Eukaryotic RNAPs:
Important Features of RNAPS:
Unlike prokaryotic genes and its RNA polymerases, eukaryotes have tremendous variations in their gene structures and enzymes that transcribe them. There is a division of labor among the RNAPs and transcribe the genes accordingly. More than 90% of the total RNA found in eukaryotic cells is rRNA which is required in large amounts for the ribosomal structures undergo turnover; similarly tRNAs, 5s rRNAs, sn/sc RNAs, sRNAs, mi/siRNAs and small molecular weight snoRNAs and a large number of non coding, but functional RNA to name only few. On the other hand structural genes which represent mRNAs, though account 2-8% of the total RNAs, their structure, their function vary so much, a majority of them are common transcripts to all cell types and many of them are tissue specific. Thus it is not very easy to comprehend them in one go. Thus these different RNAs form different categories of importance; so eukaryotic systems have developed different enzyme complexes to transcribe different class of RNAs from different but specific class of genes. First some salient features of different class of RNAPs are described. RNAPs perse don’t work, but they associate with other RNAP factors called Transcription Factors (TFs) and then RNAPs assemble on their respective promoters and initiate transcription. Promoters with their specific sequences act as the sites for the assembly of TFs and other accessory factors and RNAPs.
RNAP I:
· Transcribes all Ribosomal RNA genes.
· Very active with low ionic strength.
· It requires Mn2+ and Mg2+ for its activity.
· It is insensitive to alpha Amanitin (a-Amanitin, a drug from fungus called death-Cap, Amanita phalloides and A. bisporangia).
A list of RNAPs:
|
|
EK |
Archa (a) |
Bact |
Plastid PEP coded |
Plastid NEP coded |
Mitochondria NEP |
viral |
|
ClassI subunit |
Rpb1 |
A’A’’ |
B’ |
B’ |
110Kd |
T7~99kD= Yeast~150kD |
|
|
|
Rpb2 |
B |
B |
B” |
|
|
|
|
|
Rpb3 |
D |
a1 |
A’ |
|
|
|
|
|
Rpb6 |
K |
w |
|
|
|
|
|
|
Rpb11 |
L |
A2 |
A” |
|
|
|
|
ClassII |
Rpb4 |
f |
|
|
|
|
|
|
|
Rpb5 |
H |
|
|
|
|
|
|
|
Rpb |
E |
|
|
|
|
|
|
|
Rpb8 |
G |
|
|
|
|
|
|
|
Rpb10 |
N |
|
|
|
|
|
|
|
Rpb12 |
P |
|
|
|
|
|
|
ClassIII |
Rpb9^1 |
|
|
|
|
|
|
|
|
G down |
Rpo13 |
delta |
|
|
|
|
|
|
|
|
|
|
|
|
|
RNAP II:
· Transcribes all structural genes, in the sense mRNA genes as well as few Noncoding RNAs such as some SnRNAs, Sno-RNA and few others such as siRNA and miRNAs.
· Very active at high ionic strength.
· Have pronounced preference for Mg2+ and Mn2+ ions.
· It is very sensitive to (α-Amanitin, which binds to subunit alpha; at 1 ug/ml –50% inhibitions).
RNAP-III:
· It transcribes a variety of genes, such as 5s RNA, U6 RNA, tRNA, 7sL RNAs, 7SK RNA and in some systems it can transcribe Epstein Barr viral genes and even some genes of ADV viral genes.
· It is active at broad range of ionic strengths.
· Its activity can be inhibited at very high concentration of a-Amanitin, a fungal product.
RNAP IV and V:
- RNAP IV and V are multisubunit enzymes evolved from RNAP II. These are mostly found in plant system, which transcribe small RNAs. In plants, Pol IV and Pol V, are not strictly required for viability but are important for development, transposon taming, anti-viral and anti-bacterial defense, and inter-allelic communications mediated paramutations.
Pol IV and Pol V functions are best understood with respect to RNA-directed DNA methylation, a process in which 24nt short interfering RNAs (siRNAs) direct the cytosine methylation, and silencing, of complementary DNA sequences. Pol IV acts early in the pathway, working in partnership with RNA-dependent RNA pol2 to produce double-stranded RNAs that are diced into siRNAs and loaded (primarily) into ARGONAUTE 4(AGO4). Independent of siRNA biogenesis, Pol V generates RNA transcripts at loci that undergo RdDM; and AGO4 binds these Pol V. Pol V itself subsequently recruits chromatin modifying factors, resulting in de novo cytosine methylation and establishment of repressive histone modifications (Ek Han Tan,1,2 Todd Blevins,1 Thomas S. Ream,2,3 and Craig S. Pikaard).
DNA dependent RNAP IV transcribes Transposons and many other repeats and generates ss-RNAs. It is believed that chromosome remodeling protein CLSY may facilitate PolIV transcription. RNA-dependent RNA polymerase RDR2 converts the aberrant single-stranded RNAs to double-stranded RNAs, which are then cleaved into 24-nt siRNAs by the Dicer-like proteins DCL3.
The 24-nt siRNAs are bound by an ARGONAUTE protein AGO4, AGO6, or AGO9 tethers AGO4 to nascent polV or polII RNA transcripts to form the RNA- directed DNA methylation effector complex. The effector complex directs the de novo DNA methyltransferase DRM2 to specific chromatin regions to catalyze new DNA methylation.
DNA-dependent RNA polymerase V (Pol V) transcribes intergenic non-coding (IGN) regions, and the product is used as small interfering RNAs. PolIV and PolV play non redundant roles in small interfering RNA SiRNA–mediated pathways, including silencing Retrotransposon and endogenous repeats via si-RNA mediate methylation of chromosomal DNA.
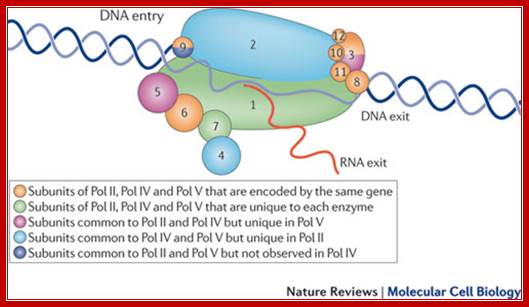
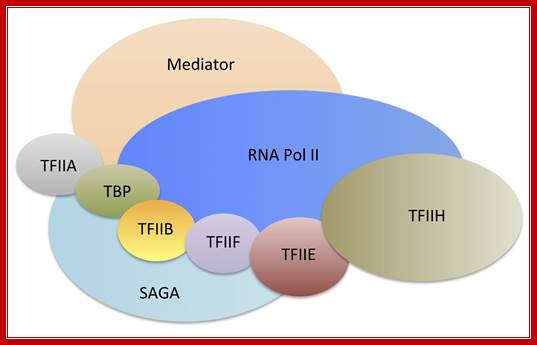
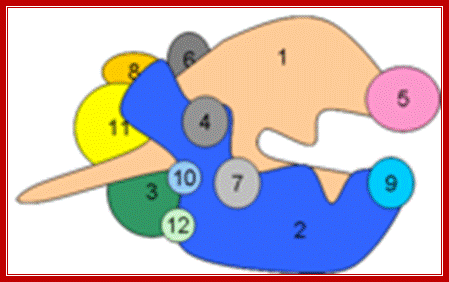
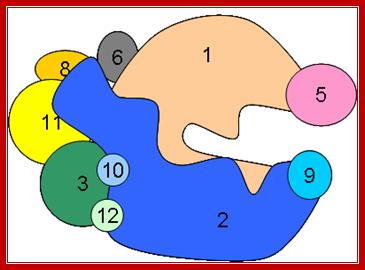
The cartoon above depicts the relative positions of RNA polymerase subunits, based on yeast RNA polymerase II (Pol II) and the relationships between Arabidopsis thaliana Pol II, Pol IV and Pol V in terms of their subunit compositions. RNA polymerase subunits are identified by their numbers and are color-coded as indicated. Subunits three and nine are shown in two colors to represent their alternative forms; Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing; Jeremy R. Haag & Craig S. Pikaard; Bottom Fig; Mechanism and Regulation of Eukaryotic transcription; The RNA Polymerase II transcription machinery is composed of RNA Polymerase, and five multi-subunit general transcription factors. Two large coactivator complexes commonly used at inducible genes are Mediator and SAGA. The transcription machinery has been conserved among all eukaryotes. Hahn Lab; https://labs.fhcrc.org
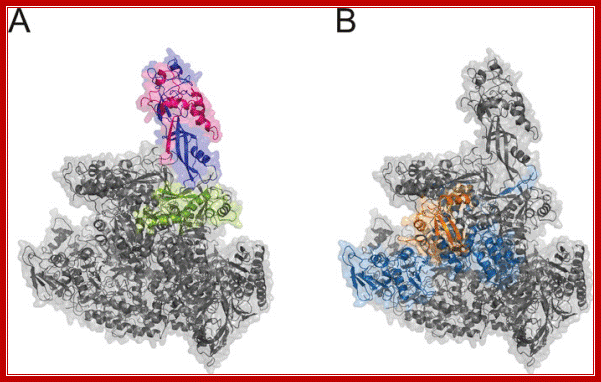
RNAP IV left and RNAP V right
RNA polymerases IV and V (Pol IV and Pol V) are plant-specific polymerases that are required for the biogenesis or function of small interfering RNAs (siRNAs) in the siRNA-directed DNA methylation pathway. This pathway silences repeated genomic sequences, including transposable elements and pericentromeric repeats. We have shown that RNA polymerase IV and/or V function is required to maintain the normal organization of the chromatin within the nucleus, such that mutation of their shared second-largest subunit (NRPD2/NRPE2) causes heterochromatin dispersal. http://www.biochemj.org/
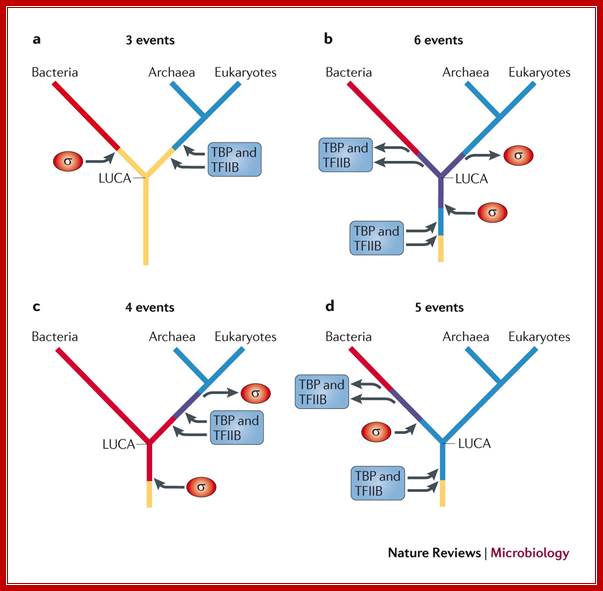
Evolution tree of multisubunit RNA polymerases in the three domains of life; Finn Werner & Dina Grohmann; Nature Reviews
The bacterial RNA polymerase (RNAP) strictly depends on σ-factors for transcription initiation, whereas archaeal and eukaryotic RNAPs require the initiation factors TATA box-binding protein (TBP) and transcription factor IIB (TFIIB). There are four potential scenarios for the evolution of these factors. As the first scenario requires the fewest independent evolutionary events, it is the simplest explanation for the occurrence of transcription initiation factors in all extant life. a | As there are no σ-factor homologues in archaea and eukaryotes, and no TBP and TFIIB homologues in bacteria, it is likely that the ancestral RNAP of the last universal common ancestor (LUCA) used neither σ-factors nor TBP or TFIIB factors, and that these factors evolved independently in the bacterial and archaeal–eukaryotic lineages, respectively, after their split. b | An alternative scenario is that the RNAP of the LUCA used both σ-factors and TBP and TFIIB factors in parallel, and then lost the relevant factors in each lineage. c | A third scenario is that the LUCA used σ-factors, and then the archaeal–eukaryotic lineage lost these factors are gained TBP and TFIIB factors. d | The final scenario is that the LUCA used TBP and TFIIB factors, and that these were then lost in the bacterial lineage and σ-factors were gained.
