF II-H onto the assembly line.
The TF II-H has multi subunits and multifunctional; two of them have helicase activity by its inherent ATPase domain. One of the subunits has kinase activity, which binds to CTD tail, and phosphorylates the CTD tail at serine residues. It is responsible for the melting of the DNA and responsible for pre initiation complex into open complex. Once the initiation succeeds it is also responsible for clearing the promoter. Yet it is involved in repairing any damage to the DNA. Associated with TF II-H are XPD, XPB, XPG, XPF and ERCC 1; all are involved in DNA repair. If the DNA is damaged, the RNAP is made to dissociate from the DNA by the TF II-H associated components, and often the large subunit of RNAP is degraded. Once the repair is done the factors and RNAP reassociate.
Activation of the RNAP perse also requires other upstream factors (they can be activators or co activators), mediator complex (multimers), whose composition differs from one species to the other and also differs from tissue to tissue. However for house keeping genes they are common. They interact with activator and co activators components found at the upstream regions of the promoter.

Assembly of basal transcrition apparatus- step by step. http://genocon.org/assignment-2012/assignment-a/basic-science-an-Eukaryotic-Promoter/:
An overview of the process of basal transcriptional apparatus formation and activation is illustrated in Fig Genocon2.
1. TFIID binds the core promoter region. In promoters containing a TATA box, the TBP subunit binds to this AT-rich region around -30 (+/- 10 bp). TFIID can also bind to additional contact sequences near the TSS, such as the INR (initiator sequence).
2. TFIIA and TFIIB help to stabilize the binding of TFIID to the promoter. TATA box containing promoters may also have a BRE (TFIIB recognition element) either just upstream and/or just downstream of the TATA box.
3. TFIIB also presents a contact to begin recruiting the RNA polymerase II.
4. RNA polymerase II makes contacts with TFIIB and TFIID. The contact between RNA polymerase II and TFIIB can be further stabilized by TFIIH.
5. TFIIF binds both RNA polymerase II and TFIIB, mediating the recruitment of RNA polymerase II.
6. TFIIE and TFIIH are recruited. These factors help to melt and unwind the promoter region, and activate the polymerase to begin RNA synthesis. Helicase activity of TFIIH allows DNA to unwind in TSS using ATP. After kinase activity of TFIIH phosphorylates RNA polymerase II, the polymerase releases from general transcription factors and can proceed in the elongation step of transcription.

Chromatin
organization: The basic unit of chromatin organization is the nucleosome, which
comprises 147 bp of DNA wrapped around a core of histone proteins. The level of
nucleosomal packaging can have profound consequences on all DNA-mediated
processes including gene regulation. Euchromatin (loose or open chromatin)
structure is permissible for transcription whereas heterochromatin (tight or
closed chromatin) is more compact and refractory to factors that need to gainACCESS![]() to
the DNA template. Nucleosome positioning and chromatin compaction can be
influenced by a wide range of processes including modification to both histones
