Mitochondrial Gene Expression:
Organelles:
1. Mitochondria:
Mitochondria are energy transducing organelles, found in all eukaryotes. The number of mitochondria per cell varies from cell type to cell type. Active muscle cells contain large numbers. Malfunction of mitochondria can lead to many diseases.
Structurally, mitochondria under electron microscope look like a rod shaped structure with two membranes, outer and inner. The outer and inner membranes contain multiplex transporters for proteins and other metabolites; these complexes are located at regions where the two membranes are found bound to each other.
The inner membrane is inwardly folded into long flat structures. This embedded with many electron transport enzyme complexes and one finds ATP synthase complex projected in to mitochondrial matrix. They are bound to inner mitochondrial membranes as nucleoids. Mitochondria, besides energy transductions, depending upon signals it release Cyt.Ox-C, which will ultimately has a cascading effect on Apoptosis.
https://themitoblog.wordpress.com/

Unbelievably, there can be hundreds or thousands of mitochondria in a single cell at any one time for example a busy liver cell can have one or two thousand mitochondria which is about a quarter of the volume of the cell; https://themitoblog.wordpress.com/
Mitochondrial DNA nucleoids; ; Nucleoids and the structure of life; In the brave new world of three-parent embryos several inherited mitochondrial diseases can potentially be solved. One slightly dubious argument used by its champions to assuage equally dubious traditional ethical objections is that a mitochondrial donor only supplies 16.6K base pairs (BPs) of mtDNA to the child—a trifling amount compared the 3.4B BPs of (nuclear) nucDNA. What this unassailable yet simplistic truth conceals is that with perhaps 10 plasmid copies per mitochondria, and 100K mitochondria per egg, we are actually talking about 16.5B BPs of mtDNA stashed in strategic fortifications throughtout the cell. Although there is considerable redundancy even in cells with a fairly heteroplasmic stock of mitochondria, that's a bit more DNA than the nucleus has. Read more at: https://phys.org/news/2014-07-nucleoids-life.html#jCp; John Hewitt; https://phys.org. https://themitoblog.wordpress.com/
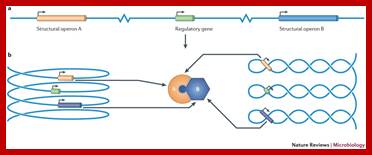
Nucleoid folding and gene regulation. A simple regulon consisting of a regulatory gene and two structural operons, A and B, is illustrated in various conformations. a |When the chromosome is represented in a one-dimensional, linear form, the three genetic loci are separated by large distances in space. b | However, when the chromosome is reorganized as a solenoid (left) or as a plectoneme (right), the periodicity of these structures brings the three genes close together, facilitating communication between the regulatory gene and its two target operons. Moreover, the products of the A and B operons are produced in close proximity, favouring their interaction. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Charles J. Dorman; http://www.nature.com

Amazingly mitochondrial DNA can only be inherited from your mother, unlike nuclear DNA which comes from both parents. Any paternal mtDNA found in the sperm is destroyed or lost after fertilization. This means that any mitochondrial diseases carried in the father’s mtDNA cannot be passed onto their offspring; https://themitoblog.wordpress.com/
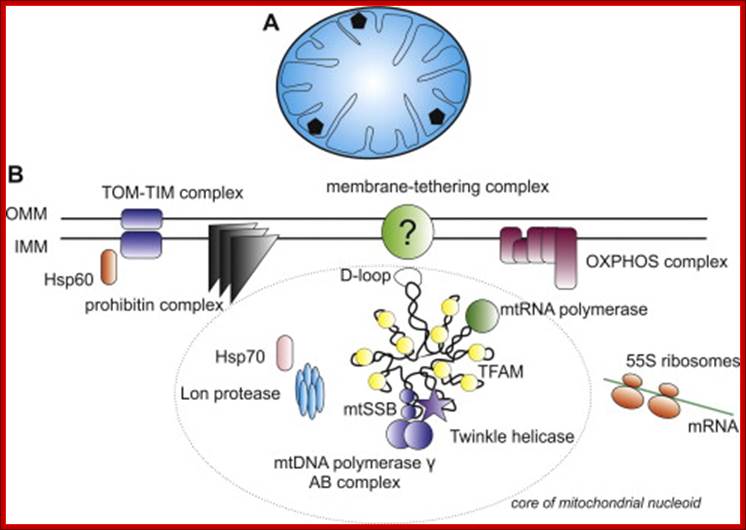
The inner space is called matrix. It contains all the required enzymes and substrate for metabolic activities, such as Krebs cycle, DNA metabolism, RNA synthesis and processing and proteins synthesis. The matrix contains a large number of 70S ribosomes for protein synthesis. The matrix also contains mitochondrial DNA, which is found in large numbers. The mtDNA exists as ds DNA in circular form associaterd with proteins called nucleoids.