Bacterial and Eukaryotic RNA polymerases-comparative account-
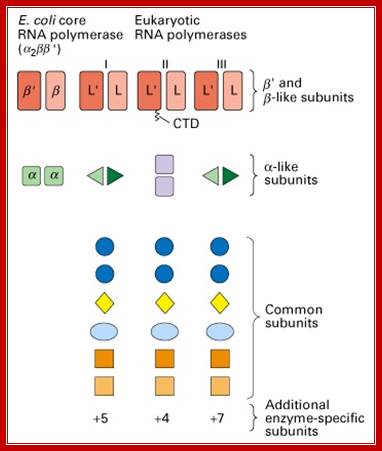
Fig: Comparison of prokaryotic and eukaryotic RNA polymerase subunit homology. (Lodish et al., 2000); http://www.cbs.dtu.dk/
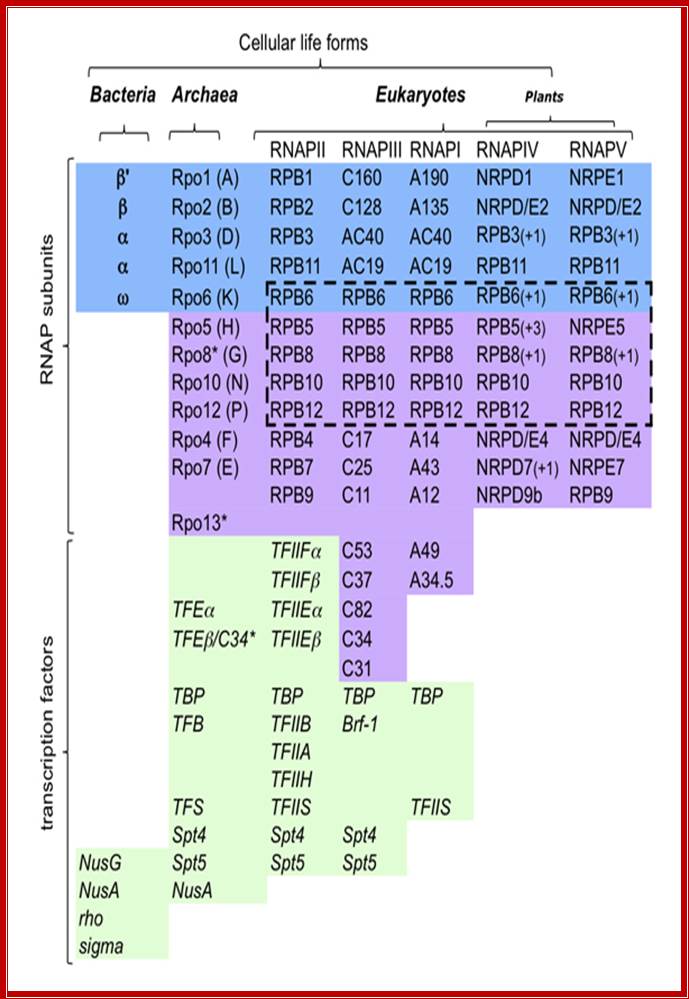
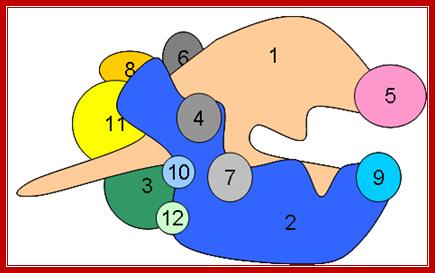
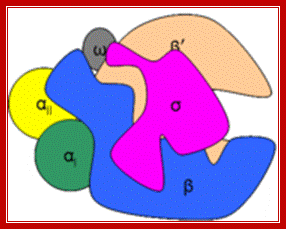
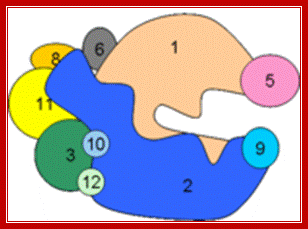
Multisubunit RNAP resemble a crab claw whose 'jaws' (Figure 1, highlighted in red) interact with duplex DNA in the direction of transcription. The DNA projects along the floor the major DNA-binding channel (blue) and is secured by the RNAP 'clamp' (green) until it encounters the active centre (yellow) at the RNAP 'wall'. The DNA-RNA hybrid is perpendicular to the downstream duplex DNA and the strands are separated by the RNAP 'lid', and the transcript is guided by interactions with the RNAP 'stalk' (orange). The NTP entry pore, or secondary channel, is located under the active site and allows access of substrates and cleavage factors to the active site and extrusion of the transcript during backtracking. All RNAP subunits can be divided into three overlapping functional classes. RNAP subunits homologous to Rpo3 (corresponding to alphaI in bacteria), 10, 11 (alphaII) and 12 form the assembly platform (deep blue), whose association nucleates RNAP assembly. The two largest subunits Rpo1 (beta') and 2 (beta) form the catalytic core that harbours the active site including the Magnesium chelating carboxylate residues, the bridge and trigger helices, the downstream DNA and DNA-RNA hybrid binding sites, the secondary NTP entry channel and loop and switch regions that are instrumental in the handling of the nucleic acids scaffold including strand separation. The combination of assembly platform and catalytic core is the minimal subunit configuration of active RNAPs. The other RNAP subunits are not strictly required for basic RNAP operations (including promoter-directed transcription) and have auxiliary functions by adding interaction sites with basal transcription factors and/or nucleic acids. Rpo5 extends the RNAP jaw's interactions with the downstream duplex DNA during transcription initiation, Rpo6 (omega) aids the folding and stability of Rpo1 and acts as anchorage point for the Rpo4/7 'stalk' complex. RNAP subunits Rpo4/7 form a stable heterodimeric sub complex, which interacts with the nascent RNA transcript during elongation and termination. http://www.ucl.ac.uk/
A List of RNAPs and RNAPs Subunits- a Comparative list:
|
|
RNAP-I |
RNAP-II |
RNAP-III |
Similar to E.coli’subunits |
Core |
A-190 |
B-220 |
C-160 |
Beta’ like |
|
Core |
A- 135 |
B-150 |
C-128 |
Beta like |
|
Core |
|
B 44.5 |
|
α like-40 |
|
|
|
B 13.5
|
|
|
|
|
|
Common to A & C |
|
|
|
|
AC 40 |
|
AC40 |
Alpha like |
|
|
AC 19 |
|
AC19 |
Alpha like |
|
|
|
Common to A,B &C |
|
|
|
|
|
ABC 27 |
|
|
|
|
|
ABC 23 |
|
=w like |
|
|
|
ABC 14.5 |
|
|
|
|
|
ABC a 10 |
|
|
|
|
|
ABC b 10 |
|
|
|
|
Specific to I |
Specific to II |
Specific to III |
|
|
|
A 49 |
B 32 |
C 82 |
|
|
|
A 43 |
B 16 |
C 53 |
|
|
|
A34.5 |
B12.6 |
C 37 |
|
|
|
A12.2 |
B12.5 |
C 34 |
|
|
|
A 40 |
|
C31 |
|
|
|
A 14 |
|
C 25 |
|
|
|
|
|
C 28 |
|
|
|
|
|
C 11 |
|
|
|
Subunits=~10 |
Subunits = ~13 |
Subunits =~12 |
|
These above subunit numbers and Mol.wt often changes with every update, because is collected from different sources.
Comparative account of Eukaryotic RNAPs mol.wt:
|
Function |
PK |
Archaea |
S.cerevisiae |
Pol II |
Pol IV |
Pol V |
|
Catalytic |
B’RPOA’ |
RPB1 |
RPB1 |
59 |
54 |
74 |
|
“ |
B |
RPOB’ ‘’ |
RPB2 |
63 |
18 |
27 |
|
“ |
ALPHA |
RPOD |
RPB3 |
57 |
28 |
45 |
|
Assembly |
a |
RPOL |
RPB11 |
75 |
56 |
68 |
|
“ |
|
RPON |
RPB10 |
55 |
54 |
55 |
|
“ |
|
RPOP |
RPB12 |
16 |
16 |
16 |
|
Auxiliary |
w |
RPOK |
RPB6 |
15/16 |
15 |
15 |
|
“ |
|
RPOG |
RPB8 |
30/30 |
18 |
|
|
“ |
|
RPOH |
RPB5 |
63 |
15 |
39 |
|
“ |
|
RPOF |
RPB4 |
61 |
13 |
8 |
|
“ |
|
RPOE |
RPB7 |
51 |
9/52 |
33 |
|
“ |
|
TFS/RPOX |
RPB9 |
22/28 |
22 |
22/22 |
Yeast RNAPs
|
RNAP-I (KD) (RPA) |
RNAP-II (KD) (RPB) |
RNAP-III (KD) (RPC) |
Similar to other RPBs |
|
190 |
220 |
160 |
|
|
135 |
150 |
128 |
|
|
49 |
- |
82 |
|
|
43 |
- |
53 |
|
|
40 |
44.5 |
40 |
|
|
34.5 |
32 |
34 |
|
|
|
|
|
|
|
C |
C |
C |
|
|
27 |
27 |
27 |
RPB-5 |
|
23 |
23 |
23 |
RPB-6 |
|
19 |
19 |
19 |
- |
|
14.5 |
14.5 |
14.5 |
RPB-8 |
|
14 |
12.6 |
22 |
|
|
12.2 |
- |
- |
RPB-10 |
|
10 |
10 |
10 |
RPB-12 |
Yeast RNAP II subunits:
|
RNAP-II (Mol.wt in KD) |
Chromosome |
Effect of chromosome deletion |
|
220 |
4 |
Inviable |
|
150 |
15 |
Inviable |
|
45 |
9 |
Inviable |
|
32 |
1`0 |
Conditional |
|
27 |
2 |
Inviable |
|
23 |
16 |
Inviable |
|
17 |
4 or12 |
In viable |
|
14 |
15 |
|
|
13 |
7 |
Conditional |
|
10 |
15 |
Inviable |
|
13 |
15 |
Inviable |
|
10 |
8 |
Inviable |
|
|
|
Inviable |
Comparative List of RNAP Components:
|
Prokaryote
|
Archaea
|
Eukaryote RNAP I |
Eukaryote RNAP II |
Eukaryote RNAP III |
|
(CORE) |
(CORE) |
Pol-I |
Pol-II |
Pol-III |
|
b’ |
A’ / A’’ |
RPA1 |
RPB1 (=β’)192kd |
RPC1 |
|
β |
B’/b’’ |
RPA2 |
RPB2(=β)139kd |
RPC2 |
|
a’ |
2D |
RPC5 |
RPB3(=α)35.3kd |
RPC5 |
|
a’’ |
2L |
RPC9 |
RPB11(=α)13.6kd |
RPC9 |
|
ω |
K |
RPB6 |
RPB6(=ω)17.9kd |
RPB6 |
|
|
+6 others |
+9 others |
RPB4 25.4= σ like |
+11 others |
|
|
|
|
RPB5 25.4kd |
|
|
|
|
|
RPB7 19.1kd |
|
|
|
|
|
RPB8 16.5kd |
|
|
|
|
|
RPB9 14.3kd |
|
|
|
|
|
RPB10&12- 8.3,7.7KD |
|
|
|
|
|
|
|
RNAP Associated Transcriptional Factors (TFs):
RNAP-I Associated TFs:
|
Factors |
Sub units (Mol.wt) |
Functions
|
|
SL-1: TBP |
TBP/ |
Binds to core sequence |
|
TAFs 1, 2 and 3 (TBP associated factors) |
TAF complex 110, 63, 48 |
Bind to core and facilitates TBP binding |
|
UBF1 (upstream binding factors) |
97/94, H3, H4 like |
UBF1 binds to both core and also to UCE region. Interaction between proteins factors brings the UCE to the core by DNA looping. This activates RNAP-I |
|
|
>5 subunits =~348KD |
|
Note: SL1 = TF IA, UBF1 = TF IB and PAFs
RNAP-II associated TFs and Co activators:
|
Factors |
Subunits (KD) |
Functions |
TF II-A |
2 subunits, 56,14 in yeast; 3subunits-12,19,35 (human) |
Binds to TBP upstream, may be required for TATA less promoters |
|
TF II-B |
35 (H) (38 yeast) |
Acts like a bridge between TATA-TPB and adjacent DNA, ; this acts as a rate limiting factor; ‘stirrup’ detects start and the orientation of promoter |
|
TF II-D: TBP-binding factor.