and DNA and ATP-dependent chromatin remodeling complexes. By Sha, K. and Boyer, L. A., stem Book 2009;
https://en.wikipedia.org
to
the DNA template. Nucleosome positioning and chromatin compaction can be
influenced by a wide range of processes including modification to both histones
and DNA and ATP-dependent chromatin remodeling complexes. By Sha, K. and Boyer, L. A., stem Book 2009;
https://en.wikipedia.org
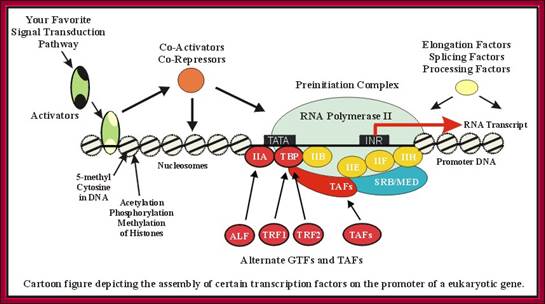
This diagram gives a wholesome picture of different components assembling in an order to initiate and clear the promoter. Certain TFII-S factors are not shown. Each of these components are interacting with one another. T. Akinyemi, B.C. Ego-Osuala, http://www.biochem.umd.edu/
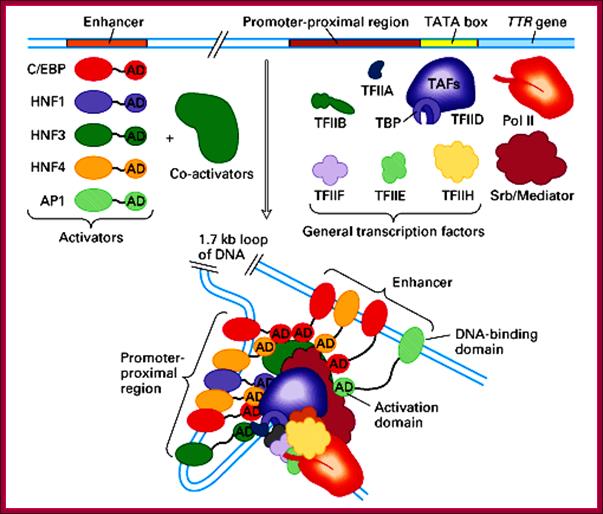
This diagram is the summation of components of RNAP II and its associated TF-II complexes, activators, co-activators and mediator factors that assemble on to their respective proximal and distal elements to initiate transcription of TTR gene. Science Blogs;PZ Myers; http://scienceblogs.com/
Holoenzyme:
TFIID is composed of TATA binding protein or TBP, monomer, plus approximately 14 additional polypeptides known as TBP Associated Factors or TAFIIs. TBP specifically binds to the TATA box, causing a sharp bend in the DNA. It is this binding of TBP with its associated factors that begins the assembly of the remaining GTFs and forms holoenzyme at the promoter.
TFIIE, TFIIF and TFIIH associate with RNA Polymerase II and another protein complex called mediator complex to form a holoenzyme. TFIIA and TFIIB are sometimes found together with the holoenzyme, and in other experiments they seem to bind independently. In any event, the holoenzyme is unable to recognize promoter sequence by itself and depends on TFIID binding at the TATA box. TFIIB finds its binding site as BRE next to TATA. Binding of TFIIB is very important.
After binding of the general transcription machinery to the promoter, TFIIE and TFIIH assist Pol II in unwinding the DNA at the start site of transcription. One of the subunits of TFIIH is a helicase responsible for the unwinding and TFIIE binds to and stabilizes the single stranded DNA.
Another subunit of TFIIH contains a kinase activity responsible for phosphorylating the CTD region of Pol II. Although this activity can be influenced by many factors, in simple systems it is believed to correlate with the initiation of transcription and that movement of RNA polymerase away from the promoter.
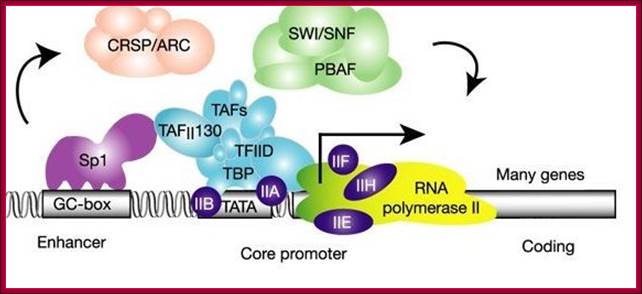
This diagram depicts the assembly of chromatin loosening components such as SWI/SNF, CRSP/ARC act to loosen the tightly coiled chromatin for the assembly of transcriptional initiation components on to proximal and upstream elements to initiate. The binding SP1 to its sequence regulates enhancer effect. www.calender-science.com

