RNAP-I Promoters:
Transcription:
The enzyme complex RNAP-I transcribes the most ubiquitously expressed genes in all eukaryotes that are ribosomal RNA genes. In fact rRNA in any cell accounts for 90% of the total cellular RNA of the cell, which means these genes have to be expressed all the times, even under severe conditions. They have to be expressed more than any other genes. That is exactly the reason why ribosomal genes are found in large numbers and organized in tandem repeats in a specialized region of the chromosome called secondary constriction or nucleolar organizer.
Most of the eukaryotic rRNA genes, specially coding for 18s, 5.8s and 28s form one unit, but 5srRNA genes are located elsewhere. Bacterial rRNA genes contain small and large and 5srRNA segments together. How 5srRNA has separated from the bacterial system and located elsewhere in eukaryotes is an enigma. In addition there are two classes of 5s 5sRNAs- cytoplasmic and nuclear.
In humans five pairs of chromosomes have such clustered tandem repeats of rRNA genes in their NOR regions. The number of rRNA genes varies one system to the other ranging from 200 to 600 genes per haploid genome. In certain development situations, like during oogenesis, the genes are amplified by rolling circle mode of replication to several thousands of copies, which is required, for the development of the egg after fertilization. In early stages of development, for nearly 6-8 generation of cell divisions, rRNAs or any other RNAs are not synthesized. That means they have to use the RNA synthesized during oogenesis and the same is stored for the use during early stages of development.
Even in certain inductive conditions, where the first indications of certain changes in the cell can be observed by the enormous increased size of the nucleolus, which is the heart and site of synthesis of cellular rRNA.
As a structure the nucleolus consists of three morphologically distinct components: the fibrillar centers (FC), which contain hundreds of rRNA genes in tandem arrays found at several chromosomal loci (termed nucleolar organizing regions (NORs) ; the dense fibrillar component (DFC), which contains actively transcribing rRNA genes and nascent rRNA transcripts; and the granular component (GC), which is the site of late processing events in the; Many Cajal body proteins are also found in nucleoli including p80 coilin, fibrillarin and Nopp140 Nucleoli; http://npd.hgu.mrc.ac.uk/
Organization of rRNA Genes and their Promoters:
Organization of promoters in the rDNA segments should be made up of efficient elements, which could and should be consistently expressed.
Most of the rRNA genes are localized in certain chromosomal secondary constriction region called nucleolar organizer region (NOR) and they are positioned in the nucleolus. There they are transcribed; in humans the chromosomes that contain rRNA genes are 13, 14, 15, 21 and 22. Their position is just behind SAT region.
Electron microscopic observation of rRNA genes, in expression mode, depicts them as Christmas tree like repeats one after another with base and the tip. Between such transcription trees non-transcribing spacers are found. The size of each tree ranges from 2.0 kbp to 9k bp, ex. Xenopus laevis has 2.7 to 9kbp, and Drosophila contains 3.7 kbp.
Again within the coding region, the genes have spacers, and they are internal transcribed spacer (ITS). Such introns are removed by specific Intron mediated splicing complexes.
Though the External non-transcribing spacer (ETS) region may be thousands of base pair long, still it has certain sequences repeated and they related to promoter elements. Many such sequences contain 60 to 81 bp repeats spread in each of the blocks.
---I rR gene I spacer I rR gene I spacer I rR gene I spacer I-
rR gene = Ribosomal RNA gene.
--I-rRNA-gene-I—----spacer------------I I-rRNA gene-I
rRNA gene = 13 KB, Non transcribing spacer = 30KB.\
Each Spacer contains many repeat sequences.
--I Rep 1—I Bam-I—Rep 2-I-Bam-I—Rep 3—61-81bpI—
Rep 1, 2 etc plus there are repeat sequences of 61/81 bp
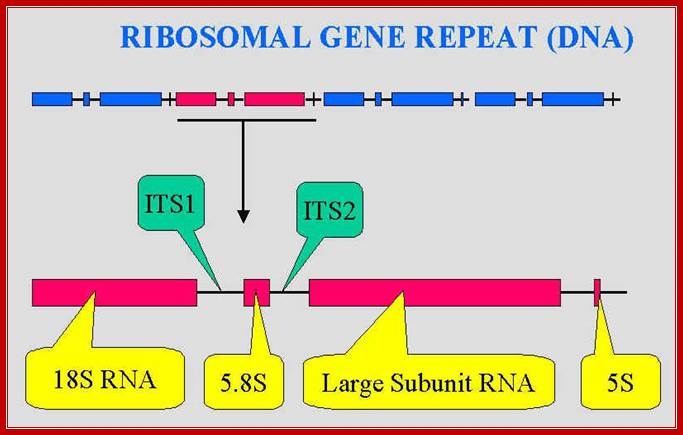
These rRNA gene repeats are found in NOR regions of chromosomes; each of the gene units are separated by NTS non transcribed spacers.; The Univ. Tennessee, Knoxville,ww.bio.utk.edu/mycology/
The rRNA genes contain internal transcribed spacers (ITS) and external transcribed spacers (ETS), between the gene units one finds NTS- non transcribed spacers.
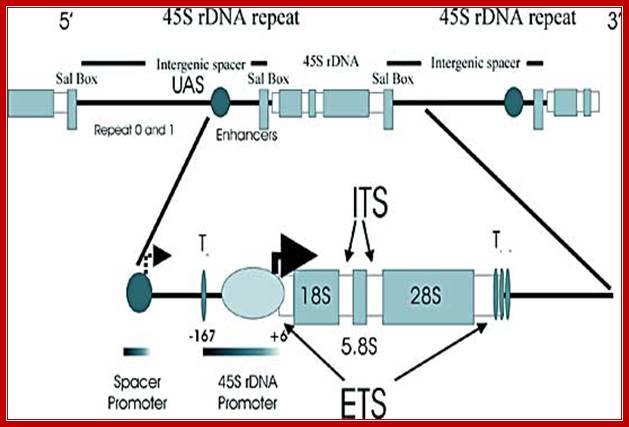
Schematic depiction of a mammalian ribosomal DNA repeat: The top portion of the cartoon depicts one and one-half ribosomal repeats in tandem, including the terminator Sal box, intergenic spacer, repetitive elements, enhancer region and the region transcribed to yield 45S rRNA. A section of the repeat is enlarged in the bottom portion of the cartoon. This section illustrates the placement of the spacer and 45S rDNA promoters, the proximal (To) and downstream promoter terminator elements (T1-7), the transcription initiation site (+6) and the external transcribed spacers (ETS).
Organization rRNA genes slightly vary from one system to another. In some the size of the gene is 45S and in yeast it is 35s. In yeast the ribosomal RNA loci also contain 5s rRNA genes as a separate entity on either side of the main 35s segments and transcribed in opposite direction.
Looking at the diagram below one can observe the 5srRNA is tucked in between rRNA genes. This is unusual for in other systems, the 5srRNA genes are located elsewhere. The 5srRNA genes in yeast cells are located in NTS (non transcribed spacers, but the transcription of 5srDNA is by RNAPIII and the direction is opposite to that of 35s rDNA.