TAF-II (10-12 subunits, species and tissue specific) |
38-50- monomer (H), (27-yeast)
250,150,110,80,60,40,30(a), 30(b) ? ?TAF interacts with co activators and core subunits |
TBP selects and binds to TATA box, disrupts the helix, bends the DNA, it further facilitates the binding of TAFs; TAFs in turn facilitate the binding of other factors and RNAP, bind to InR and DPE; They vary species to species and tissue to tissue |
TF II-E |
56kd , 34kd. H(2)=165 Y(2)=184
|
Binds to RNAP II CTD-tail; They interact with SP1 factors’ help in recruiting TFII-H
|
|
TF II-F: RAP |
RAP 38 (2) like sigma RAP 74 (2)like helicase |
Like sigma factors they bind to RNAP and interacts with IIB, brings RNAP on to TBP ; -Like an helicase, binds to RNAP-II and facilitates to restart of RNAP passage to elongation |
|
TF II-J |
43 41 35 |
Kinase |
|
TFII-H 9-12subunits), Y=9=512
|
XPG (XPD and XPB), XPF,Ercc1) XPC (C=RAD) binds to damaged DNA |
Specific subunits: Helicase and CTD Kinase required for unwinding and clearing the promoter, also involved in excision-DNA repair, during transcription. |
|
TF II-S |
SII 38KD S III 15KD, 18KD,110 KD’ HSPT5-required for elongation |
Stimulates elongation and limits RNAP pausing ? involved in proof reading- mismatch repair |
|
|
~28 subunits=~1625 KD |
|
ELL1 |
PTEFb1-80KD |
Recruits TAT-SF which recruit splicing components |
|
|
PTEFb2-43KD,124KD HSPT5-required for elongation |
Phosphorylates serine in CTD tail, p.lates HSPT5 |
|
|
TAT-SF1 |
Recruits splicing components |
|
|
|
|
Comparative Account of Mol.wt of TBPs &TAFs:
|
|
Drosophila |
Human |
S.cerviciae |
Remarks |
TBPs |
38 |
43 |
27 |
Single subunit binds minor groove to TATA box |
|
TAF1 |
250/230 |
250 |
130/145 |
Binds TBP |
|
TAF2 |
150/110 |
150 |
150 (TSMI) |
|
|
TAF3 |
80/85 |
100 |
|
|
|
TAF4 |
60/62 |
70/80 |
90 |
H3 like |
|
TAF5 |
40/42 |
31/32 |
60 |
H4 like |
|
TAF6 |
30/28 |
20 |
17 |
H3 like |
|
TAF7 |
22 |
15 |
68/61 |
H2B like |
|
TAF830B |
30B |
28,55, 30, 18 (25/23,47, 30,19), |
40, 67,23,19 |
Has histone fold |
|
|
|
|
|
|
RNAP-III associated TFs- 5s rRNA genes:
|
Factors |
Mol.wt |
Functions |
|
TF IIIA |
>300kd |
Binds to A box, which facilitates the binding of IIIC |
|
TF III C |
600KD, >six subunits |
Binds to IIIA and C facilitates the binding of IIIB and RNAP III |
|
TF IIIB |
B=3 subunit TBP/TAF (BRF) complex;90, 72
|
Identifies the start site and facilitates the positioning of RNAPIII via TFIIID |
RNAP-III associated TFs: tRNA Genes:
|
Factors |
Mol wt |
Functions |
TF IIIB |
TBP, BRFs- B=90 TAF=172, TAF-L
|
Facilitates RNAPIII to bind and initiate transcription |
TF IIIC |
600kd, six subunits |
Initially binds to both boxes and recruits IIIB and RNAP III |
|
|
|
|
|
|
|
|
RNAP associated Factors for 7SLRNA genes:
|
Factors |
Mol.wt |
Functions |
TF-III B |
TBP, BRFs, SNAPs |
Positioning of RNAP to the promoters |
|
PSE, OCTA, GC binding factors |
|
|
|
|
|
|
|
|
|
|
Note: the kinds of factors and their molecular weights given are the collection from different sources and made it more uniform. Different authors and different books give the kind, number and Mol.wt differently; this leads to lot confusion to both teachers and students.
A symphony of transcription factors for gene control:
|
Types |
Kinds |
Examples |
|
|
I |
Activator and repressor targets to inherent core machinery, promoter recognition and enzyme functions |
TAFs, TFIIA, NC2, PC4 |
|
|
II |
Activator and repressor adapters, modulate DNA binding, target other co-regulators |
OCA-B/OBF-1, Groucho, Notch, CtBP, HCF, E1A, VP16 |
|
|
III |
Multifunctional structurally related but highly divergent co-regulators: some interact with RNA Pol II and/or multiple types of activators, some also appear to have inherent enzymatic functions or chromatin-selective properties |
yeast: Mediator, SRBs human a: CRSP, PC2 human b: ARC/DRIP/TRAP human c: NAT, SMCC, Srb/Mediator |
|
|
IV |
Chromatin-modifying activator and repressor adapters, acetyltransferase or deacetylase activities with multiple substrates: histones, histone-related proteins, activators, other co-regulators and the core machinery |
CBP/p300, GCN5, P/CAF, p160s (SRC1, TIF2, p/CIP, etc.), HDAC-1 and HDAC-2 (rpd3), Sir2 |
|
|
V |
ATP-dependent chromatin remodeling activities |
SNF2-ATPase (SWI/SNF, RSC) and ISWI-ATPase (NURF, ACF, |
|
|
|
|
|
|
Composition of RNA polymerase II holoenzyme:
The holoenzymes contains the core RNA pol II, the general transcription factors (GTFs: TFIIH, TFIIE, TFIIF, TFIIB), the core Srb-mediator complex, the Srb10p cyclin-dependent kinase (CDK) complex, and the SNF-SNF complex.
|
Gene |
Synonyms |
ORF |
Size of subunits (kDa) |
Features**) |
|
RPB1 |
RPO21 SUA8 RPB220 |
YDL140C |
220 |
Heptapeptide repeat |
|
RPB2 |
RPB150 SIT2 SOH2 |
YOR151C |
150 |
. |
|
RPB3 |
. |
YIL021W |
45 |
. |
|
RPB4 |
. |
YJL140W |
32 |
. |
|
RPB5 |
SPP51 |
YBR154C |
27 |
Shared by PolI, II; and III |
|
RPB6 |
RPO26 |
YPR187W |
23 |
Shared by PolI, II; and III |
|
RPB7 |
. |
YDR404C |
17 |
. |
|
RPB8 |
. |
YOR224C |
14 |
Shared by PolI, II; and III |
|
RPB9 |
SSU73 |
YGL070C |
13 |
. |
|
RPB10 |
. |
YOR210W |
10 |
Shared by PolI, II; and III |
|
RPB11 |
. |
YOL005C |
13 |
. |
|
RPB12 |
RPC10 |
YHR143W-A |
9 |
Shared by PolI, II; and III |
|
|
|
|
|
|
|
SWI/SNF Complex in S. cerevisiae: activator complex; chromatin remodeling complexes |
|
Gene |
Synonym |
ORF |
Size of subunit (kDa) |
Features**) |
|
. |
YOR290C |
194 |
DNA dependent ATPase |
|
|
SWI1 |
. |
YPL016W |
148 |
acts to assist gene-specific activators through chromatin remodeling |
|
SNF5 |
. |
YBR289W |
103 |
acts to assist gene-specific activators through chromatin remodeling |
|
SWI3 |
. |
. |
93 |
. |
|
SWp82p |
. |
. |
~82 |
. |
|
SNF12 / SWP73 |
. |
. |
64 |
. |
|
ARP7 |
. |
YPR034W |
54 |
acts to assist gene-specific activators through chromatin remodeling |
|
ARP9 |
. |
YMR033W |
53 |
. |
|
SNF6 |
. |
. |
38 |
. |
|
ANC1 / TFG3 |
. |
. |
27 |
. |
|
SNF11 |
. |
. |
19 |
. |
|
SRB Mediator Complex: activators |
|||||
|
Subunits |
Gene |
Synonym |
ORF |
Size of subunit (kDa) |
Features **) |
|
Srb's |
SRB2 |
HRS2 |
YHR041C |
23 |
. |
|
. |
SRB4 |
. |
YER022W |
78 |
Target for Gal4 activator |
|
. |
SRB5 |
. |
YGR104C |
34 |
Unknown function |
|
. |
SRB6 |
. |
YBR253W |
14 |
. |
|
. |
SRB7 |
. |
YDR308C |
16 |
. |
|
Med's |
MED1 |
. |
YPR070W |
64 |
. |
|
. |
MED2 |
. |
YDL005C |
48 |
. |
|
. |
MED3 |
PGD1 HRS1 |
YGL025C |
47 |
. |
|
. |
MED4 |
. |
YOR174W |
32 |
. |
|
. |
MED6 |
. |
YHR058C |
32 |
Role in activation of some genes |
|
. |
MED7 |
. |
YOL135C |
32 |
. |
|
. |
MED8 |
. |
YBR193C |
25 |
. |
|
. |
MED9 |
CSE2 |
YNR010W |
17 |
. |
|
. |
MED10 |
NUT2 |
YPR168W |
18 |
. |
|
. |
MED11 |
. |
YMR112C |
15 |
. |
|
Gal11p |
GAL11 |
RAR3 SDS4 SPT13 |
YOL051W |
120 |
. |
|
Rgr1p |
RGR1 |
. |
YLR071 |
123 |
. |
|
Sin4p |
SIN4 |
GAL22 TSF3 BEL2 SSN4 SDI3 |
YNL236W |
111 |
. |
|
Rox3p |
ROX3 |
SSN7 NUT3 ARE3 |
YBL093C |
25 |
. |
Taken from: FCP Holstege et al.: Dissecting the Regulatory Circuitry of a Eukaryotic Genome (1998), Cell 95, 717-728
|
Gene |
Synonym |
ORF |
Size of subunit (kDa) |
Features |
|
SRB8 |
NUT6 SSN5 |
YCR080W |
144 kDa |
. |
|
SRB9 |
NUT8) SCA1 |
YDR443C |
160 kDa |
. |
|
SSN3 UME5; |
YPL042C |
61 kDa |
Cyclin dependent CTD kinase |
|
|
SRB11 |
SSN8 UME3 |
YNL025C |
38 kDa |
Srb10p cyclin partner |
|
General Transcription Factors |
|||||
|
Gene |
ORF |
Size of subunit (kDa) |
Features**) |
Shared |
|
|
TFIID |
. |
||||
|
. |
TAF150 / TSM1 |
YCR042C |
161 |
Required for cell cycle progression |
TFIID spec. |
|
. |
TAF145 / TAF130 |
YGR274C |
121 |
Histone acetyltransferase |
TFIID spec. |
|
. |
TAF90 |
YBR1410 |
89 |
Required for cell cycle progression |
SAGA |
|
. |
TAF67 |
YMR227C |
67 |
. |
. |
|
. |
TAF61 / TAF68 |