- Chromatin remodeling and assembly of Transcriptional complex; The mammalian molecular clock comprises a complex network of transcriptional programs that integrates environmental signals with physiological pathways in a tissue-specific manner. Emerging technologies are extending knowledge of basic clock features by uncovering their underlying molecular mechanisms, thus setting the stage for a 'systems' view of the molecular clock. Here we discuss how recent data from genome-wide genetic and epigenetic studies have informed the understanding of clock function. In addition to its importance in human physiology and disease, the clock mechanism provides an ideal model to assess general principles of dynamic transcription regulation in vivo. Romeo Papazyan, Yuxiang Zhang, & Mitchell A Lazar, http://www.nature.com

https://www.researchgate.net Components of Transcription complex; the assembly on promoter involves various factors such as GTF, activators, co activators, and many chromosomal modulators. https://www.researchgate.net
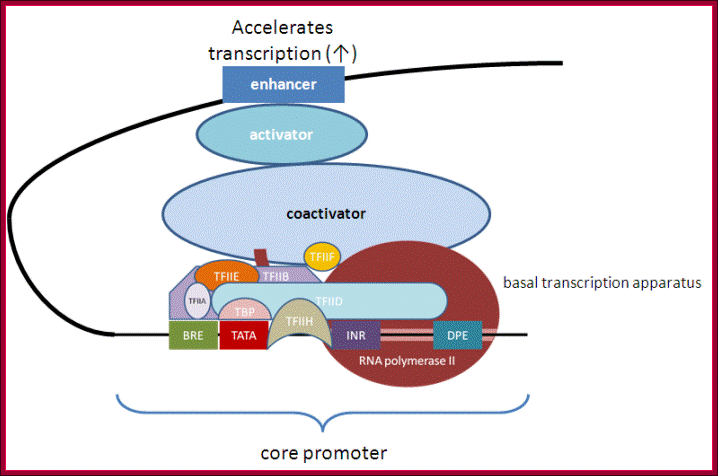
Tissue specific TFs- accelerators and repressors; acceleration of transcription by the assembly of components as enhanceosome. http://genocon.org/assignment-2012/assignment-a/basic-science-a-eukaryotic-promoter/
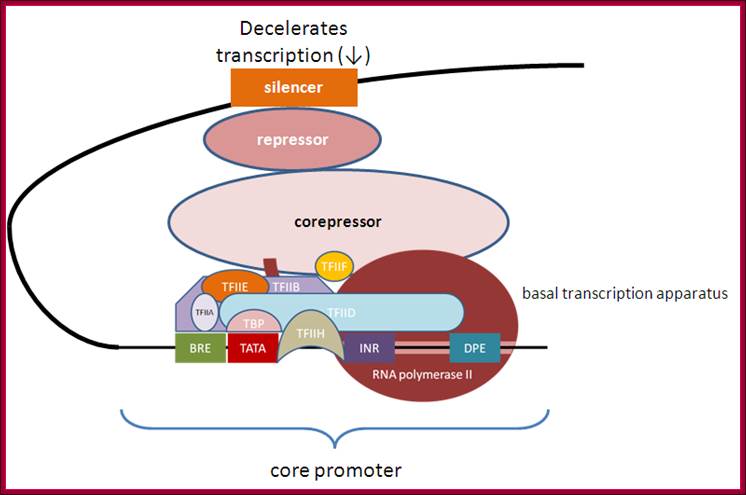
Deceleration of transcription with the assembly of corepressors, repressors and siencer components, opposite to enhaneosomal assembly. http://genocon.org/assignment-2012/assignment-a/basic-science-a-eukaryotic-promoter/
Tissue specific repressors; http://genocon.org/assignment
General transcription factors (green ovals) bind to core promoter regions through recognition of common elements such as TATA boxes and initiators (INR). However, these elements on their own provide very low levels of transcriptional activity owing to unstable interactions of the general factors with the promoter region. Promoter activity can be increased (represented by +) by site-specific DNA-binding factors (red trapezoid) interacting with cis elements (dark blue box) in the proximal promoter region and stabilizing the recruitment of the transcriptional machinery through direct interaction of the site-specific factor and the general factors (step 1). Promoter activity can be further stimulated to higher levels by site-specific factors (orange octagon) binding to enhancers (step 2). The enhancer factors can stimulate transcription by (bottom left) recruiting a histone-modifying enzyme (for example, a histone acetyltransferase (HAT)) to create a more favourable chromatin environment for transcription (for example, by histone acetylation (Ac)) or by (bottom right) recruiting a kinase that can phosphorylate (P) the carboxy-terminal domain of RNA polymeraseII and stimulate elongation.
Activation is initiated by interaction of RNAP with TAFs, and regulated by CTD tail kinase of TF II-H. The mediator complex in human and yeast is made up of 20 or more subunits of which seven of them show significant interaction with RNAP-II. By its name indicates that it is mediator transcriptional initiation. Among them the function of Srb4 is important. There are different modulators containing different subsets of factors and they regulate different sets of genes. Most of the mediator complexes associate at promoter regions.
Chromosomal modulators can be histone acetylases or deacetylases; they can be demethylases or methylases and phosphorylases or dephosphorylases; all of them play a significant role in modulating chromatin either blocking the assembly of transcriptional factors or allowing the assembly of the same.