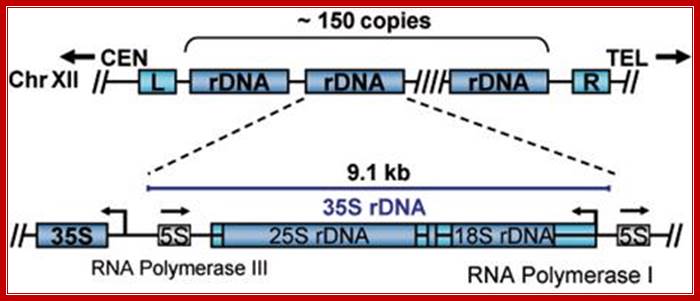
Joachim Griesenbeck; http://www.biologie.uni-regensburg.de/Biochemie;

pre‑rRNA; In E. coli, the rrn operon is transcribed into a 30S precursor RNA, containing 3 rRNAs and 2 tRNAs. The segment containing 16S rRNA (small ribosomal subunit) and the one containing 23S rRNA (large ribosomal subunit) are flanked by inverted repeats that form stem structure in the RNA. http://www. bx.psu.edu
The stems are cleaved by RNase III. There is no apparent single sequence at which RNase III cleaves ‑ perhaps it recognizes a particular stem structure. This plus subsequent cleavage events (by an activity called M16) generates the mature 16S and 23S rRNAs. The rRNAs are also methylated.
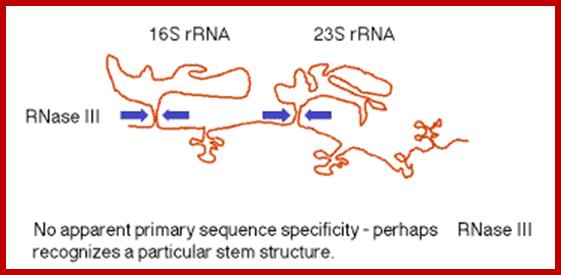
http://www.bx.psu.edu
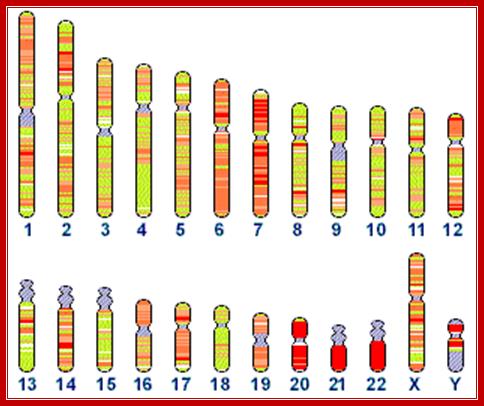
Human chromosomes, and the status of their sequencing; http://www.ncbi.nlm.nih.gov/genome/seq/
Distribution
of 5s rRNA genes on chromosome 1 , and 45srRNA genes in human chromosomes 13,
14 14 21 and 22: Total ribosomal RNA genes in the genome:
5S: 100 copies of 2.2 kb repeats = 220 kb. (estimate 100 kb essential, 120 kb
junk); 45S: 98 copies of 43 kb repeats = 4214 kb. (estimate 1500 kb
essential,2714 junk)
A look at the promoter regions of main rRNA genes, one finds that they lack TATA segments, which is so ubiquitous in structural genes.
In most of the observed systems, the start nucleotide is A, other wise it is G. The start point A or G is surrounded on either side by pyrimidines, -2py A +4py, which is named as InR or what is called Initiator Region.
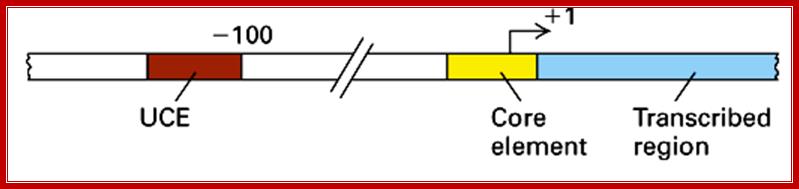
But in the upstream of the start there is a consensus sequence between –45 to -20, considered to be a core component (CE). This region is fairly GC rich.
In the upstream –160 to –110 there is another GC rich sequence, which is termed as upstream control elements (UCE).
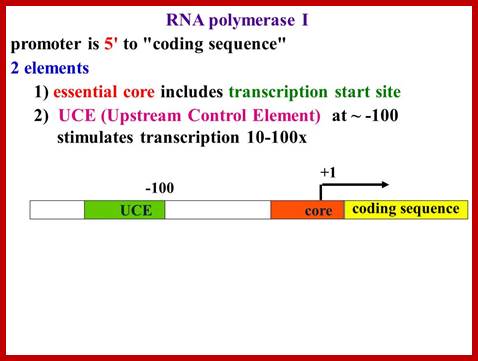
www.Slide player.com
This is simple representation of RNAP-I promoter elements, ttp://blog.sciencenet.cn/

http://slideplayer.com
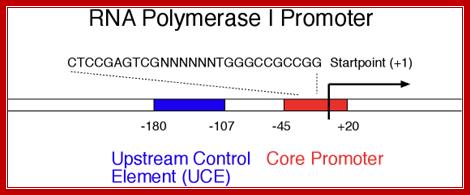
I-_180 ----UCE-(–)107---------------CPE-(–)45—InR-+1>>--
Core sequence- from -45 to +20
CTCCGAGTCGNTTTTTGGGCCGCCGG,
UCE = Upstream Control Elements, (-) 160 sequence- is also GC rich, it is bound by UBF1 factor (upstream binding factors).
InR = (initiator) sequence is – TACTGACACG
InR =AT rich.
CPE = Core Promoter Elements, GC rich .
http://slideplayer.com
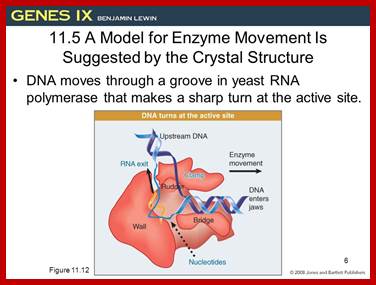
This picture is a 3-D Model of RNA Polymerase –I
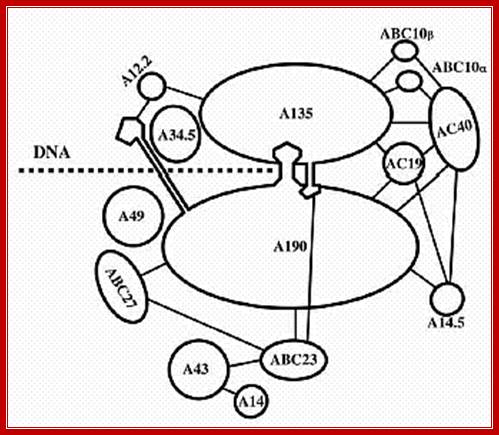
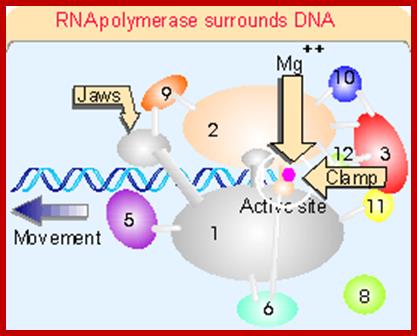
Ten subunits of RNA polymerase are placed in position from the crystal structure. The color of the subunits is the same as the color of the corresponding parts of the crystal structure. Note that movement is from right to left. The colors of the subunits are the same as in the crystal structures of the following figures. The numbers identify the individual subunits.;http://www.biodados.icb.ufmg.br
Mechanism of Transcription:
Pol I transcription cycle:
In the process of transcription (by any polymerase), there are three main stages:
- Initiation: the construction of the RNA polymerase complex on the gene's promoter with the help of transcription factors
- Elongation: the actual transcription of the majority of the gene into a corresponding RNA sequence
- Termination: the cessation of RNA transcription and the disassembly of the RNA polymerase complex.