YDR145W |
61 |
Similar to histone H2B |
SAGA |
|
. |
TAF60 |
YGL112C |
58 |
Similar to histone H4 |
SAGA |
|
. |
TAF47 |
YPL011C |
40 |
. |
. |
|
. |
TAF40 |
YML015C |
41 |
. |
. |
|
. |
TAF30 / ANC1 / TFG3 / SWP29 |
YPL129W |
27 |
Component of TFIIF |
. |
|
. |
TAF25 / TAF23 |
YDR167W |
23 |
. |
SAGA |
|
. |
TAF19 / FUN81 |
YML098W |
19 |
. |
. |
|
. |
TAF17 / TAF20 |
YMR236W |
17 |
Similar to histone H3 |
SAGA |
|
. |
TBP1 / SPT15 |
YER148W |
27 |
. |
. |
|
. |
SUA7 |
YPR086W |
. |
Functions in selection of site for transcription |
. |
|
. |
SSU71 / TFG1 |
YGR186W |
82 |
RNA polymerase II transcription initiation factor, factor g subunit |
. |
|
. |
TFG2 |
YGR005C |
47 |
RNA polymerase II transcription initiation factor, factor g subunit |
. |
|
. |
ANC1 / TFG3 |
YPL129W |
27 |
RNA polymerase II transcription initiation factor, factor g subunit |
. |
|
. |
YKL028W |
55 |
RNA polymerase II transcription initiation factor |
. |
|
|
. |
TFA2 |
YKR062W |
37 |
RNA polymerase II transcription initiation factor |
. |
|
. |
TFB1 |
YDR311W |
73 |
Nucleotide excision repair |
. |
|
. |
TFB2 |
YPL122C |
59 |
Nucleotide excision repair |
. |
|
. |
TFB3 |
YDR460W |
32 |
Nucleotide excision repair |
. |
|
. |
TFB4 |
YPR056W |
37 |
. |
. |
|
. |
RAD3 |
YER171 |
90 |
DNA helicase/ATPase of RNA polymerase II transcription initiation factor TFIIH |
. |
|
. |
SSL1 |
YLR005W |
52 |
TFIIH subunit
(transcription initiation factor), factor B |
. |
|
. |
SSL2 / RAD25 |
YIL143C |
95 |
DNA helicase component of RNA polymerase II |
. |
|
. |
YDL108 |
35 |
Cyclin dependent CTD ser/thr protein kinase |
. |
|
|
. |
CCL1 |
YPR025 |
45 |
TFIIH subunit (transcription initiation factor), |
. |
|
SAGA Factors in S. cerevisiae |
|
Gene |
Synonym |
ORF |
Size of subunit (kDa) |
Features**) |
Shared with |
|
ADA4 SWI9 |
. |
51 |
Histone acetyltransferase |
SAGA spec |
|
|
SPT3 |
. |
YDR392W |
38 |
. |
SAGA spec. |
|
SPT7 |
GIT2 |
YBR081C |
153 |
. |
. |
|
SPT8 |
. |
YLR055C |
66 |
. |
. |
|
SPT20 |
ADA5 |
YOL148C |
68 |
. |
SAGA spec |
|
ADA1 |
HFI1 SUP110 |
YPL254W |
54 |
. |
. |
|
ADA2 |
SWI8 |
YDR448W |
51 |
. |
. |
|
ADA3 |
NGG1 SWI7 |
YDR176W |
79 |
. |
. |
|
TAF90 |
. |
YBR1410 |
89 |
Required for cell cycle progression |
TFIID |
|
TAF61 |
. |
YDR145W |
61 |
Similar to histone H2B |
TFIID |
|
TAF60 |
. |
YGL112C |
58 |
Similar to histone H4 |
TFIID |
|
TAF25 |
. |
YDR167W |
23 |
. |
TFIID |
|
. |
YMR236W |
17 |
Similar to histone H3 |
TFIID |
Additional panels: Chromatin remodeling; http://labcoulombe.usherb.ca
|
|
|
Comparison of TAFIIs (TFIID) |
|||||
|
Yeast Gene |
Size |
Drosophila |
Human |
Features |
Contacts in POL II |
|
TBP |
27 |
dTBP |
hTBP |
. |
TFIIA and TFIIB |
|
TAF145/130 |
121 |
dTAF250 |
hTAF50/230 |
Cell cycle progression (G1/S); HMG Box, bromodomains; HAT activity |
TFIIF kinase |
|
TAF150 |
161 |
dTAF150 |
hTAF150/CIF150 |
Cell cycle progression |
TBP |
|
. |
. |
dTAF110 |
hTAF135/130 |
. |
TFIIA |
|
. |
. |
. |
hTAF105 |
Cell-specific hTAF135/130 variant |
. |
|
TAF90 |
89 |
dTAF85/80 |
hTAF100/95 |
. |
. |
|
TAF67 |
67 |
. |
hTAF55 |
. |
. |
|
. |
. |
. |
hTAF43 |
. |
. |
|
TAF25/23 |
23 |
. |
hTAF30 |
. |
TBP |
|
TAF47 |
40 |
. |
. |
. |
. |
|
TAF30 |
27 |
. |
AF-9,ENL |
. |
TFIIF; SWI/SNF complex |
|
. |
. |
. |
. |
. |
. |
|
TAF68/61 |
61 |
dTAF30a/28/22 |
hTAF20/15 |
Similarity to H2B histone |
TBP |
|
TAF60 |
58 |
dTAF62/60 |
hTAF80/70 |
Similarity to H4 histone |
TFIIF, TFIIE56, TBP |
|
TAF20/17 |
17 |
dTAF42/40 |
hTAF32/31 |
Similarity to H3 histone |
TFIIB |
|
TAF40 |
41 |
dTAF30b |
hTAF28 |
SImilarity to SPT3 (SAGA); histone fold |
. |
|
TAF19 |
19 |
. |
hTAF18 |
SImilarity to SPT3 (SAGA); histone fold |
. |
Among all the RNAPs, the largest subunit (220kd) is found in RNAP-II; contains a common sequence of seven amino acids at its C-terminal. The repeats are Tyr (Y), Ser (S), Pro (P), Thr (T), Ser (S), Pro (p), Ser(S) = YSPTSPS (TSPYSPS). This region is called C-terminal domain (CTD).
· This sequence is repeated 20 times in yeasts and 50-52 times or more in mammalian systems.
· Predominance of hydroxyl amino acids indicates, perhaps these are subjected phosphorylation and dephosphorylation during the course of transcriptional activity and it is a fact that they are phosphorylated at specific positions during processing of transcripts.
· Some of the mRNA processing protein complexes are Cap adding, Splicing, and poly-Adenylation.
· Five out seven are hydroxyl amino acids, each of the hydroxyl groups can be phosphorylated, and mostly serine, threonine and tyrosine are phosphorylated by one of the RNAP accessory complex.
· The said pol-II large subunit can exist either in non-phosphorylated form called II-O or phosphorylated form called II-a, and each of them have specific function(s).
Phosphorylated form of II-a may be essential during transcriptional initiation. But the II-O form i.e. non-phosphorylated form is absolutely required for promoter clearance and for elongation (contradicts the role of TFII H).
· Except for the two large subunits in each of the classes, other subunits constitution especially of specific subunits can vary from one class of organisms to another.
The core components of RNAP-II are RPB1, RPB-2 and RPB-3. Others are accessory.
· RPB 5, 6,8,10 and 12 are common for all RNAPs. RPB 4 and RPB9 are nonessential.
Near the carboxy end of Pol II’s largest subunit a unique region is found (CTD, carboxy terminal domain). This region contains a stretch of 7 amino acids that is repeated between 26 and 52 times (differences in the number of repeats occur in a species specific manner). During transcription initiation several amino acids in the repeat becomes phosphorylated;A Cartoon- RNAP II with its subunits- number of subunits showed 12; http://www.conservapedia.com/
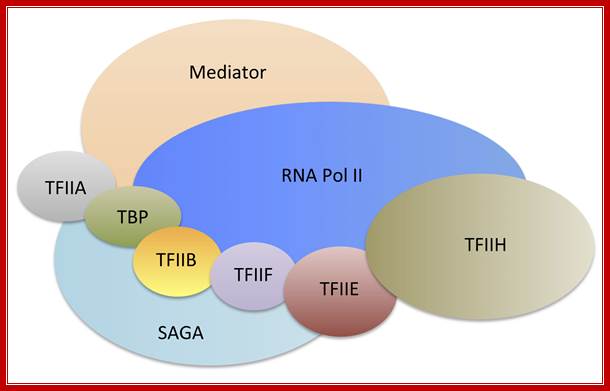
The RNA Polymerase II transcription machinery is composed of RNA polymerase, and five multi-subunit general transcription factors. Two large coactivator complexes commonly used at inducible genes are Mediator and SAGA. The transcription machinery has been conserved among all eukaryotes. https://labs.fhcrc.org
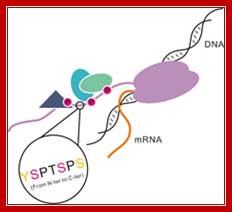
Model of the CTD of RNA polymerase II: RNAP II has CTD tail which gets associated with several splicing and mRNA modifying subunits. The RNA polymerase II is colored with purple. Different shapes bound to the CTD indicate various proteins that are recruited by the CTD.
Coordinately regulated phosphorylation and dephosphorylation of the CTD plays an essential role not only in the recruitment and assembly of transcription complexes but also in temporal control of transcription and mRNA processing., http://www.nano-reviews.net/
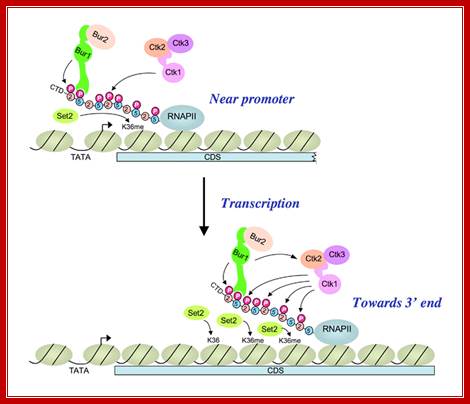
A cascade of cyclin-dependent kinases regulates Pol II CTD phosphorylation in the transcription elongation cycle;
(Upper) Kinase BUR1/BUR2 is recruited to Pol II phosphorylated on Ser5 of the heptad repeats in the CTD by KIN28 of TFIIH near the promoter. Recruited BUR1/BUR2 phosphorylates Ser2 of the CTD repeats, enhancing histone H3-Lys36 methylation by SET2. BUR1/BUR2 and CTK1 make similar contributions to phosphorylated Ser2 (Ser2P) at this location. (Lower) As elongation proceeds downstream, kinase CTK1 makes an increasingly larger contribution to Ser2P and attendant H3-Lys36 methylation. Because the occupancies of both kinases remain high at the 3′ end, either CTK1 becomes more active, or BUR1/BUR2 activity declines, as elongation proceeds. Our results suggest that CTK1 function is stimulated by BUR1/BUR2 distal to the promoter. http://2009annualreport.nichd.nih.gov/
Lack of RPB (?) results in the loss of RPB-7; however mutants in both genes don’t affect the function of the enzyme.