The enzyme is a huge complex of proteins and its 3-D structure resembles that of crabs’ claw, where one of the two strands that act as template is held by the subunit that has catalytic activity, it also has the CTD tail, which is located at the RNA exit site.

Near the carboxy end of Pol II’s largest subunit a unique region is found (CTD, carboxy terminal domain). This region contains a stretch of 7 amino acids that is repeated between 26 and 52 times (differences in the number of repeats occur in a species specific manner). During transcription initiation several amino acids in the repeat becomes phosphorylated; http://www.conservapedia.com/
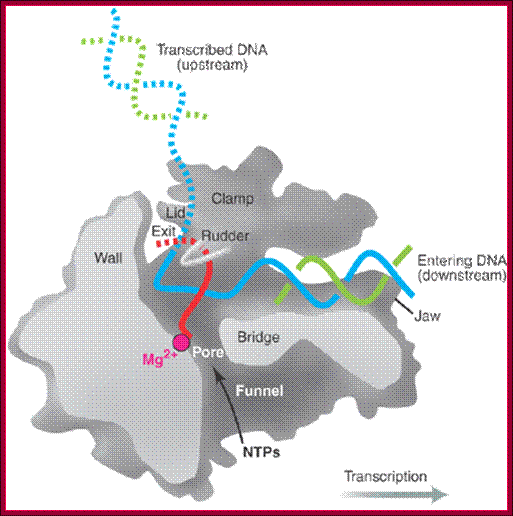
Side (cutaway) view of the RNA polymerase II transcribing complex, showing the paths of the nucleic acids and the locations of some functional elements of the enzyme: Cut surfaces of the protein, in the front, are lightly shaded and the remainder, at the back, is darkly shaded. By convention, the polymerase is moving on the DNA from left to right (direction of large arrow, bottom). This view exposes the DNA duplex entering the enzyme: The template strand coding for RNA is in blue, the non-template strand is in green, and the RNA in the DNA-RNA hybrid in the active center region is in red. DNA entering the enzyme is gripped by protein "jaws" (the upper jaw is not seen in the cutaway view). The 3' (growing) end of the RNA is located adjacent to an active site Mg 2+ ion (pink sphere). A "wall" of protein blocks the straight passage of nucleic acids through the enzyme, as a result of which the axis of the DNA-RNA makes almost a right angle with the axis of the entering DNA. The bend exposes the end of the DNA-RNA hybrid for addition of substrate nucleoside triphosphates (NTPs). The NTPs may enter through a funnel-shaped opening on the underside of the enzyme and gain access to the active center through a pore. The 5' end of the RNA abuts a loop of protein (the rudder), which prevents extension of the DNA-RNA hybrid beyond 9 base pairs, separating DNA from RNA. The exit path of the RNA passes beneath the rudder and beneath another loop of protein (the lid). Exiting RNA, DNA, and also the non-template strand of the DNA, are not seen in the electron density map (dashed red, blue, and green lines). The rudder and lid emanate from a massive clamp that swings over the active center region (from back to front in the view shown), restraining nucleic acids and likely contributing to the high processivity of transcription; A simplified view showing half of the protein complex showing internal features, http://xray.bmc.uu.se/
Most remarkable of all, and unexpected at this stage of the analysis, the authors, from whom the material is taken, are able to propose a convincing model for the translocation step that must follow the addition of nucleotides to the elongating RNA chain (see the figure, below). They propose that translocation is accomplished with the help of a protein helix (the "bridge helix") that spans the cleft between Rpb1 and Rpb2. This helix is also present in the structure of the bacterial RNA polymerase (which is homologous to the eukaryotic polymerase II), whose structure has been determined at somewhat lower resolution by Darst and colleagues. Amino acid side chains from the bridge helix (threonine and alanine) make hydrophobic contacts with the base of the coding nucleotide in the template strand at the active site. This region is straight in the yeast polymerase II structure, but bent in the bacterial version by about 3 angstroms along the direction of the template strand. Kornberg and colleagues, therefore suggest that the bridge helix acts as a ratchet, allowing the release of the DNA and RNA strands for translocation but maintaining its grip on the growing end of the hybrid, thus enabling the next step in the elongation cycle to take place.