TEM, Cristae-https://epiehonorsbiology.wikispaces.com
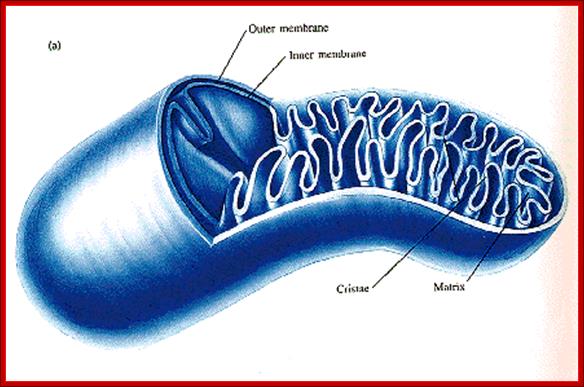
Mitochondrion; http://teachline.ls.huji.ac.il/
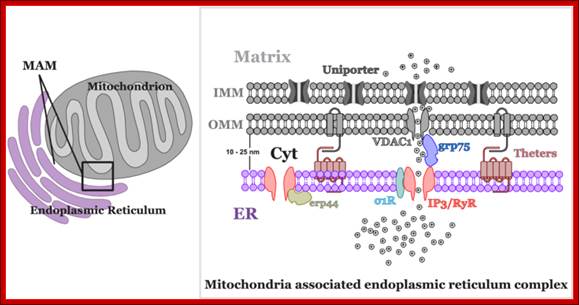
Mitochondria not only govern energy production, but are also involved in crucial cellular signalling processes. They are one of the most important organelles determining the Ca2+ regulatory pathway in the cell ; Piotr Szopa et al; http://m.iopscience.iop.org/
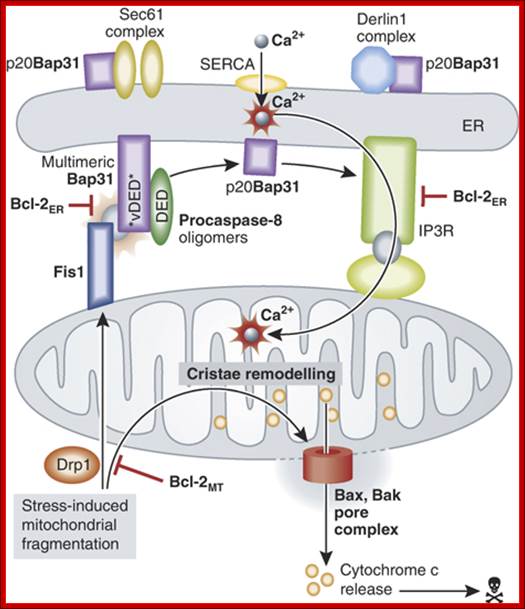
Model for the involvement of the Fis1/Bap31/procaspase‐8 platform in Bax/Bak‐mediated permeabilization of the mitochondrial outer membrane. Additional opportunities for regulation involve prosurvival members of the Bcl‐2 family operating at the ER, as well as other targets of p20Bap31 that might influence the ER membrane protein trafficking machinery.;http://emboj.embopress.org/
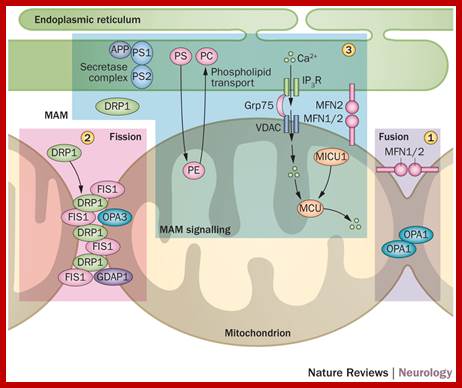
Mitochondria form a highly interconnected tubular network throughout the cell via a dynamic process, with mitochondrial segments fusing and breaking apart continuously. Strong evidence has emerged to implicate disturbed mitochondrial fusion and fission as central pathological components underpinning a number of childhood and adult-onset neurodegenerative disorders. Several proteins that regulate the morphology of the mitochondrial network have been identified, the most widely studied of which are optic atrophy 1 and mitofusin 2. Pathogenic mutations that disrupt these two pro-fusion proteins cause autosomal dominant optic atrophy and axonal Charcot–Marie–Tooth disease type 2A, respectively. These disorders predominantly affect specialized neurons that require precise shuttling of mitochondria over long axonal distances. Considerable insight has also been gained by carefully dissecting the deleterious consequences of imbalances in mitochondrial fusion and fission on respiratory chain function, mitochondrial quality control (mitophagy), and programmed cell death. Interestingly, these cellular processes are also implicated in more-common complex neurodegenerative disorders, such as Alzheimer disease and