Initiation:
Initiation: The construction of the polymerase complex on the promoter. Pol I requires no TATA box in the promoter, instead relies on a UCS (Upstream Control Sequence).
- UBF (Upstream Binding Factor) binds the UCE.
- CPE recruits and binds a protein complex incorporating TBP (TATA Binding Protein) and three TAFs (TBP Associated Factors) called SL1 or TIF-IB. The TBP is forced to bind to the initiator region but no-sequence specific.
- Rrn3/TIF-IA gets phosphorylated and binds Pol I.
- The UBF factors interact with core complexes, results in looping of the DNA so as to facilitate the activation of RNAP complex.
- Pol I binds to the UBF/SL1 complex via Rrn3/TIF-IA, and transcription starts.
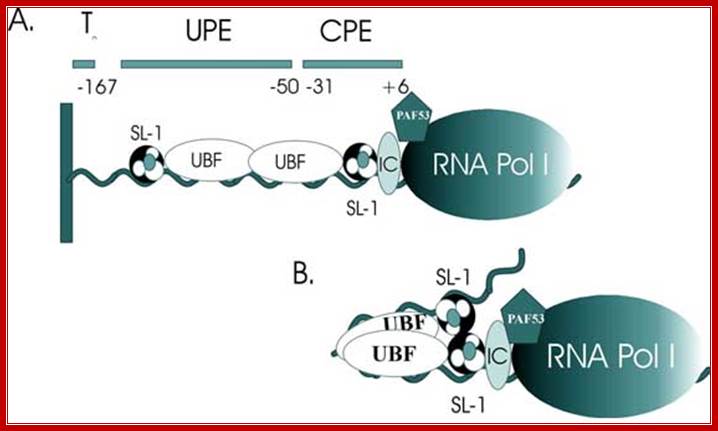
Upstream elements and core elements of rRNA gene promoters are depicted: Binding of UBF and SL1 on either sides of UBF leads to DNA looping and activating transcription by RNAPI; almost similar to enhancer mode;
Frontiers in Bioscience 3, d376-398, March 26, 98] http://www.bioscience.org/
Schematic depiction of the factors interacting with the 45S rRNA promoter. The factors include SL-1, UBF, TFIC (IC), PAF53 and RNA polymerase I (RNA Pol I). A. Illustration of the DNA binding sites with which the factors interact. These include the upstream promoter element (UPE) and the core promoter element (CPE). B. Illustration of a model where UBF bends the rRNA promoter bringing two molecules of SL-1 in contact. Note that the stoichiometry of SL-1 to the promoter is not known.
TBP binds as a monomer, but when it binds it binds to minor groove; very few DNA binding proteins bind to minor groove. By binding it induces structural deformation in the DNA where it is bound, and it affects the major grove in such a way, it leads to the melting of the DNA to about 6-8 base pairs long, this is exactly what is required for the RNAP to initiate the process.
As earlier said that TBP is a TATA binding protein but here there is no TATA sequence. As TBP is associated with TAFs; this complex acts as positional factor for RNAP-I. When RNAP binds to this complex, it initiates transcription at predefined nucleotide.
Initiation at predefined position is remarkably precise and incorporates a specific nucleotide; a change of space of 3.4 Å in this region makes a hell of a difference for the transcript.
This is an extraordinary process of finding positions in a jungle of millions and even billions of nucleotide sequences. Identification of sequences of similar nature, and sorting out specific sequence segments delineates the identity of the gene to be transcribed. Transcription generates 45s long precursor rRNA, which is further processed to 5.8s, 18s and 28s RNAs. In yeast the transcription generates 35s precursor rRNA.
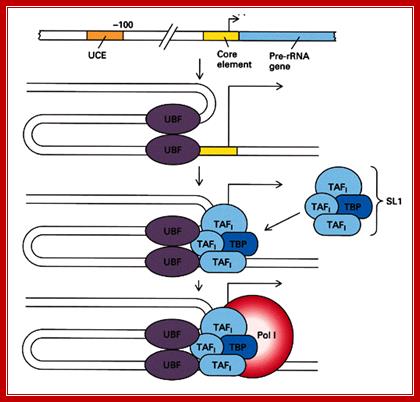
The diagram depicts events of assembly of RNAP-I and its associated TF complexes. Note in a figure above this you find some differences in factors and their kinds associated at promoters. http://blog.sciencenet.cn/
Transcription Initiation on the rRNA Genes and the Role of UBF in rRNA Transcription:
In eukaryotes, a dedicated set of proteins is used to transcribe the rRNA genes, allowing the cell to regulate these genes independently of the rest of the genome. The α-amanitin-resistant DNA-dependent RPI (RNA polymerase I) is dedicated solely to the transcription of the rRNA genes. In mammals, formation of an RPI pre-initiation complex requires SL1 (selectivity factor 1; or mTIF-IB), HMG1-box (High Mobility Group) DNA-binding proteins and UBF (upstream binding factor). Originally it was believed that UBF specifically bind to the upstream and/or core promoter regions, creating a situation propitious for the SL1 complex to bind and form a ‘stable’ pre-initiation complex. In vitro, this complex is able to recruit RPI and, in the presence of nucleotide triphosphates, to initiate transcription. When it was discovered that the N-terminal half of UBF (core UBF) could form the ‘enhanceosome’, a nucleoprotein structure that somewhat resembles the nucleosome, the finding led to a speculative explanation for the co-operativity between UBF and SL1. In this model, UBF juxtaposes the two promoter domains, presenting the SL1-binding sites on the same side of a DNA super helix, see Figure below. However, it was later found that UBF does not restrict itself to the RPI promoter but is found throughout the rDNA repeat. This suggests that UBF defines a specialized rRNA gene chromatin and is consistent with its indiscriminate DNA-binding properties.
Schematic with RNAP I and UBF depiction of the transcription initiation complex on the rRNA gene promoter. The factors include UBF, SL-1, and promoter. Additionally, UBF binds SL-1 and brings it to the core element, leading to transcriptional activation and PAF53 interacts with UBF and RNAPI. The UBF binds to the promoter as a dimer and bends the rRNA gene promoter. Additionally, UBF binds SL1 and brings it to the core element, leading to Transcriptional activation. The roee of HMG is not shown here. HMG proteins when bind to DNA they bend the DNA to 80-90 ^o thus factors bound to promoter element interact and activate RNAP1. A cartoon above showing the now classic paradigm of RP-I initiation in mammals: Cell, 109, T. Moss and V.Y. Stefanovsky, at the center of eukaryotic life.’ 545–548, © 2002.