Molecular mechanism of gene activation and repression. Gene activation and repression are regulated by acetylation of core histones. Histone acetylation is mediated by coactivators that have intrinsic HAT activity, opening up the chromatin structure to allow binding of RNA polymerase II and transcription factors that were unable to bind DNA in the closed chromatin configuration. This is reversed by corepressors, which include histone deacetylases (HDACs) and other associated proteins that reverse this acetylation, thereby causing gene silencing. STATs, signal transduction activated transcription factors. Assembled Pic with associated activator complexes- a model
https://www.researchgate.net
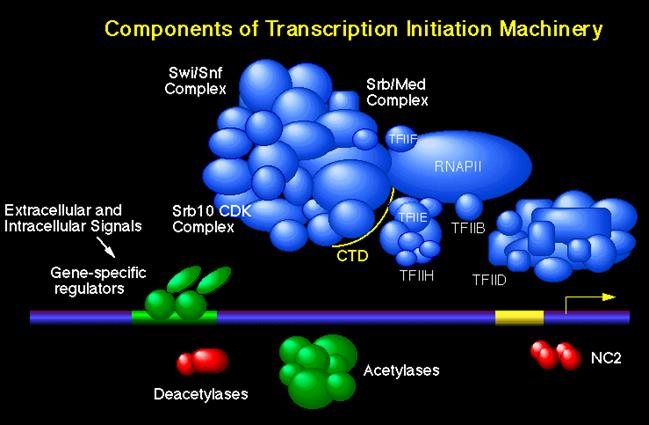
This is a picture of the components of the transcription initiation machinery. The mediator complex is the Med/Srb Complex. It has been created by Young Lab.http://legacy.earlham.edu/; www.slideplayer.com/slide
http://jcs.biologists.org/content; Another assembled complex-another model
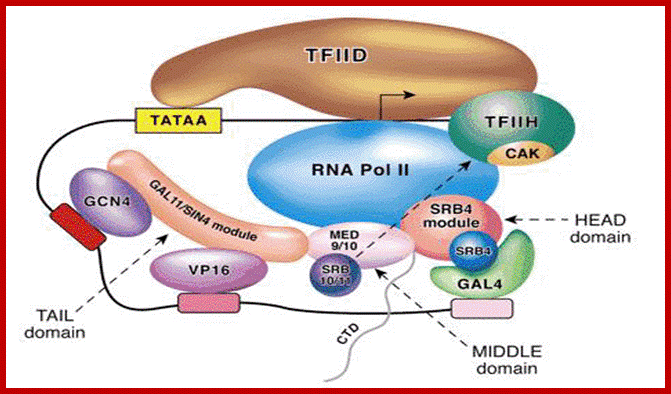
The yeast mediator model of activator-dependent transcription. Shown is a hypothetical promoter containing a canonical TATA box and interacting with the TFIID complex. Additional TFIID contacts are made with the initiator element (at the start site) and downstream of the start site. The arrow within the TFIID complex represents the start site of transcription. The three activators (GCN4, VP16 and GAL4) are shown binding to their DNA sites and recruiting yeast mediator to the promoter via a physical interaction with a mediator module (indicated by the overlap between the activator and the respective mediator module) (Koh et al., 1998; Lee et al., 1999b). Additional overlap among the three yeast mediator modules represents physical interactions between the three modules. Lastly, overlap/physical interactions between the MED9/MED10 and SRB4 modules and RNA polymerase II are illustrated (Kang et al., 2001). The HEAD, MIDDLE and TAIL domains are indicated and are based on recent structural data (Asturias et al., 1999; Davis et al., 2002; Dotson et al., 2000b). Note that the negative regulation of TFIIH CAK (cyclin H/cdk7) by the SRB10/SRB11 (cyclin-C-CDK8) is specific to human and has not been observed in yeast (Akoulitchev et al., 2000).