· Using mutations, all the subunits of yeast’s RNAP-II have been identified and to some extent characterized.
And all genes have been cloned and sequenced.
· Using epitope-tagging technique the whole intact complexes have been isolated.
Architectural co activators bend the DNA to assist activators and co activators to synergize.
TBP, HMG and LEF-1 bend DNA to facilitate the binding of activators and enhancers near to each other.
Activator TFIID/A recruit Pol-II holoenzyme and interact with same.
· TAF-250 may interact with TFIIF,
· TBP interact with TD tail
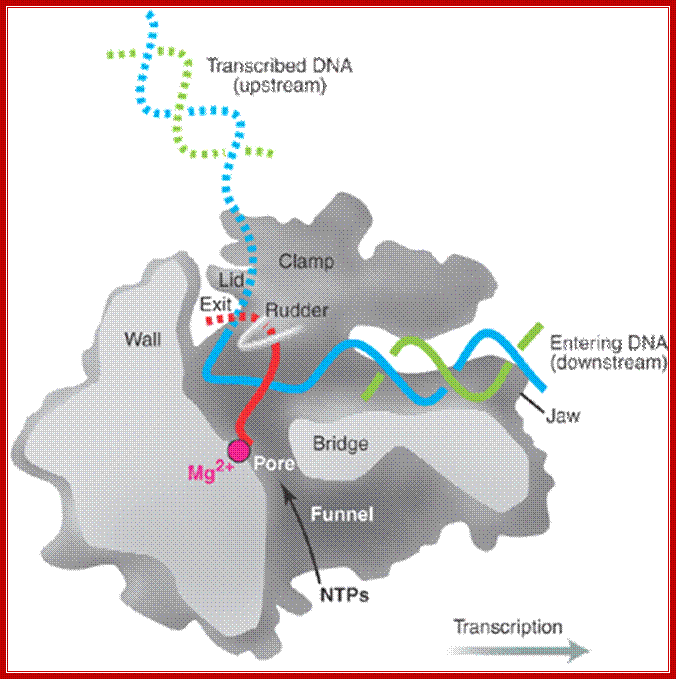
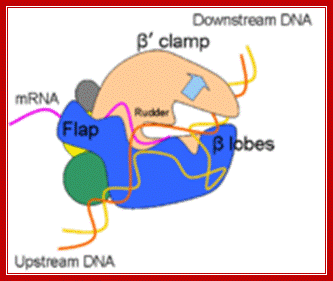
“Side (cutaway) view of the RNA polymerase II transcribing complex, showing the paths of the nucleic acids and the locations of some functional elements of the enzyme. Cut surfaces of the protein, in the front, are lightly shaded and the remainders at the back and darkly shaded”. http://xray.bmc.uu.se/
“By convention, the polymerase is moving on the DNA from left to right (direction of large arrow, bottom). This view exposes the DNA duplex entering the enzyme: The template strand coding for RNA is in blue, the nontemplate strand is in green, and the RNA in the DNA-RNA hybrid in the active center region is in red. DNA entering the enzyme is gripped by protein "jaws" (the upper jaw is not seen in the cutaway view). The 3' (growing) end of the RNA is located adjacent to an active site Mg 2+ ion (pink sphere). A "wall" of protein blocks the straight passage of nucleic acids through the enzyme, as a result of which the axis of the DNA-RNA makes almost a right angle with the axis of the entering DNA. The bend exposes the end of the DNA-RNA hybrid for addition of substrate nucleoside triphosphates (NTPs). The NTPs may enter through a funnel-shaped opening on the underside of the enzyme and gain access to the active center through a pore. The 5' end of the RNA abuts a loop of protein (the rudder), which prevents extension of the DNA-RNA hybrid beyond 9 base pairs, separating DNA from RNA. The exit path of the RNA passes beneath the rudder and beneath another loop of protein (the lid). Exiting RNA, DNA, and also the nontemplate strand of the DNA, are not seen in the electron density map (dashed red, blue, and green lines). The rudder and lid emanate from a massive clamp that swings over the active center region (from back to front in the view shown), restraining nucleic acids and likely contributing high processivity of transcription.
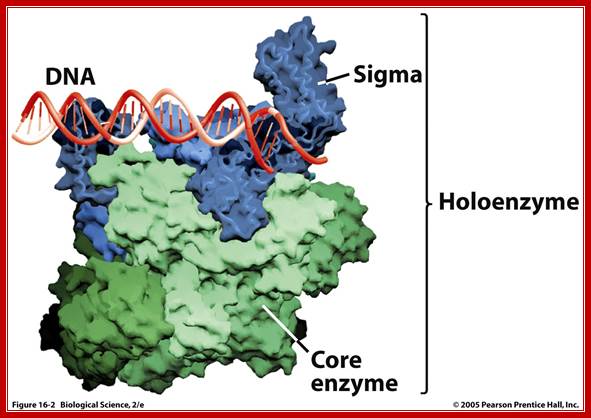
Prokaryotic; holozymes; http://www.uic.edu
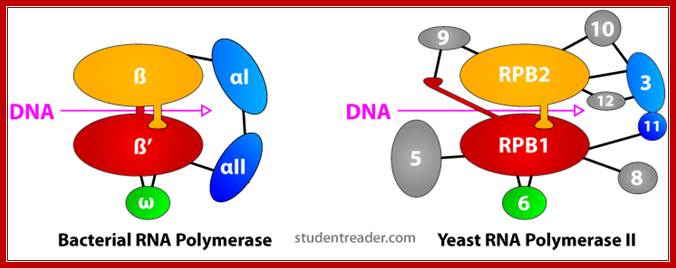
Comparison between bacterial and eukaryotic RNAPs; WIKI; edited figure from tutorial material. http://lh5.ggpht.com/

|
Comparison of interaction
networks in the archaeal and eukaryotic RNAPs.
Left-RNA pol bacteria and Right RNA pol Eukaryotic; http://rationalwiki.org/
Left-Bacterial RNAP involved in Transcription; Right- Eukaryotic RNA pol with CTD tail; http://rationalwiki.org/ |
“Bacterial RNAP is approximately 150 A long, 115 A in height and 100 A wide. The X-ray crystal structure of Thermus aquaticus core RNAP revealed that the enzyme has a shape similar to a crab claw, with the two arms of the claw defining a wide internal cleft between them (Zhang 1999). This cleft is about 27 A in wide, which can accommodate a double stranded DNA molecule and contains the Mg2+ bound active center. The RPB1 and RPB2 subunit makes up two arms of the claw and the base of the cleft (Ebright-2000).
Like bacterial RNAP, RNAP II has a shape similar to a crab claw, with the two arms of the claw forming a deep cleft between them (Cramer 2000). The Fig. below shows, the relative positions of the conserved subunits in bacterial RNA and eukaryotic RNAP II (Ebright 2000). Hence, RPB1 forms one arm of the claw and a portion of the base of the cleft, while RPB2 forms the other arm of the claw and a portion of the base of the cleft.”

Image of yeast RNA polymerase II (Weaver 2002). http://www.biochem.umd.edu
“A 25-Ǻ channel goes through RNAP with mainly Rpb1 on one side and Rpb2 on the other (Weaver 2002). The opening of the channel is composed of Rpb1, Rpb9, and the Rpb5 subunits, which acts to grasp DNA (Weaver 2002). Studies by Roger Kornberg and colleagues show that DNA and RNA bind in the central cleft between Rpb1 and Rpb2 subunits. The surface charge distribution of yeast at 2.8 Ǻ resolution shows that hundreds of positively charged amino acid side chains line the cleft appropriately for interaction with negatively charged DNA and RNA (Kornberg 2001). Duplex DNA enters RNAP II and unwinds 3 bases before the active site. The template strand makes a 90˚ bend and points downward toward the floor of the cleft for readout at the active site (Kornberg 2001). The ribonucleotide on the RNA chain is then base paired to the DNA strand forming a RNA-DNA hybrid (Kornberg 2001). The structure determination of a crystallized transcribing RNAP proved that the RNA and DNA interact in the RNAP cleft (Kornberg 2001).”
Comparison: Bacteria and Eukaryotes:
|
Bacterial RNAP |
Eukaryotic RNAP II |
|
β’ |
RPB1 |
|
β |
RPB2 |
|
άI |
RPB3 |
|
aII |
RPB11 |
|
ώ |
RPB6 |
Table . Conserved subunits of bacterial RNAP and eukaryotic RNAP II
Structural
similarities eukaryotic RNAPs
The eukaryotic RNA polymerases Pol I, Pol II, and Pol III are the central multiprotein machines. Detailed structural studies of Pol I and Pol III were reported recently and showed that the active center region and core enzymes are similar to Pol II and those strong structural differences on the surfaces account for gene class-specific functions.
RNA polymerases I and III ? The number of subunits in RNAP III is more. http://rationalwiki.org/
Organelle RNAPs:
· Mitochondria and plastids are symbionts of free living bacterial systems, which long time back (billions of years ago) entered eukaryotic cells and became symbionts. In the process they have transferred most of their genome into host nuclear genome and retained only a small number of genes as their own. But for their biogenesis and function nuclear genome and symbionts genome express and coordinate to function.
Mitochondria:
· Human mitochondrial genome is about 16.569 Kbp, the genome size varies among other members. It contains 2ribosomal RNA genes, 21-22 tRNA genes and 13 protein coding genes and 7-8 unknown cistrons (URFs). In some one or two nuclear coded tRNAs are imported into mitochondria. Mitochondria hosts about ~615(?) different types of proteins (wiki) but only 13 are coded by mt.genome. Often mitochondrial DNA damage is a possible cause for many degenerative diseases including Parkinson’s, Alzheimer’s, and coronary artery disease.
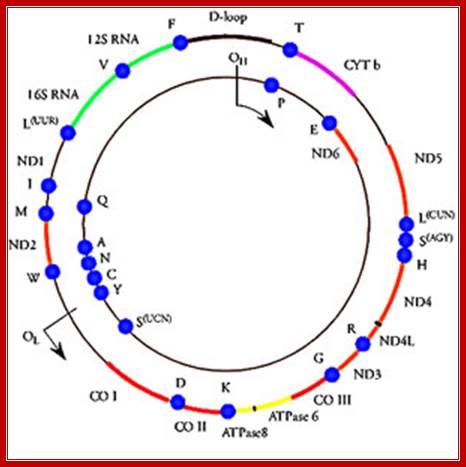
The above diagram shows human mt-genome- 16569 nucleotideas long(1981 sequence). The genes are shown and even replication origin and direction is shown. http://cellbiology.med.unsw.edu.au/
Except for 37 mt-gene products coded for by the mt-genome, the rest of the proteins 151 (361 (dataset)) or more are nuclear coded and the translated products are transported into mitochondria. Yeast 584 (750) and human 734 mit proteins.
· Mitochondrial genome is transcribed by specific mitochondrial RNAPs.
· The mtRNAP is more or less like T7 phage RNAPs consisting of single subunit. But it to be effective in transcription requires two additional factors called TFAM and TFB2M; the TFAM binds and recruits RNAP and TFB2M to its promoter heavy strand promoter HSP1/HSP2. It requires TFB2M factor. Light strand promoter LSP requires TFAM.