A hypothetical model of the promoter structure showing two consecutive UBF–DNA enhanceosome complexes; juxtaposing the upstream control elements (UCE) and core promoter elements. Data are based on the low-resolution enhanceosome structure. This is a new paradigm for RNAP-I transcription regulation. Multiple adjacent enhanceosomes arrest elongating RPI. ERK and CBP combine to unfold the enhancesome, allowing passage to the polymerase. The DNA is shown in red, UBF in dark blue, RPI and associated factors in light blue and SL1 in orange. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling; TBP, TATA-box-binding protein; TAF, TBP-associated factor. Adapted from Molecular Cell, 21, V.Y. Stefanovsky, F. Langlois, T. Gagnon-Kugler, L.I. Rothblum and T. Moss,
A new paradigm for the regulation of the mammalian ribosomal RNA genes
T. Moss1, V. Stefanovsky, F. Langlois and T. Gagnon-Kugler
Ribosome assembly occurs co-transcriptionally on the rRNA genes. This process requires the co-ordinated expression and assembly of many hundreds of proteins and is finely tuned to cell and organism growth. Co-ordinate regulation of the rRNA genes and the ribosomal protein genes is therefore essential for high-fidelity ribosome assembly. Recent work shows that rRNA gene transcription is regulated at the level of elongation via the mitogen-activated protein kinase pathway. We argue that this may provide an explanation for the high fidelity of ribosome assembly.
Elongation:
As Pol I clear the promoter, UBF and SL1 remain-promoter bound, ready to recruit another Pol I complex. Indeed, each active rDNA gene can be transcribed multiple times simultaneously, as opposed to Pol II-transcribed genes, which associate with only one complex at a time. While elongation proceeds unimpeded in vitro, it is unclear at this point whether this process happens in a cell, given the presence of nucleosomes. Pol I do seem to transcribe through nucleosomes, either bypassing or disrupting them, perhaps assisted by chromatin-remodeling activities. In addition, UBF might also act as positive feedback, enhancing Pol I elongation through an anti-repressor function. An additional factor, TIF-IC, can also stimulate the overall rate of transcription and suppress pausing of Pol I. As Pol I proceed along the rDNA, super coils form both ahead and behind the complex. These are unwound by topoisomerase I or II at regular interval, similar to what is seen in Pol II-mediated transcription.
Elongation is likely to be interrupted at sites of DNA damage. Transcription-coupled repair occurs similarly to Pol II-transcribed genes and require the presence of several DNA repair proteins, such as TFIIH, CSB, and XPG.
Transcription Termination:
Chain termination takes place down stream from the defined end of mature RNA. It requires an 18 nucleotide long sequence to which ancillary factor binds while RNAP is still associated with the template. Termination of transcription by RNA polymerase I involve pausing of transcription by Transcription Termination Factor (TTF1), and the dissociation of the transcription complex, releasing pre-rRNA and RNA polymerase I from the template. Polymerase1 Transcription Release Factor (PTRF) is required for dissociation of the ternary transcription complex (By similarity)
Transcription Termination Factor (TTF-I) when binds it bends the termination site at the 3' end of the transcribed region. This will force Pol I to pause. TTF-I, with the help of transcript-release factor PTRF and the T-rich region induce Pol I into terminating transcription mode and it dissociate from the DNA. Evidence suggests that termination might be rate-limiting in cases of high rRNA production. TTF-I and PTRF will then indirectly stimulate the reinitiation of transcription by Pol I at the same rDNA gene. Evidence suggests that termination might be rate-limiting in cases of high rRNA production. TTF-I and PTRF will then indirectly stimulate the reinitiation of transcription by Pol I at the same rDNA gene.
The rRNA produced in this process is a large precursor RNA, which later gets processed to their specific segments.
Mammalian RNAP I termination:
Transcription terminates at the transcript release element composed of a stretch of Ts, upstream of the TTF-I-mediated pause site. The release factor PTRF, which interacts with RNAPI and TTF-I, recognizes the transcript release element and most likely binds the stretch of Us on the nascent RNA. The RNAPI-specific subunit Rpa12 also plays a role in yeast RNAPI termination that is possibly conserved in mammals. After cleavage of the precursor downstream from the 28S gene, Xrn2 degrades the downstream RNA, “catches up” with RNAPI, and participates in RNAPI release, possibly in conjunction with SETX. An unidentified 3′–5′ exonuclease might be involved in processing of the rRNA precursor.
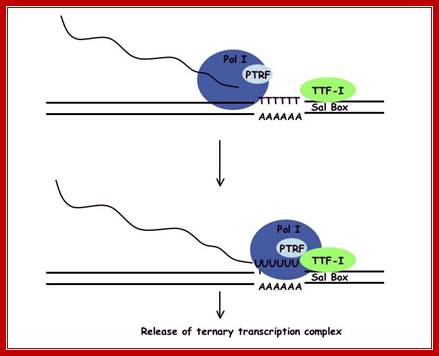
Termination of transcription by RNA polymerase I is a 4 step process. Initially TTF-1 binds the template rDNA. This complex pauses polymerase I allowing PTRF to interact with the quaternary complex releasing both pre-rRNA and Pol I from the template and TTF-1; http://gallus.reactome.org/
Mammalian RNAPI termination: Transcription terminates at the transcript release element composed of a stretch of Ts, upstream of the TTF-I-mediated pause site. The release factor PTRF, which interacts with RNAPI and TTF-I, recognizes the transcript release element and most likely binds the stretch of Us on the nascent RNA. The RNAPI-specific subunit Rpa12 also plays a role in yeast RNAPI termination that is possibly conserved in mammals. After cleavage of the precursor downstream from the 28S gene, Xrn2 degrades the downstream RNA, “catches up” with RNAPI, and participates in RNAPI release, possibly in conjunction with SETX. An unidentified 3′–5′ exonuclease might be involved in processing of the rRNA precursor; https://www.google.co.in

Quantification of Gene Expression; www.gene-quantification.com/strategy.html
Quantification of rRNA Transcription in a ‘Given Time’, per enzyme per Unit:
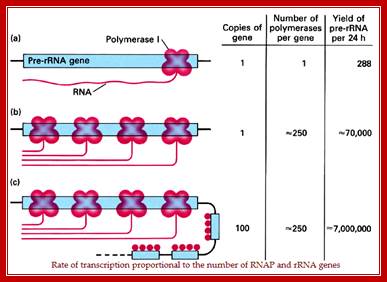
This diagram shows the rate of transcription is proportional to the number of Genes with their promoters and the number of RNA polymerases available for transcription. REF? https://images.search.yahoo.com
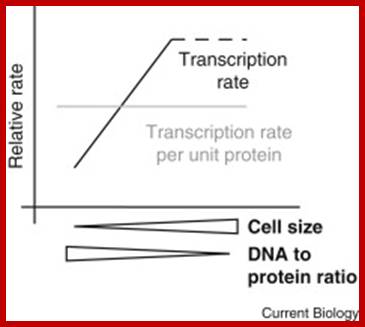
Is bigger better? Scientists have long puzzled over the potential relationship between cell size and the rate of mRNA production. A recent report builds a strong case that global transcription rates scale with size; http://www.cell.com

CDC6 id the part of the RNA POL1;http://jcs.biologists.org
rRNA Synthesis and Processing:
The
genes coding for rRNA (except 5S rRNA) are located in the nucleolar part of the
nucleus. The rRNA genes are highly repetitious and mammalian cells contain 100
to 2000 copies of the rRNA genes per cell. The genes are organised in
transcription units separated by non-transcribed spacers. Each transcription
unit contains sequences coding for 18S, 5.8S and 28S rRNA.