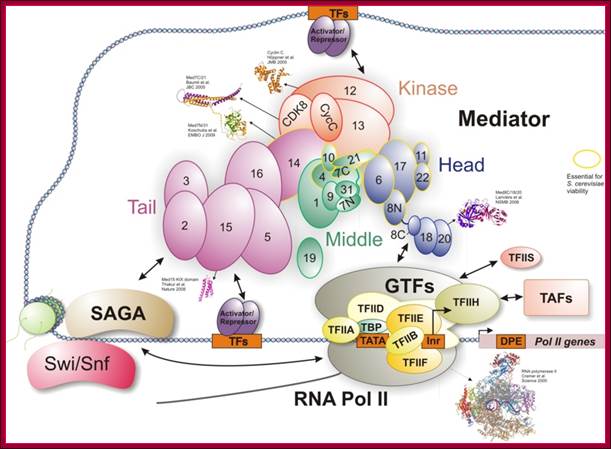
Tentative organization of the S. pombe Mediator complex: Subunits unique to S. pombe are shown in yellow. The green subunits define essential Mediator components identified in both S. cerevisiae and S. pombe. The dark green components also have homologs in metazoan Mediator and may represent a conserved Mediator core. The exact placement of individual subunits is based on previous studies of S.cerevisiae. This diagram shows the assembly of so many components at the site of transcription of a gene; more or less this is a general scenario in most of the cases. www.cramer.genzentrum.lmu.de
Transcriptional Initiation and Elongation:
This preinitiation complex assembled at the core promoter elements, by itself capable of initiating transcription, but the rate of initiation is very slow. For proper and efficient assembly and initiation requires a host of other factors such as activating factors, co activators, mediator complex and enhancers and others (mentioned earlier). These may be located just upstream of the core promoter elements or they may be located far away from the core promoter locus. When these modulate and contact the components of PIC, it is then the enzyme is transformed from loose binding to the DNA as closed complex into an active tight bound–open complex. During initial steps the enzyme goes through few isomerization steps and then stabilizes as tight open complex and starts assembling ribonucleotide triphosphates at predefined position. In this process RNAP does not require any primers as DNA Pols. Remarkable feature of the enzyme is that it initiates exactly at the start nucleotide.
It is important to remember that all the GTFs by themselves are capable of self-assembly, yet activator members, mediator members and other upstream factors facilitate GTF assembly on to a particular gene. Whether the transcription of a gene to be initiated or blocked depends upon the accessory factors that operate on GTFs or PICs. The interaction is always mediate by Mediator Complex (MC) of proteins. The MC mediates interaction with the CTD tail of the RNAP of the PIC on one side, and on the other side it interacts with activators, which are bound to their sequence elements elsewhere.
At the initiating site nucleosomal modifiers and remodeling factors are also required; if the chromosomal sites are not open for enzyme assembly they are made to open. The composition of mediator complex varies from tissue to tissue and gene to gene. They are not same for all genes and for all tissues and to all organisms.
When the PIC is activated, it initiates the assembly of rNTPs at predefined position and assembles few nucleotides only to abort and reinitiates. This process takes place several times but when it assembles more than 9-10 nucleotides, the enzyme leaves all other TFs in their place and clears the promoter. The TF II-H, which phosphorylates serine, threonine and serine residues of CTD activates to clearance of the promoter and by its ATP dependent Helicase action clears the promoter that leads the polymerase into elongation mode. For elongation factors such as TF II-S and HspT5 are required for they are actually elongation factors. TF II components associated with the RNAP complex are also involved in proof reading the transcript for the RNAP itself has 3’ to 5’ exonuclease activity. The rate of RNA synthesis is approximately 2000 to 3000 nucleotides per minute; it is 10 to 15 times slower than prokaryotes.
The CTD tail is about 800 Ĺ and it acts as a site for the assembly for a host of proteins. The other elongation factors that are involved are P-TEF-b (Kinase), hsp-T5 and TAT-SF1. Even TF II-S is also an elongation factor stimulates elongation. P-TEF-b is recruited by activator proteins. This kinase phosphorylates two serines in the CTD tail. It also phosphorylates HspT5 and TAT-SF1, so elongation is activated by different components. The HspT5 overcomes pausing during elongation and also involved in recruiting enzymes involved in Cap addition to the 5’ end of the emerging transcript. TAT-SAF1 factor recruits splicing machinery. Thus the CTD tail and its associated component have various functions related not only to the elongation the transcript but also the termination and poly- adenylation and splicing of the transcript.