Parkinson disease, indicating a common pathological thread and a close relationship with mitochondrial structure, function and localization. Understanding how these fundamental processes become disrupted will prove crucial to the development of therapies for the growing number of neurodegenerative disorders linked to disturbed mitochondrial dynamics; Florence Burté et al-http://www.nature.com

Involvement of BCl2 family of proteins in Apoptosis; Biochemistry Blog, Science writing;, Christie Cade.
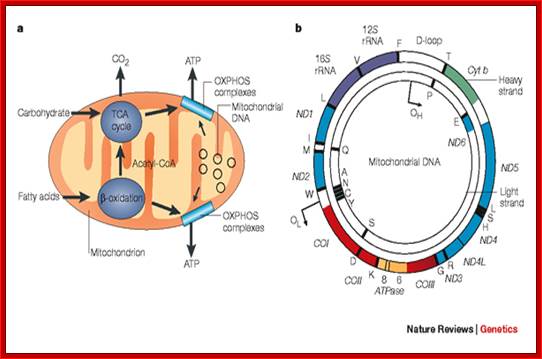
The role of the mitochondrial genome in
energy generation.;This highlights the
importance of the mitochondrial genome in contributing polypeptide subunits to
the five enzyme complexes that comprise the oxidative phosphorylation (OXPHOS)
system within the inner mitochondrial membrane — the site of ATP synthesis. The
reoxidation of reducing equivalents (NADH (reduced nicotinamide adenine
dinucleotide) and FADH2 (reduced
flavin adenine dinucleotide)) that are produced by the oxidation of
carbohydrates (the tricarboxylic acid (TCA) cycle) and fatty acids (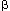 -oxidation) is coupled to the
generation of an electrochemical gradient across the inner mitochondrial
membrane, which is harnessed by the ATP synthase to drive the formation of ATP. b | A map of the human mitochondrial
genome. The genes that encode the subunits of complex I (ND1–ND6 and ND4L) are shown in blue; cytochrome c oxidase (COI–COIII) is shown in red; cytochrome b of complex III is shown in green; and
the subunits of the ATP synthase (ATPase 6 and 8) are shown in yellow. The two ribosomal RNAs (rRNAs; 12S and 16S, shown in purple) and 22 tRNAs, indicated by black
lines and denoted by their single letter code, which are required for
mitochondrial protein synthesis are also shown. The displacement loop (D-loop),
or non-coding control region, contains sequences that are vital for the
initiation of both mtDNA replication and transcription, including the proposed
origin of heavy-strand replication (shown as OH). The origin of light-strand
replication is shown as OL.