The transcription units are transcribed by RNA polymerase I into giant RNA
molecules, primary transcripts, that in addition to the sequences corresponding
to 18S, 5.8S and 28S rRNA contains external and internal transcribed spacer
sequences. The rate of nucleolar transcription is very high and many
polymerases operate on the same transcription unit. The transcriptionally
active DNA therefore has a Christmas tree-like appearance on electron
microscopic pictures.
The primary
transcript is processed into the mature 18S, 5.8S and 28S rRNAs. The processing
involves exo- and endo-nucleolytic cleavages guided by snoRNA (small nucleolar
RNAs) in complex with proteins. The mature rRNAs contain modified nucleotides
which are added after transcription by a snoRNA-dependent mechanism.
5S ribosomal RNA is transcribed by RNA
polymerase III in the nucleoplasm. Each eukaryotic cell contains a high number
of copies of the 5S coding gene (up to 20 000 copies per cell). 5S rRNA
contains overlapping binding sites for two different proteins, ribosomal
protein L5 and transcription factor TFIIIA. The mutual exclusive binding of
these two proteins to 5S rRNA is important for coordinating the expression of
5S rRNA to the production of the other rRNAs.
While transcripts are produced they go through folding required for processing. As the RNAPs successively associates with the 5’ end of the rRNA gene and transcribe the gene they generate a series of transcripts, from the smallest (at the start site) to the longest (at the end site); and they look like an inverted Christmas tree. The transcripts immediately associate with snoRNAs and snoRNPs and other proteins. They start processing to generate final products such as 18s, 5.8s and 28S. The 5srRNA is synthesized and processed elsewhere, but joins the main ribosomal RNA fragments in nucleolus. In some cases the 5sRNA is tucked in between main rRNA genes (yeast).
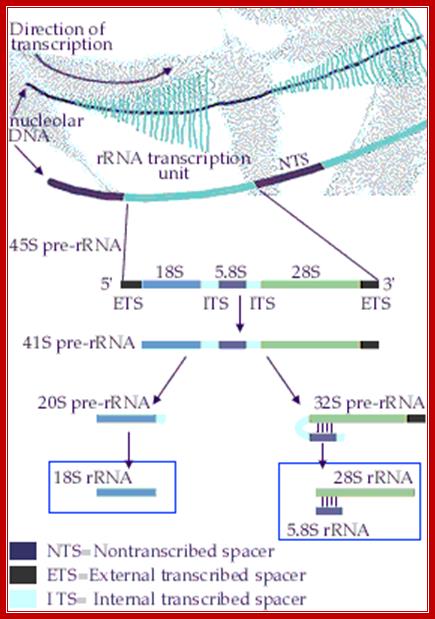
rRNA Synthesis and Processing;
The genes coding for rRNA (except 5S rRNA) are located in the
nucleolar part of the nucleus. The rRNA genes are highly repetitious and
mammalian cells contain 100 to 2000 copies of the rRNA genes per cell. The
genes are organised in transcription units separated by non-transcribed
spacers. Each transcription unit contains sequences coding for 18S, 5.8S and
28S rRNA.
The transcription units are transcribed by RNA polymerase I into giant RNA
molecules, primary transcripts, that in addition to the sequences corresponding
to 18S, 5.8S and 28S rRNA contains external and internal transcribed spacer
sequences. The rate of nucleolar transcription is very high and many
polymerases operate on the same transcription unit. The transciptionally active
DNA therefore has a Christmas tree-like appearance on electron microscopic
pictures; https://www.nobelprize.org/
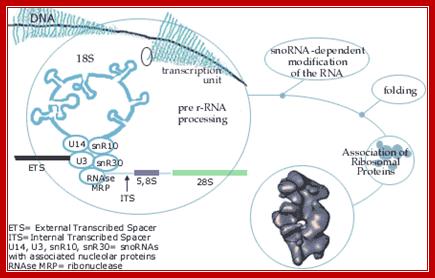
The genes
coding for rRNA (except 5S rRNA) are located in the nucleolar part of the
nucleus. The rRNA genes are highly repetitious and mammalian cells contain 100
to 2000 copies of the rRNA genes per cell. The genes are organised in
transcription units separated by non-transcribed spacers. Each transcription
unit contains sequences coding for 18S, 5.8S and 28S rRNA.
The transcription units are transcribed by RNA
polymerase I into giant RNA molecules, primary transcripts, that in addition to
the sequences corresponding to 18S, 5.8S and 28S rRNA contains external and
internal transcribed spacer sequences. The rate of nucleolar transcription is very
high and many polymerases operate on the same transcription unit. The
transciptionally active DNA therefore has a Christmas tree-like appearance on
electron microscopic pictures. http://www.nobelprize.org/
As rRNAs transcribed they are bound by snoRNPs, RNase and process them
into functional rRNA; this happens in Nucleolus, cajal body also facilitates the process. http://www.nobelprize.org/
Regulation rRNA transcription:
Cells go through several regulatory events like cell division, differentiation and stationary phase; accordingly synthesis of rRNA for ribosome production is regulated. As shown below cell cycle events are triggered by mitogenic signals that are transduced ultimately leading to activation of UBF and TIF1-IA, by the activity of CDK2/4 and cyclin D/A/E (phoshorylation rely); this leads to the transcription rRNA to the needs of the cell.