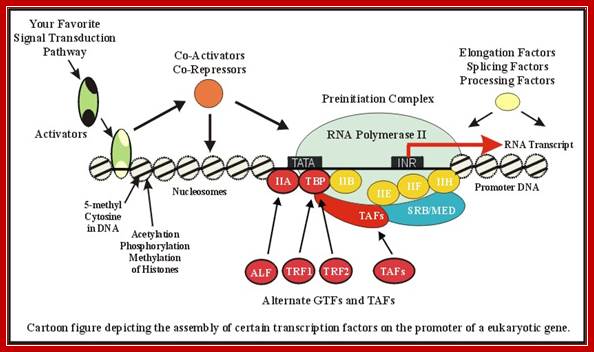
RNA Polymerase II is the major enzyme responsible for transcription of mRNA from a DNA template strand. The transcription machinery pictured above illustrates the mechanism that works in concert with activators, repressors, cofactors(mediators), general transcription factors (GTF), and the RNA Polymerase II (RNAP II); Remodeling of chromatin and assembly of transcriptional complex. http://www.biochem.umd.edu/
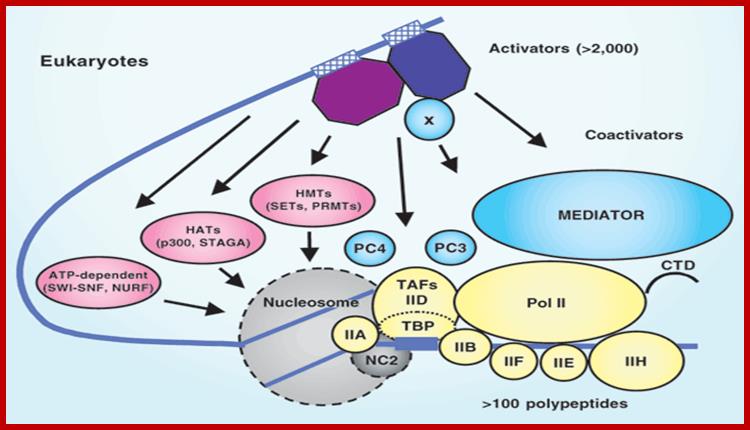
The eukaryotic transcriptional machinery: complexities and mechanisms unforeseen;
The temporal and spatial expression of specific genes is central to processes such as development, differentiation and homeostasis in eukaryotes, and is regulated primarily at the level of transcription. An understanding of the molecular basis for this regulation has presented a major challenge for the past 40 years.
Assembly of Transcriptional complex, Robert G Roeder; http://www.nature.com
Promoter clearance is absolutely required for the next initiation step. When RNAP moves out of the promoter most of the factors are left behind on the template at initiation point or released from the RNAP complex. Phoshorylation of CDT tail is essential for promoter clearance. But how the factors dissociate from RNAP is yet to be understood (?).
Inspite of its ability to initiate transcription, for its efficiency, it often requires the interaction of few ubiquitously produced transcriptional factors like CAAT binding factors or GC box binding factors such as SP1 or OCTA, whose locations are generally very close by in the upstream (from –60, -80 and –100) of BTA bound region.
· Interaction of the said factors with BTA through TAFs or other factors activates the transcriptional apparatus. The activators in the neighborhood of the start site or found at a distance in Enhancers, they are brought close to the BTA either directly or with the help of co activators or mediator complexes, the activator domain by interacting with TAF and other components of BTA, initiates transcription.
TF II-H subjects the CTD tail with YSPTSPS to phosophorylation. Phoshorylation of CTD tails has many effects, one RNAP moves out of the promoter leaving other TFs, second various proteins assemble at the CDT tail, and they are cap adding enzyme, splicing complex and transcription termination components.
· The CTD tail is involved in processing of mRNA.
Guanylyl transferase is also bound to the tail.
· Some splicing factors and poly (A) adding factors are also associated with CTD tail.
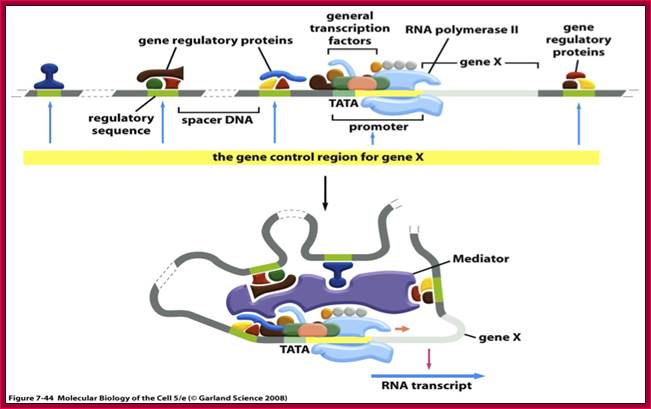
DNA
looping and interaction of general and specific transcription
factors with RNA polymerase in eukaryotic cells during transcription; https://www.studyblue.com
Transcriptional Termination:
Unlike prokaryotic mode of chain termination, there is neither fixed position nor any sequences that determine the position of chain termination by RNAPII. The process of chain elongation by RNAP can continue 100 to 1000 nucleotides after the TER codons. But the pre-mRNAs are released from this long terminal region by a host of factors responsible for cleaving the chain and poly-adenylation of the same (the process is described in the chapter RNA processing).