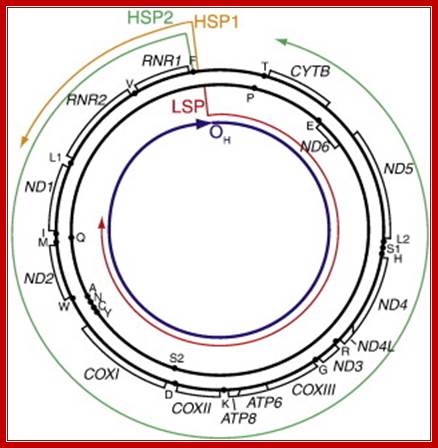
Diagram of mitochondrial genome organization, promoters, and pre-mRNA. Gene organization is adapted from Taanman, 1999 [33] with pre-mRNA from Montoya et al., 1982 [38], and the location of first strand replication initiation interpreted from various publications , and . Shown are the light strand promoter (LSP) and pre-mRNA generated from that promoter (red); the heavy strand promoter 1 (HSP1) and its pre-mRNA (orange); the heavy strand promoter 2 (HSP2) and its pre-mRNA (green); and the location of first-strand DNA replication initiation (OH), which is primed from LSP-derived transcripts; Christopher T. Campbell, Jill E. Kolesar, Brett A. Kaufman
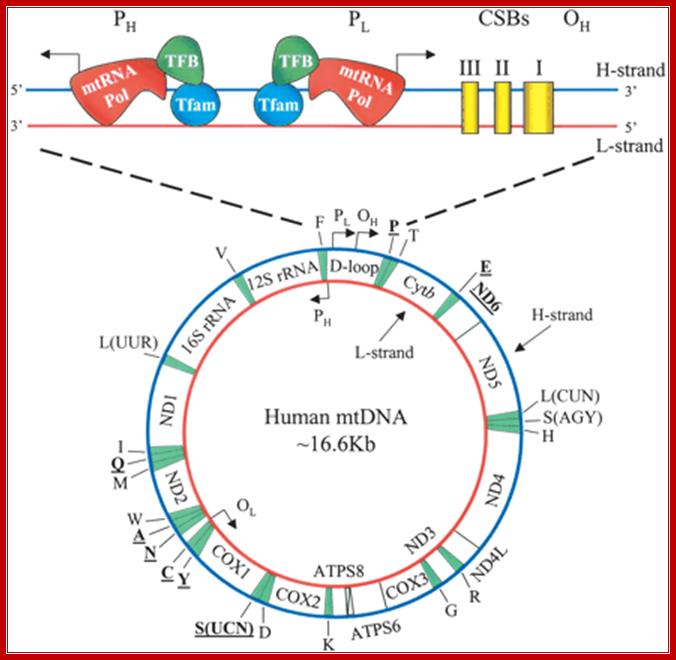
Human mitochondrial DNA (mtDNA). The genomic organization and structural features of human mtDNA are depicted in a circular genomic map. The D-loop regulatory region is expanded and shown above. Protein coding and rRNA genes are interspersed with 22 tRNA genes (denoted by the single-letter amino acid code). The D-loop regulatory region contains the L- and H-strand promoters (PL and PH, respectively) along with the origin of H-strand replication (OH). mtDNA transcription complexes containing mitochondrial RNA polymerase, Tfam, andTFB are depicted in the expanded D-loop along with the conserved sequence blocks (CSB I, II, and III). The origin of L-strand replication (OL) is displaced by approximately two-thirds of the genome within a cluster of five tRNA genes. Protein-coding genes include cytochrome oxidase (COX) subunits 1, 2, and 3; NADH dehydrogenase (ND) subunits 1, 2, 3, 4, 4L, 5, and 6; ATP synthase (ATPS) subunits 6 and 8; and cytochrome b(Cytb). ND6 and the eight tRNA genes encoded on the L-strand are in bold type and underlined; all other genes are encoded on the H-strand. http://genesdev.cshlp.org/

ABC of mitochondria transcription :Two forms of a transcription factor, similar in sequence to an rRNA modifying enzyme, have been identified as the missing components of the transcription initiation machinery for human mitochondrial DNA (mtDNA). Transcription from mtDNA promoters can now be reconstituted in vitro with three recombinant proteins: mitochondrial transcription factors A and B and the core mitochondrial RNA polymerase.
Human mtDNA is a small circular genome encoding a handful of proteins that are essential components of the enzyme complexes of oxidative phosphorylation. Mutations in mtDNA cause a wide spectrum of multisystem human disorders with varying degrees of tissue specificity and severity. Eric A. Shoubridge; http://www.nature.com/
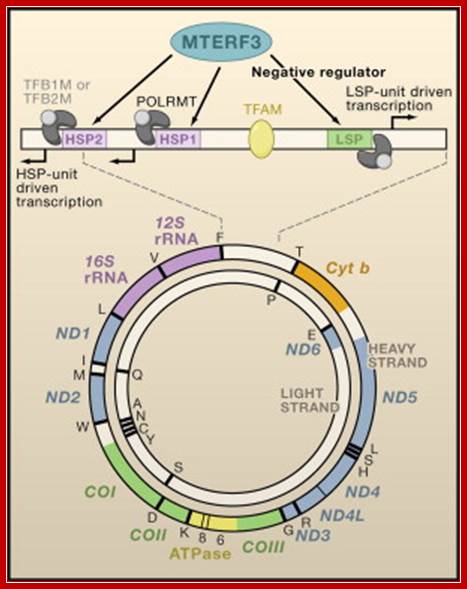
MTERF3 Is a Regulator of Mitochondrial DNA Transcription: Shown is a map of the 16.6 kb circular, double-stranded mitochondrial genome encoding 13 proteins, 22 tRNAs, and 2 rRNAs. The 1.1 kb noncoding control region is shown in linear form. Transcription of the heavy strand is initiated either from the heavy-strand promoter 1 (HSP1), generating a short transcript that terminates at the 16S rRNA, or from HSP2, generating a polycistronic message including both rRNA genes, 12 mRNA genes, and 14 tRNA genes. Light-strand transcription from the light-strand promoter (LSP) generates the ND6 mRNA and 8 tRNAs. Transcription from all promoters requires the involvement of the transcriptional activator TFAM that binds upstream of the promoters, together with a single subunit RNA polymerase (POLRMT), which forms a heterodimeric complex with one of two transcription factors, TFB1M or TFB2M. Together with as yet unidentified interacting partners that are crucial for its function, MTERF3 acts as a negative regulator of transcription by binding to mitochondrial DNA at or near the light-strand promoter and heavy-strand promoters. Chan Bae Park, Jorge Asin-Cayuela etal
The mt RNAP is related to T3 and SP6 RNA polymerases and similar to T7 RNAP (99kD). The T7 RNAP is not inhibited by Rifamycin. In the case of human mitochondrial genome mt RNAP starts transcription from HSP1 and HSP2 promoters and transcribes rRNA genes long polycistronic mRNAs from HSP2. The start point is located at 569-685 bp position. The mitochondrial RNAP is called POLARMT. For its activity it requires a transcription factor TFB2M. This mode of transcription is inhibited by another TF called TFAM which is required for the transcription of light strand from LSP promoter.

http://www.cn.anoword.com
Yeast mt 150kD (RPO41), and the protein 70kD acts as selective factors. The single RNA polymerase (RNAP) of Saccharomyces cerevisiae mitochondria is composed of only two nuclear-encoded protein subunits—the core enzyme and a transcription initiation factor encoded by the MTF1 gene. There are 13 primary multicistronic transcripts initiated at nanonucleotide promoter sequences. The transcripts undergo processing to form mature RNAs.
TTATTTATTATTATATAAGT +1> AATAAATAATAGTTTTATAT
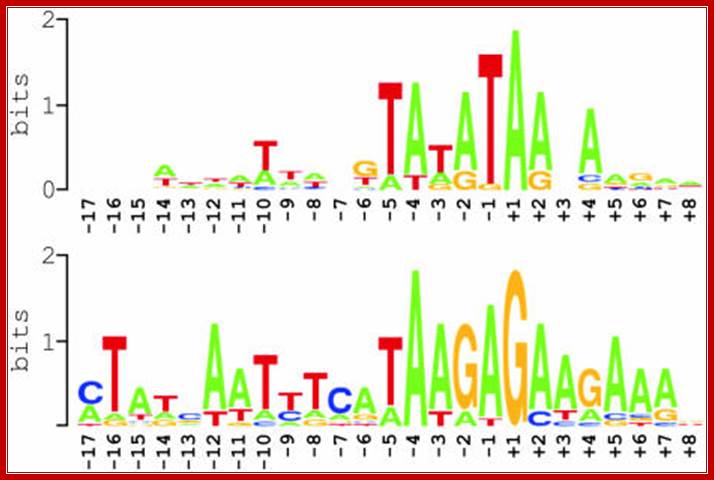
Summary of nucleotide sequences around experimentally defined transcription initiation sites in Arabidopsis mitochondria, as displayed in the above figure1. Two sequence logos are shown that were generated using WebLogo; http://weblogo.berkeley.edu/logo.cgi,
![]()
Transcription in human mitochondria is carried out by a single-subunit, T7-like RNA polymerase assisted by several auxiliary factors. We demonstrate that an essential initiation factor, TFB2, forms a network of interactions with DNA near the transcription start site and facilitates promoter melting but may not be essential for promoter recognition- Marina Sologub1, 3, Dmitry Litonin1, 3, Michael Anikin1, Arkady Mustaev2 and Dmitry Temiakov1,
Mitochondria from plasmodia species are heterogeneous with respect to size, shape, behavior and internal structure. Human P. falciparum contains unusual mitochondria. Its Mt. genome consists of 3 protein coding genes. The rest codes for small RNAs of 23 to 190ntds. Transcription is by nuclear coded mtRNAP assisted by a transcriptional factor.
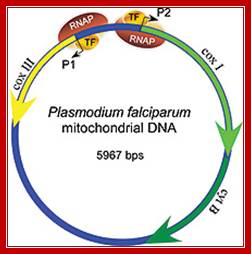
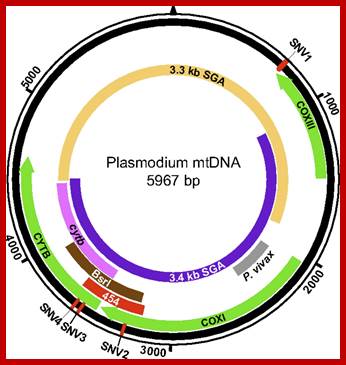
Mitochondria of the parasite contains the genome that encodes three proteins only, its transcription is performed by mtRNAP assisted by at least one TF and uses two promoter elements. Schematic representation of the Plasmodium mitochondrial genome. DNA fragments amplified for diagnostic purposes (cytb, BsrI, P. vivax), 454 sequencing (454), and SGA (mtDNA-3.3 kb; mtDNA-3.4 kb) are shown in relation to cytochrome b (cytb), cytochrome c oxidase subunit I (coxI), and cytochrome c oxidase subunit III (coxIII) coding regions, respectively. The positions of four SNVs (SNV1–SNV4), which distinguish human P. falciparum from ape Laverania parasites, are shown in red http://www.pnas.org
Plastids:
Plastids features vary from the stage to stage and cell types of plant cells. Kinds of plastids are Pro-plastids, leucoplasts, amyloplasts, elaioplasts, chromoplasts and chloroplasts. All of the above forms develop from pro-plastids.