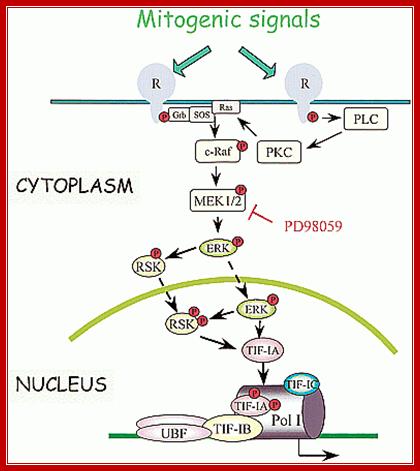
Mitogenic signals transduce kinases cascade resulting in activation of rRNA gene transcription; www.slideplayer.com

Model Depicting Contribution of DksA to Regulation of rRNA Expression DksA (blue dots) binds to RNAP (light blue oval), decreases the half-life of the open complex (RP o ), thereby increasing the concentration of the iNTP (gold) required for transcription initiation, and amplifies inhibition by ppGpp (green). rRNA promoters are sensitive to changes in the concentrations of the iNTP and ppGpp because DksA brings the lifetime of the open complex into a range where it is rate limiting for transcription in vivo. Two feedback loops control rRNA transcription (Gaal et al., 1997; Murray et al., 2003b): ppGpp is synthesized by RelA in response to uncharged tRNAs in the ribosomal A site, and translation consumes ATP and GTP. In a ⌬ dksA mutant (red), the rRNA promoter open complex is longer lived, making it insensitive (double red slashes) to physiologically relevant changes in iNTP and ppGpp concentrations, eliminating regulation of rRNA transcription. ;https://www.researchgate.net
Signaling pathway in response to nutrients:
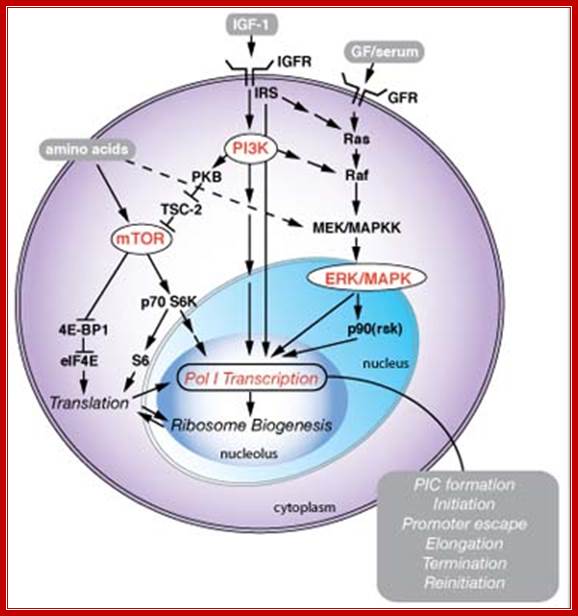
Signaling pathways associated with nutrients, cell growth and proliferation regulate Pol I transcription in the nucleolus. Growth factors (GF), for example Insulin-like growth factor 1 (IGF-1), serum, and nutrients and amino acids, regulate Pol I transcription coordinately through several signaling cascades involving reversible phosphorylation events. Pol I transcription drives ribosome biogenesis, which is additionally dependent on ribosome protein translation, and coordinated with Pol II (mRNAs encoding ribosomal proteins) and Pol III (5S rRNA) transcription. The regulation of Pol I transcription possibly could influence any of the steps of the transcription cycle, from preinitiation complex (PIC) formation, through to termination and reinitiation of transcription. Pol I transcription is additionally subject to cell-cycle control, but this is not included here. See James and Zomerdijk, J Biol. Chem. (2005 (Dr J Zomerdijk)
Repression of rDNA transcription by the tumor suppressor proteins pRb and p53:
The discovery that pRb represses Pol I transcription is very interesting. UBF is the target for pRb-induced repression of Pol I transcription. pRb accumulates in the nucleoli of differentiated or cell-cycle-arrested cells and has been shown to repress rDNA transcription in vitro and in vivo. Transcriptional repression is brought about by interaction of the C-terminal part of pRb with HMG boxes 1 and 2 of UBF. Thus, inactivation of UBF appears to be a most effective way for pRb to shut down rRNA synthesis and inhibit cell growth, (Cavanaugh et al. 1995; Voit et al. 1997; Hannan et al. 2000).
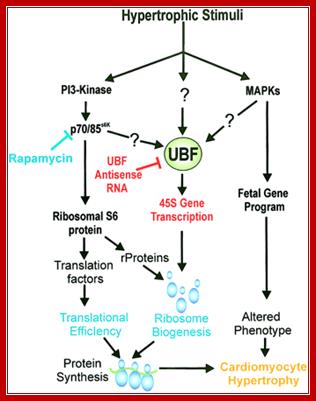
Signaling pathway in response hypertrophic stimuli:
Schematic diagram depicting the proposed pivotal role UBF plays in regulating cardiac rRNA synthesis and growth. Hypertrophic stimuli, by yet undefined pathways, increase UBF cellular activity resulting in accelerated rates of the 45S ribosomal gene (rDNA) transcription and ribosomal RNA (rRNA) synthesis. The increased pool of rRNA together with an increased translation of ribosomal proteins (rProtein) results in more functional ribosomes (increased translational capacity). UBF antisense RNA (red text) blocks UBF expression after hypertrophic stimuli and thus prevents synthesis of new ribosomes required for the growth response. Blue lines and text indicate pathways blocked by the 70 kDa S6 ribosomal protein kinase (p70S6k) inhibitor rapamycin.
Epigenetic silencing of RNA polymerase I transcription:
Ingrid Grummet and Craig S. Picard.

A | Binding of transcription termination factor TTF-I and UBF to unmethylated nucleosomal rRNA genes allows transcription initiation. b | Methylation of the cytosine at position -133 impairs binding of UBF to nucleosomal rRNA genes, thereby preventing transcription-initiation-complex formation. Mutation of the cytosine at position -133 abolishes methylation-dependent transcriptional repression on nucleosomal templates. c | This part of the ribosomal transcription unit comprises the mouse rRNA gene promoter and the approximate position of the upstream terminator T0, the upstream control element (UCE) and the core element (CORE) relative to the transcription initiation site (arrow), and the three CpG residues (at -143, -133 and +8).Nature Reviews; http://www.nature.com

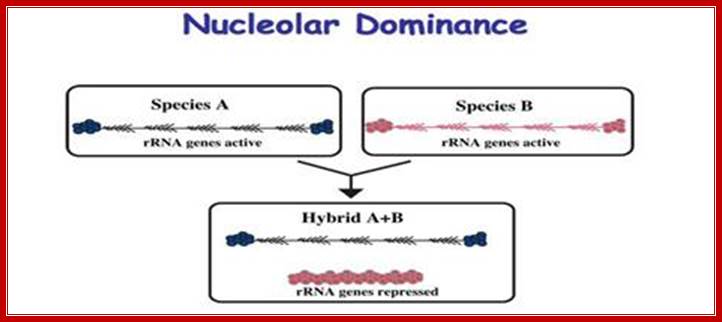
Ribosomal RNA genes encoded in chromatin undergo silencing and activation by deacetylation and methylation of histone tails and activation by demethylation and acetylation respectively. In genetic hybrids it is often the case that the rRNA genes from one parent (species A) are transcribed, and as a result assemble a nucleolus, but the rRNA genes inherited from the other parent are repressed (species B). This reversible, epigenetic phenomenon occurs in diverse eukaryotes and is known as nucleolar dominance. Although the mechanisms that initially discriminate the parental sets of rRNA genes are unclear, silencing is known to involve covalent modifications of both DNA and associated histones. Chemicals or genetic mutations that inhibit DNA methylation or histone deacetylation can de-repress the silent set of rRNA genes. http://www.nature.com/
Model of NoRC-mediated rRNA gene silencing:
Epigenetic silencing of RNA polymerase I transcription: Ingrid Grummet and Craig S. Picard
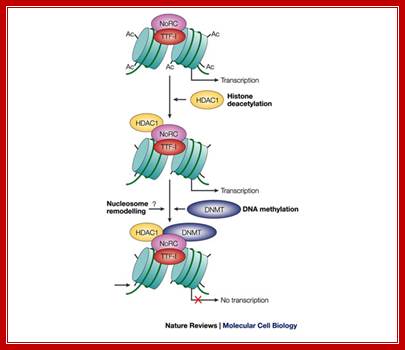