In most of the cases, termination occurs, nearly 1000 base pairs or more, down stream of the mature RNA’s end. There is no distinct sequence or structural features for termination as found in bacterial system.
There are multiple sites, in a rather long terminator region, where the RNAP-II ceases RNA synthesis. With the exception of histone mRNA all mRNAs have poly-A tail.

Mammalian RNAPII termination at protein-coding genes: Poly(A) site recognition leads to changes in the EC. Some factors associated with RNAPII through elongation that may function as anti-terminators are released (Paf1C, PC4) upon passage through the poly(A) site. At the same time, other factors, such as Xrn2 (Xrn2 is a nuclease), are recruited to the EC. After cleavage, Xrn2, most likely recruited by p54nrb/PSF, degrades the downstream RNA, “catches up” with RNAPII, and, perhaps with the aid of the helicase SETX, terminates transcription by releasing RNAPII from the template DNA. Involvement of chromatin remodeling factors and pausing sequences and factors are depicted. The RNAPII subunits Rpb3 and Rpb11 are also shown to play a role in termination by perhaps transducing a “termination signal.” The possible involvement of SR proteins in termination is also indicated. http://genesdev.cshlp.org
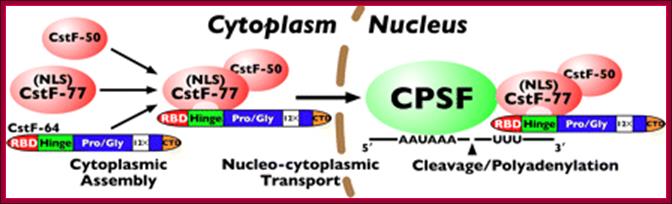
Model for CstF assembly, nuclear translocation, and polyadenylation: CstF assembles in the cytoplasm through the interaction of CstF-64 with CstF-77; the assembled complex is then transported to the nucleus via the nuclear localization signal in CstF-77. Finally, CstF associates with CPSF through a CstF-77-CPSF-160 interaction, allowing for cleavage and polyadenylation of pre-mRNA. http://www.jbc.org/content/285/1/e99903/F1.medium.gif
Cleavage and polyadenylation occur ∼10–30 nucleotides (nt) downstream from a conserved hexanucleotide, AAUAAA, and ∼30 nt upstream of a less conserved U- or GU-rich region. The AAUAAA signal is recognized by the cleavage and polyadenylation specificity factor (CPSF), which contains five subunits: CPSF-30, CPSF-73, CPSF-100, CPSF-160, and Fip1.

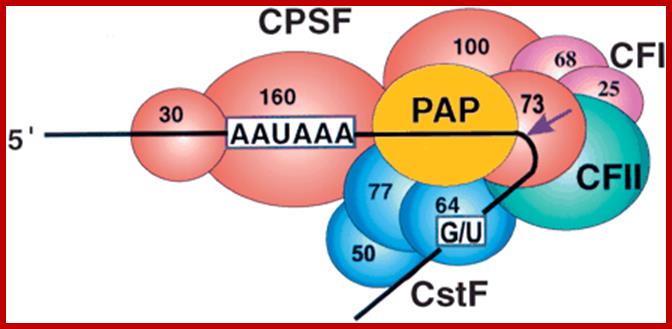
CPSF-73 as the 3′ processing nuclease. The core polyadenylation machinery (e.g., RNAP II is not included) is illustrated as it would assemble in a precleavage complex on an mRNA precursor, with CPSF-73 at the site of cleavage. The AAAUAA and GU-rich signal sequences are indicated, and the arrow denotes the cleavage site. The protein–protein and protein–RNA interactions depicted are consistent with available data. See text for details of protein factors.;Polyadenylation complex; www.hixonparvo.info; http://rnajournal.cshlp.org/
Note- Included more number of diagrams, so students can pick whichever they like; at certain points some; Information is repeated so as emphasize the point, for the benefit of students.