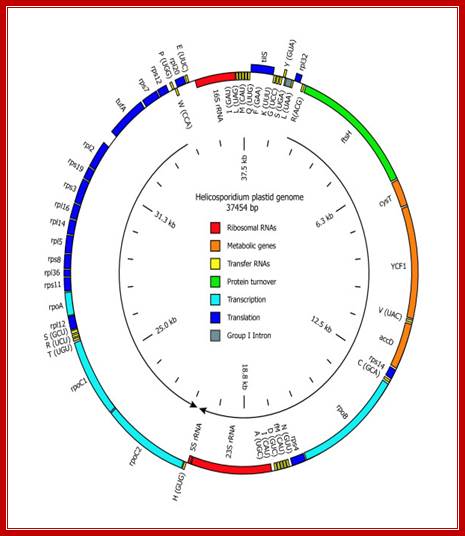
Gene map of Helicosporidium plastid DNA -37454 bp long; http://www.antibodyreview.com/
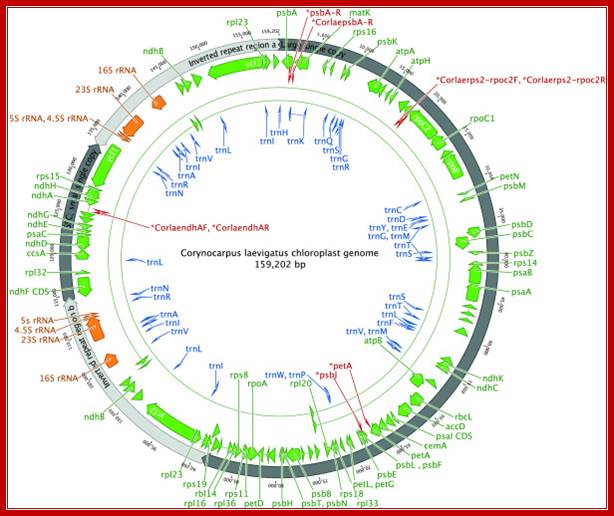
Complete gene map of the entire Corynocarpus laevigatus chloroplast genome. Arrowheads depict the direction of transcription. Binding sites for the primers used to generate the PCR products for resolving homopolymers are indicated in red. This figure was produced using Geneious; http://www.plantmethods.com
Plastogenome varies among the plant species. The genome is circular and contains 120 or more genes; they include 2x rRNA genes, as inverted repeats (not in all), tRNA genes and protein coding genes.
Two types of plastid RNAPs are found in plant systems; they are derived from the nucleus and plastid genomes. The RNAP from plastid genome (PEP) is more like cyanobacterial RNAPs ~900kD consisting beta- rpoB’, beta-rpoA’ and 2alpha rpoC1/2 subunits. The plastid genome has specific promoters for each of these kinds of RNAPs. For example rRNA operon (PrrnP1) is transcribed by PEP enzyme and uses the promoter elements such as conserved −35 (TTGACG) –17-19 ntds −10 (TATAATA) (17 ntds apart) promoter elements. It also contains (GTGGGA) upstream of -35 activator RUA.
−35 (TTGACG) –17-19 ntds −10 (TATAATA) (17 ntds apart)—5-7ntds-+1>
.
NEP RNAP-: 10-nucleotide consensus, ATAGAATA/GAA, overlapping the transcription-initiation site
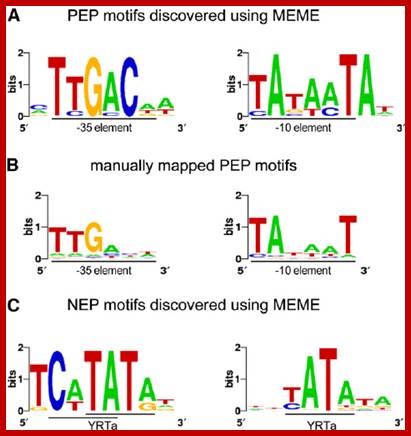
Sequence Logos of Promoter Motifs Detected in Green and White Plastids.
In green plastids, MEME analysis discovered a -10 (right) and -35 (left) PEP consensus element upstream of 44 and 20 TSSs, respectively. The motifs were found to be significantly enriched in green pre-TSS sequences. (A) A manual search for the PEP promoter elements detected the -10 box (right) in 156 TSSs and the -35 box (left) in 109 of the TSSs with mapped 10 elements. (C) Two versions of the YRTa motif were discovered by MEME in white plastids. A TCaTATat motif (left) was found upstream of 22 of the white TSSs and Gene expression in plastids of higher plants is dependent on two different transcription machineries, a plastid-encoded bacterial-type RNA polymerase (PEP) and a nuclear- encoded phage-type RNA polymerase (NEP) 110kDa, which recognize distinct types of promoters. YRTata (right) upstream of 151 (62%). https://www.researchgate.net/
Interaction between factors produced by Nucleus and Plastids and Mitochondria.
Localization of organellar phage-type RNA polymerases are found in different organisms. Genes in the nucleus (N) of eukaryotic organisms code for T3/T7 phage-type RNA polymerases, which are imported into mitochondria (M) and plastids (P) as indicated by arrows. Green algae such as Chlamydomonas and the lycophyte Selaginella possess only one nuclear RpoT gene probably encoding the mitochondrial RNA polymerase (RpoTm). Nuclear genomes of cereals contain two RpoT genes, one encoding the mitochondrial RNA polymerase (RpoTm), the other a plastid RNA polymerase (RpoTp) representing NEP. Arabidopsis and other eudicots additionally acquired RpoTmp, which contributes to transcription in mitochondria and plastids. The moss Physcomitrella patens possesses three RpoT genes encoding polymerases, two of which are dually targeted into both mitochondria and plastids (RpoTmp1, RpoTmp2) and an enzyme solely dedicated to mitochondria (RpoTm).
Algae, Selaginella: RPO Tmà Mitochondria.
Monocots: RPoTM moves to Mitochondraia and RpoTp is transported to plastids.
PHYSCOmitrella- Rpo Tm moves to Mitochondria, RPoTmp1 and Tmp2 move to mitochondria and plastids respectively.
Arabidopsis, and Nicotiana- RpoTm and RpoTmp move to mitochondria, but RPO Tmp and RpoTp move to plastids.
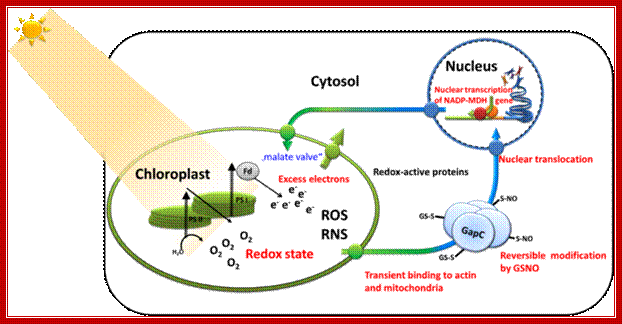
Retrograde signaling of chloroplast redox-state to nucleus. Working model for the stress-induced expression of nuclear encoded NADP-MDH: Excess electron pressure in the chloroplasts leads to ROS/RNS formation, generating nitroso-glutathione (GSNO) that readily modifies sensitive thiols such as those of GapC. This reaction and its reversal are most likely catalyzed by "redoxins". Changed properties of the modified cytosolic enzymes is supposed lead to altered microcompartmentation, and possibly to new functions, namely in gene expression, e.g. of NADP-MDH as part of the malate valve relieving electron pressure in the chloroplasts. Its increased expression will be used as a read-out for the investigated redox-signaling cascade. In this proposal, we therefore ask the question: What is the sequence of events in vivo and what are the particular functions of GapC in this process? Renate Scheibe; http://www.biologie.uni-osnabrueck.de/
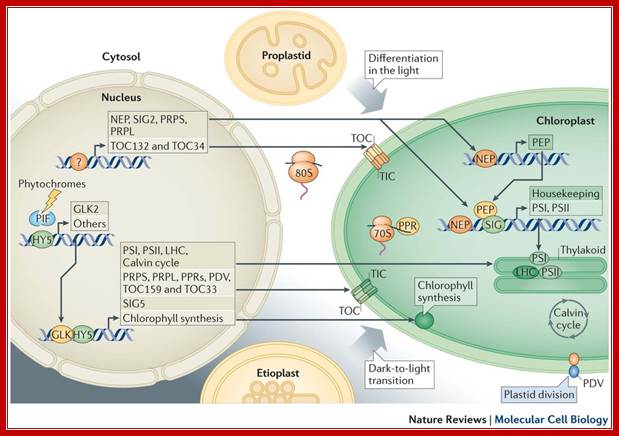
Developmentally, the bulk of information flows from the nucleus to plastids. Non-photosynthetic plant cells require basal operation of plastid housekeeping proteins that have been synthesized in the cytosol (by 80S ribosomes) and that have been imported through a housekeeping translocator (TOC132–TOC34, orange). Basal production of plastid ribosomal proteins (PRPS and PRPL, which are constituents of the 70S ribosome) also takes place. Factors driving and determining the extent of this initial plastid biogenesis are unknown (as indicated by the question mark). Plastid DNA expression is initiated by nucleus-encoded RNA polymerase (NEP); subsequently, plastid-encoded polymerase (PEP) becomes active. Proplastid-to-chloroplast differentiation during leaf development involves the expression of plastid genes that encode core components of the photosystems PSI and PSII, which is driven by PEP and involves regulators such as pentatricopeptide repeat proteins (PPRs) and abundant translational activity. Nucleus-encoded subunits, including those of the light-harvesting antenna complexes (LHC), assemble around these core components. Their import, in large quantities, uses a photosynthetic translocator (TOC159–TOC33; green), and is accompanied by extensive chloroplast growth and division (represented by the plastid division component PDV). Similar phenomena occur when chloroplasts develop from etioplasts following prolonged growth in darkness. Photosynthesis-related nuclear genes often possess G-box promoter elements. In the dark, the basic Leu zipper (bZIP) transcription factor HY5 is ubiquitylated and degraded, whereas basic helix–loop–helix (bHLH) PHYTOCHROME-INTERACTING FACTOR (PIF) regulators bind G-box elements and repress transcription. Photoreceptor activation causes PIF turnover and represses HY5 degradation; this enables its G-box binding and transcriptional activation. GLKs, which are Golden 2-like MYB transcription factors, are also major drivers of photosynthetic genes, with one (GLK2) being light-induced. Light- or circadian-mediated nuclear control of plastid DNA expression is exerted through σ-factors (SIG), which control PEP specificity (for example, SIG5).Light mediated anterograde control of chloroplast development; Paul Jarvis
& Enrique López-Juez: http://www.nature.com/
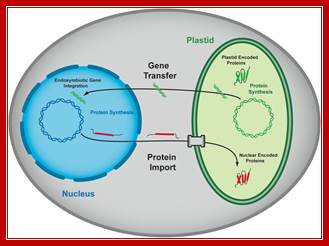
Interaction factors between plastids and nucleus.