Model of NoRC-mediated rRNA gene silencing; Epignetic silencing of RNA pol I transcription; Transcription termination factor TTF-I is bound to the terminator T0 of transcribed rRNA genes, and the tails of histone H4 are acetylated (Ac). To establish the silent state, the nucleolar remodeling complex (NoRC) is recruited to the rRNA gene promoter by specific interaction between the amino-terminal region of TTF-I and an internal region of TTF-I interacting protein 5 (TIP5) (not shown). In subsequent steps, histone deacetylase 1 (HDAC1) and DNA methyltransferase DNMT1 and/or DNMT3b activities are recruited to the rRNA gene promoter, resulting in histone deacetylation and de novo DNA methylation. As a consequence, UBF binding and transcription-complex formation is impaired. The question mark denotes the lack of information as to whether NoRC remodels the promoter-bound nucleosome before or after DNA methylation. Also, we do not yet know at which step histone H3 becomes methylated on lysine 9; therefore, histone methylation is required for the binding ofHP1 protein. Ingrid Grummt and Craig S. Pikaardhttp://www.nature.com/
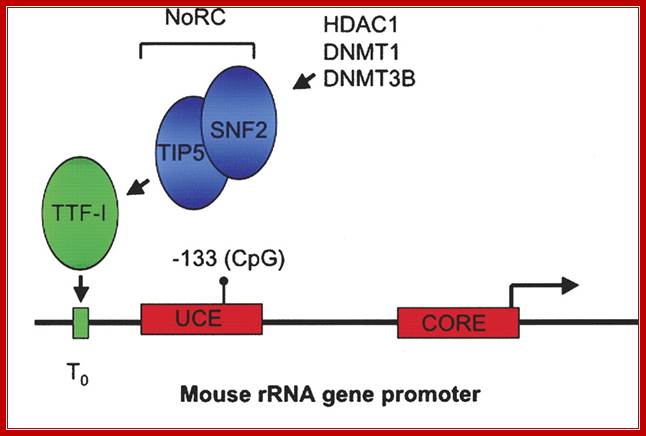
Recruitment of NoRC (Nucleolar repressing complex) represses rRNA gene transcription in mammals. The mouse rRNA gene promoter contains two promoter elements: a CORE element centered over the transcription initiation site, and an upstream control element (UCE). Binding of the Pol I transcription factor UBF to the UCE is inhibited by CpG methylation at position −133. A promoter-proximal transcription terminator, T0, binds the factor TTF-I that in turn recruits the Nucleolar remodeling complex NoRC, resulting in transcriptional repression of the linked promoter. NoRC is comprised of TIP5 and the ATP-dependent remodeling factor SNF2. NoRC in turn recruits DNA methyltransferases DNMT1 and DNMT3b and the histone deacetylase HDAC1 that methylate promoter DNA and deacetylate promoter-bound histones (not shown), respectively. The domains of TIP5 that specify its various interactions is shown below.
Distribution of silent and active rRNA genes in A. suecica nuclei. In the hybrid A.suecica, rRNA genes from the A. arenosa parental species are dominant over those from A. thaliana. Active genes within the A. arenosa NOR are decondensed, DNA hypomethylated, and associated with the histone modifications shown on the right. Silent genes within the A. arenosa. NOR and the entire underdominant/silent A. thaliana NOR are condensed, DNA-hypermethylated, and associated with the histone modifications shown on the left. The nucleus is depicted in dark blue, with two nucleoli in pale blue. Histone deacetylase HDA6, a presumptive deacetylase HDT1, unidentified DMTs and MBD proteins are required for maintaining silence.

Recruitment of NoRC represses rRNA gene transcription in mammals. The mouse rRNA gene promoter contains two promoter elements: a CORE element centered over the transcription initiation site, and an upstream control element (UCE). Binding of the Pol I transcription factor UBF to the UCE is inhibited by CpG methylation at position −133. A promoter-proximal transcription terminator, T0, binds the factor TTF-I that in turn recruits the Nucleolar remodeling complex NoRC, resulting in transcriptional repression of the linked promoter. NoRC is comprised of TIP5 and the ATP-dependent remodeling factor SNF2. NoRC in turn recruits DNA methyltransferases DNMT1 and DNMT3b and the histone deacetylase HDAC1 that methylate promoter DNA and deacetylate promoter-bound histones (not shown), respectively. The domains of TIP5 that specify its various interactions; ;Nucleolar dominance: a model for rRNA gene silencing; http://genesdev.cshlp.org
Regulation of rRNA Synthesis by TATA-Binding Protein-Associated
Factor Mot1![]()
Arindam Dasgupta,1† Rebekka O. Sprouse,1† Sarah French,2† Pavel Aprikian,3 Robert Hontz,1 Sarah A. Juedes,1 Jeffrey S. Smith,1 Ann L. Beyer,2 and David T. Auble1*
Mot1 is an essential, conserved, TATA-binding protein (TBP)-associated factor in Saccharomyces cerevisiae with well-established roles in the global control of RNA polymerase II (Pol II) transcription. Previous results have suggested that Mot1 functions exclusively in Pol II transcription, but here we report a novel role for Mot1 in regulating transcription by RNA polymerase I (Pol I). In vivo, Mot1 is associated with the ribosomal DNA and loss of Mot1 results in decreased rRNA synthesis. Consistent with a direct role for Mot1 in Pol I transcription, Mot1 also associates with the Pol I promoter in vitro in a reaction that depends on components of the Pol I general transcription machinery. Remarkably, in addition to Mot1's role in initiation, rRNA processing is delayed in mot1 cells. Taken together, these results support a model in which Mot1 affects the rate and efficiency of rRNA synthesis by both direct and indirect mechanisms; with resulting effects on transcription activation and the coupling of rRNA synthesis to processing.
Budding yeast cells contain ~140 copies of the ribosomal DNA (rDNA) repeat, approximately half of which are transcriptionally active during exponential growth. Recent evidence supports the hypothesis that the yeast regulatory response to the growth state is mediated by regulation of the number of active RNA polymerase I (Pol I) molecules rather than the number of active gene repeats.
In Saccharomyces cerevisiae, the Pol I preinitiation complex (PIC) assembles via cooperative interactions between TATA-binding protein (TBP), Rrn3, the multisubunit upstream activating factor (UAF), the core factor (CF), and Pol I itself. UAF associates stably with the Pol I promoter and recruits CF and TBP, which provide a substrate for the Pol I-Rrn3 complex, which is the competent form of the polymerase. Association of Pol I-Rrn3 with the PIC also stabilizes the association of CF with the promoter, suggesting a model in which the CFs dissociates from the promoter following departure of Pol I and Rrn3 as a result of productive initiation. In this way, UAF has been proposed to facilitate multiple rounds of transcription characterized by cycles of association and dissociation of the basal machinery with the promoter.