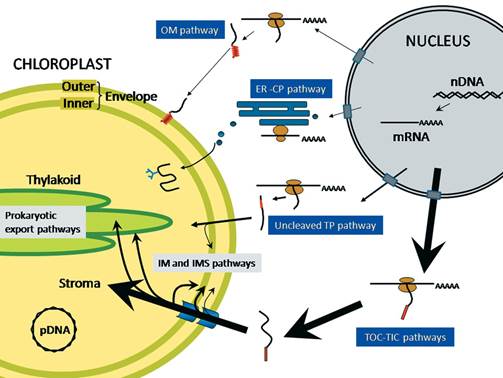
Protein trafficking to plastids: one theme, many variations; Plastids are a diverse group of essential organelles in plants that include chloroplasts. The biogenesis and maintenance of these organelles relies on the import of thousands of nucleus-encoded proteins. The complexity of plastid structure has resulted in the evolution of at least four general import pathways that target proteins into and across the double membrane of the plastid envelope. Several of these pathways can be further divided into specialty pathways that mediate and regulate the import of specific classes of proteins. The co-ordination of import by these specialized pathways with changes in gene expression is critical for plastid and plant development. Moreover, protein import is acutely regulated in response to physiological and metabolic changes within the cell. In the present review we summarize the current knowledge of the mechanism of import via these pathways and highlight the regulatory mechanisms that integrate the plastid protein-trafficking pathways with the developmental and metabolic state of the plant. Takehito Inaba Danny J. Schnell; http://www.biochemj.org/
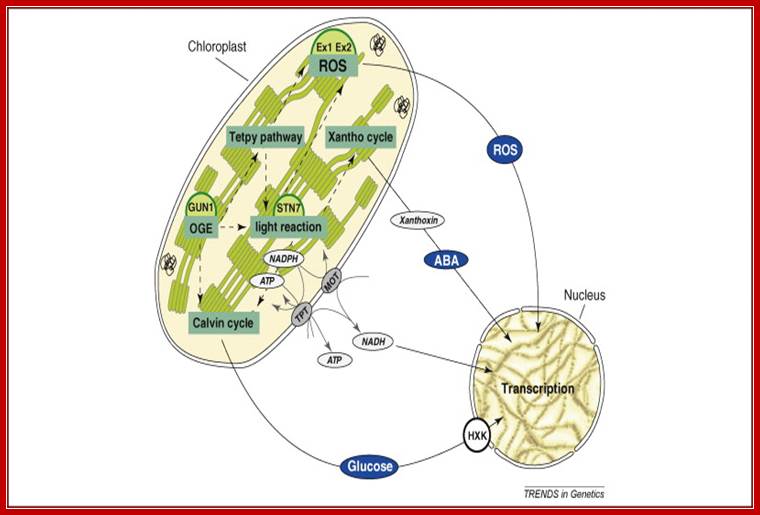
The concept of plastid signalling posits that signals originating from chloroplasts modulate nuclear gene expression (NGE). Put simply, it claims that signalling factors are exported from the chloroplast, traverse the cytosol, and act in the nucleus. Pertinent signals are thought to derive from various sources, including the tetrapyrrole pathway, protein synthesis, reactive oxygen species, or the redox state of the organelle. Recent studies have cast doubt on the most popular candidate signalling molecule, the tetrapyrrole pathway intermediate Mg-protoporphyrin IX, indicating that chloroplast activity might control NGE indirectly by affecting cytosolic metabolite levels or redox states (metabolic signalling). Here, we focus on recent developments and confusions in the field of plastid signalling research and highlight alternative scenarios of plastid–nucleus signal transduction. Future analyses of chloroplast–nucleus communication should focus on providing an integrated view of plastid signalling under physiologically relevant conditions; Tatjana Kleine, Christian Voigt and Dario Leister
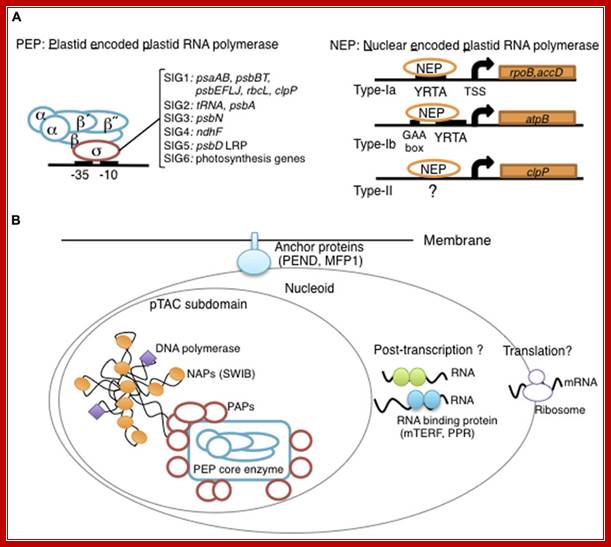
Overview of chloroplast transcription. (A) Basic transcriptional machinery in higher plants. Higher plants have two distinct types of chloroplast RNA polymerase: plastid-encoded plastid RNA polymerase (PEP; left panel) and nucleus-encoded plastid RNA polymerase (NEP; right panel). PEP is a bacterial-type multi-subunit RNA polymerase composed of the core enzymatic subunits α, β, β′, β″ (blue) and a sigma subunit (red) that is responsible for promoter recognition. Plastid sigma factors are divided into six subgroups, SIG1–SIG6, and selectively recognize bacterial-type promoters in the plastid. NEP (right panel) is a monomeric enzyme that resembles mitochondrial T7-type RNA polymerases. NEP is involved in the transcription of housekeeping genes such as rpo genes for PEP core subunits, and ribosomal protein-coding genes. Positioned upstream of genes transcribed by NEP are three distinct types of promoter structures (Type-Ia, Type-Ib, and Type-II). (B)The chloroplast nucleoid subdomain and its components. Chloroplast nucleoids are attached to the membrane (envelope or thylakoid) by anchor proteins (PEND and MFP1). The plastid transcription active chromosome (pTAC) is one of the nucleoid subdomains, which contains the transcription factory. Chloroplast genomic DNA is packed by chloroplast-specific nucleoid-associated proteins (NAPs; orange circle). The mature chloroplast contains a large PEP complex with several PEP associate proteins (PAPs; red circles). Recent proteome analysis suggested that chloroplast nucleoids contain additional subdomains, which regulate post-transcriptional RNA maturation and translation; Yusuke Yagi and Takasshi Shiina; *http://journal.frontiersin.org/
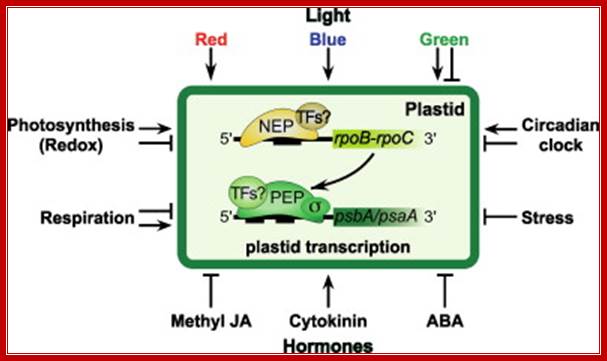
Exogenous and endogenous factors such as light, hormones, photosynthesis, plastid types and chloroplast development differentially modulate transcription of plastid genes. Cues with repressing action on plastid transcription are depicted with lines terminated with a bar while arrows represent promoting signals (see text for references; effects of respiration on chloroplast transcription: Database screening helped to identify regulatory proteins of chloroplast RNA polymerase, including a novel putative RNA-binding protein (RBP).The previously identified transcription kinase (cpCK2) is likely to play a common role in number of plant species.? Model on the role of nuclear-encoded phage-type RNA polymerases in regulating plastid gene expression (updated and modified after Liere and Börner, 2007b). The nuclear RpoTp gene encodes in part the NEP transcription activity. NEP transcribes and therefore may regulate the expression of the plastid rpoB operon encoding subunits of the plastid-encoded RNA-polymerase (PEP). PEP, however, transcribes genes encoding components of the photosynthetic complexes (PSI, PSII) that modulate nuclear transcription by generating various ‘plastid signals’ (e.g. ROS, reactive oxygen species). PEP also transcribes trnE encoding a tRNA (trnA Glu ). https://www.researchgate.net/ T. Potapova, Y. Zubo, Y. Konstantinov, T. Börner, unpublished data).

Fig. Model on the role of nuclear-encoded phage-type RNA polymerases in regulating plastid gene expression (updated and modified after Liere and Börner, 2007b). The nuclear RpoTp gene encodes in part the NEP transcription activity. NEP transcribes and therefore may regulate the expression of the plastid rpoB operon encoding subunits of the plastid-encoded RNA-polymerase (PEP). PEP, however, transcribes genes encoding components of the photosynthetic complexes (PSI, PSII) that modulate nuclear transcription by generating various ‘plastid signals’ (e.g. ROS, reactive oxygen species). PEP also transcribes trnE encoding a tRNA (trnAGlu). The tRNA is required for the synthesis of δ-aminolevulinic acid (ALA), the precursor of the tetrapyrrole (TP) biosynthesis (chlorophyll and heme), which too is thought to provide ‘plastid signals’. Thus, the regulatory network of the nuclear and plastid transcription machineries may be a key element for adjusting the expression of genes located within different compartments of the plant cell in response to exogenous and endogenous factors
The RNAP derived from the nucleus is made up of single subunit called NEP (110kD) similar to mt and phage T3/T7 polymerases. Therefore, rpoA, rpoB, rpoC1, and rpoC2 encode PEP subunits that are not essential components of the NEP transcription machinery. However tobacco plastids lack PEP enzymes? Th NEP binds to consensus -35 TCaTATA -10 sequence ATAGAATA/GAA, overlapping the transcription-initiation site, require plastid sigma factors.
Most of the plastid housekeeping genes are coded for by both NEP and PEPs. Both genes for PSI and PSII are dependent on PEP. Some housekeeping genes are transcribed by NEP RNAPs.
Eighty nine of the 113 chloroplast genes are part of 20 operons (polycistronic gene clusters), while 24 genes are transcribed monocistronically in barley plastids.
The plastid genome (plastome) contains functional rpo genes coding for homologs of the cyanobacterial RNA polymerase subunits α, β, β’, and β’’, which form the core of the plastid-encoded plastid RNA polymerase (PEP). NEP promoters were found to be more active in early leaf development, while transcription activity of PEP is reported to increase during chloroplast maturation. The RNAP is like T phage RNAPs.

Apicoplast:
Phylum Apicomplexa including Plasmodium falciparum contain a plastid called Apicoplast. It lacks photosynthetic functions, it is thought derived from secondary endosymbiosis from red-algal plastids. It carries type-II fatty acid synthesis, haem and isoprenoids. The apicoplast resides very close to mitochondria It has 35kb genome which codes just for 30 proteins. Rest of the proteins required is nuclear coded and imported. The apicoplast exists in close proximity to the mitochondrion of Plasmodium and attains an intricate branched structure in the trophozoite stage. It is maternally inherited. It has 4 membranes-secondary symbiont first involving cyanobacteriium and the second by endosymbiosis of a red alga and eukaryote host.
The plastid genome replicates at the late trophozoite stage of the parasite intraerythocytic cycle. It proceeds predominantly via a D-loop/bi-directional ori mechanism with replication ori localized within the inverted repeat region. The process of replication involves a nuclear-encoded DnaJ homolog that binds to the ori site.
Viral RNAPs: There are RNAPs that transcribe viral DNA and RNAs. They are viral genome coded RNAPs.
T7 RNAP is 883a.a long (96-99 kDa). Most of them are monomeric proteins and perhaps assisted by cofactors. It uses dsDNA and it binds to its promoter elements such as ‘TATAGTGAGTCGTATTA’ on template strand. It requires a cofactor-Mg2+. Related members of T7 are T3 and Sp6 RNA polymerases.
Rhabdo viruses produce their own RNAPs and their transcribed mRNAs are capped and poly adenylated. The same enzyme is also used for its genome replication. Even influenza orthomyxovirus and paramyxoviruses produce their own viral RNAPs which transcribe each of the genomic segments.
Though the viral coded RNAPs are monomeric proteins, but by themselves are incapable of positioning on to their respective gene promoters. For which they require guided assistance from other accessory protein complexes. Each class of RNAPs have their own set of accessory transcriptional factors (TFs), they are named after their main RNAPs; virTFs etc. Each of the complexes contains one or more subunits and they have their own structures and functions. Not all characters of all virTFs are known. Many of the components of complexes are specific to RNAPs, but the individual components may vary.