In mammalian cells, growth factor-mediated activation of rDNA transcription occurs by stimulation of transcriptional elongation rather than enhanced recruitment of Pol I. In yeast, the histone deacetylase Rpd3 participates in “closing” active rDNA repeats as cells enter stationary phase
Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development:
Savraj S. Grewal1, Ling Li1, Amir Orian1, Robert N. Eisenman1 & Bruce A. Edgar1
Regulating ribosome number is thought to control cellular growth. Synthesis of ribosomal RNA (rRNA) is a limiting step in ribosome biogenesis and rates of rRNA synthesis are generally altered depending on the growth status of a cell. Although studies in unicellular systems have addressed the mechanisms by which this occurs, few studies have applied a genetic approach to examine growth-dependent control of rRNA synthesis in metazoans. Here, we show that in Drosophila melanogaster Myc (dMyc) is a regulator of rRNA synthesis. Expression of dMyc is both necessary and sufficient to control rRNA synthesis and ribosome biogenesis during larval development. Stimulation of rRNA synthesis by dMyc is mediated through a rapid, coordinated increase in the levels of the Pol I transcriptional machinery. In addition, the growth effects of dMyc in larval wing imaginal discs require de novo rRNA synthesis. We suggest that during animal development, the control of rRNA synthesis and ribosome biogenesis is an essential Myc function.
Different steps in the pathway to ribosome production that can lead to cancer, when deregulated; Davide Ruggero & Pier Paolo Pandolfi, Nature Reviews Cancer 3, 179-192 (March 2003)
The regulation of ribosome biogenesis is a highly coordinated process that leads to accurate initiation and regulation of protein synthesis. There are three steps in this process, which, when deregulated, can contribute to increased tumorigenesis. The first step in ribosome production requires the synthesis of the 45S rRNA precursor. The transcription of this rRNA gene is negatively regulated by tumor suppressors such as p53 and retinoblastoma (RB) and augmented on mitogenic stimuli by several kinases that phosphorylate components of the transcription complex that are responsible for 45S synthesis. The accurate regulation of rRNA synthesis can be lost in cancer cells through inactivating mutations in tumor suppressors or upregulation of these kinases.
Another
step in ribosome biogenesis that maintains accurate cellular function involves
the modification of rRNA. One type of modification, which is catalyzed by an
enzyme known as dyskerin, converts uridine into pseudouridine (![]() ). Mutations in the gene encoding dyskerin, DKC1,
result in increased tumor susceptibility. Animal models that have lost DKC1
function show a marked increase in tumor incidence associated with a decrease
in rRNA processing.
). Mutations in the gene encoding dyskerin, DKC1,
result in increased tumor susceptibility. Animal models that have lost DKC1
function show a marked increase in tumor incidence associated with a decrease
in rRNA processing.
Ribosome assembly that involves the association of rRNA with more than 70 ribosomal proteins (made in the cytoplasm) is a coordinated process. The rRNA and proteins are assembled into the large subunit (60S) and small subunit (40S) of the ribosome. An increase in ribosomal protein production and activity has been observed in many cancer types. Mutations in ribosomal proteins such as S19 have also been associated with a human syndrome that is characterized by increased tumor susceptibility.

Each of the three highlighted steps (blue boxes) might have deleterious effects on the cell that could contribute to tumor initiation or cancer progression, or both, through aberrant regulation of protein synthesis. This can be manifested either by an increase in ribosome production, thereby leading to an upregulation in total translation, or by alterations in translation of specific mRNAs, which are involved in the regulation of cell proliferation. Furthermore, when key checkpoints in the cell that are important in coordinating ribosome production with accurate cell-cycle progression are lost, 'nucleolar stress' can result, and subsequently unrestrained cellular proliferation occurs. The advent of proteomics will aid in identifying protein targets that are deregulated as a result of perturbations in these pathways. Nature Reviews Cancer
Ribosomal RNA gene expression and State of chromatin during expression
All eukaryotic cells irrespective of species, do exhibit rRNA synthesis all the time in the nucleolar region of the nucleus. Nucleolar size changes, when cells are activated, in the case of IBA induced root initiation in Phaseolus vulgaris, the size of the nucleolus is so large it occupies ¾ of the nucleus. This shows the requirement of rRNA for pericyclic cells not for others. Requirement of rRNA for any cell is very high and so requires the transcription of rRNA all the time. To supply such high quantity, rRNA genes in the secondary constriction region, looped out as naked DNA consisting of 100ds of rRNA genes in tandem repeats or more, depending upon the species and the stage of development. In humans chromosome 13, 14, 15, 21 and 22 contain rRNA genes in a region called NOR or nucleolar organizer region. Yeast contains rRNA genes on chromosome XII. After telophase as the nuclear membrane reconstitutes, chromosomes relax and open; at this point of time rRNA coding DNA open out in the form of loops of various sizes and organize into nucleolus, where the DNA is freed from all chromatin proteins gets associated with transcription complex and transcription goes on.
Pikaard Lab Research , Indiana University; ttp://sites.bio.indiana.edu/
And one can visualize each of the rRNA genes that are in the process of transcription, on the open DNA, and one can observe Christmas tree like transcripts arranged-from the nascent to old transcript. In this the chromosomal DNA is totally devoid of any histones, and the only proteins associated are RNAP I and its associated factors are found. This goes on 24 hrs a day and 365 days a year. This implies for massive transcription the DNA should be free for the transcription complex to operate.
This is true for Arabidopsis thaliana (A.t.), Arabidopsis arenosa (A.a) and their hybrid, A. suecica (A.s.). However, some genes are expressed from the chromosomes inherited from only one parent. An example is nucleolar dominance an epigenetic phenomenon in hybrids which describes the formation of a nucleolus (or nucleoli) on the chromosomes inherited from only one of the progenitors, regardless of whether that progenitor is the maternal or the paternal parent. Nucleolar dominance occurs in both the plant and animal kingdoms and is due to the expression of only one parental set of rRNA genes.
In 1997 plant mol. biologists found that nucleolar dominance in Brassica hybrids or A. suecica can be reversed by chemical inhibitors of DNA methylation or histone deacetylation which led to the realization that nucleolar dominance is due to the selective silencing of one set of rRNA genes rather than the selective activation of the other. Because rRNA genes are clustered by the hundreds, spanning millions of base pairs of chromosomal DNA, nucleolar dominance is one of the most extensive gene silencing phenomena known.
Further study has shown that rRNA gene silencing involves concerted changes in DNA methylation and histone modification and a model has been proposed whereby DNA and histone modifications are each upstream of one another in a self-reinforcing, circular pathway. Changes in DNA methylation, histone acetylation and histone methylation are critical to the on-off switch mechanism that controls the number of active rRNA genes, both in hybrids displaying nucleolar dominance and in non-hybrids that regulate the effective dosage of their rRNA genes in response to the physiological needs of the cell.
It has been observed nucleolar loci exist in certain chromosomes and not in all. They are inherited by the same parent, i.e. but pollen-male and egg-female are contributors; it is virtually similar to biparental contribution. It so happens, the nucleolar DNA that opens up for transcription is either from one parent or the other. So one set of rRNA genes are kept silent and the other is expressed; which is similar to X chromosome inactivation in humans.