Robert W. Taylor & Doug M. Turnbull; http://www.nature.com/nrg/
-oxidation) is coupled to the
generation of an electrochemical gradient across the inner mitochondrial
membrane, which is harnessed by the ATP synthase to drive the formation of ATP. b | A map of the human mitochondrial
genome. The genes that encode the subunits of complex I (ND1–ND6 and ND4L) are shown in blue; cytochrome c oxidase (COI–COIII) is shown in red; cytochrome b of complex III is shown in green; and
the subunits of the ATP synthase (ATPase 6 and 8) are shown in yellow. The two ribosomal RNAs (rRNAs; 12S and 16S, shown in purple) and 22 tRNAs, indicated by black
lines and denoted by their single letter code, which are required for
mitochondrial protein synthesis are also shown. The displacement loop (D-loop),
or non-coding control region, contains sequences that are vital for the
initiation of both mtDNA replication and transcription, including the proposed
origin of heavy-strand replication (shown as OH). The origin of light-strand
replication is shown as OL.
Robert W. Taylor & Doug M. Turnbull; http://www.nature.com/nrg/
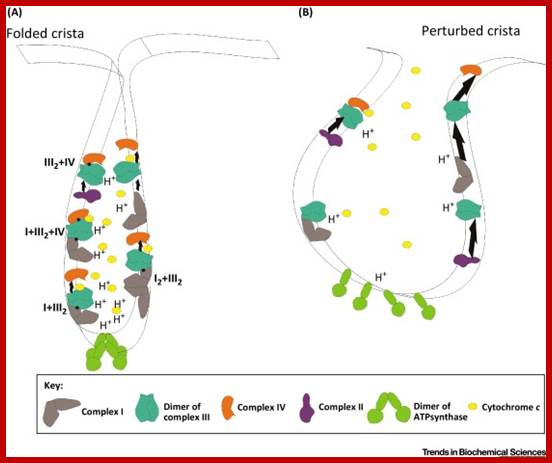
The Supercomplexes Are Disorganized when Cristae Are Unfolded. Schematic representation of the oxidative phosphorylation (OXPHOS) system in a folded crista (A). The different complexes are assembled in different supercomplexes of very dynamic compositions along the cristae membrane. The dimers of ATP synthase are organized in long rows on the edge of the cristae and cytochrome c is maintained inside the cristae. In this situation, the electron transport chain works efficiently (represented by short arrows between the complexes) and the proton gradient accumulates at the edge of the cristae, where the ATP synthase can use it. Upon perturbation of cristae structure (B), supercomplexes and dimers of ATP synthase disassemble and cytochrome c is mobilized. As a result, the electron transport chain is less efficient (longer arrows) and, thus, the mitochondrial respiratory performance decreases. (The amount of H+, and the localization and amount of complexes and supercomplexes are only representative and do not correspond to the real stoichiometry.) http://www.cell.com
Disease of mitochondria results due to the failure of mitochondria. Dysfunction in the mitochondria fails to produce energy that is needed for the sustainment of life and growth of an organism. Injury in the cell or even cell death results in the production of less energy. If the process happens throughout the body, the whole system begins to fail. The disease primarily affects young. The mitochondrial disease causes most of the damage to the cells of brain, heart, liver, muscles, kidney, respiratory and the endocrine systems. The symptoms may be as follows depending upon the cells that are affected:
- Loss of motor control,
- Muscle weakness and pain,
- Gastro-intestinal disorders,
- Swallowing difficulties,
- Poor growth,
- Cardiac disease,
- Liver disease,
- Respiratory illness,
- Seizures,
- Visual/hearing problems,
- Lactic acidosis,
- Developmental delays and
- Susceptibility to infection.

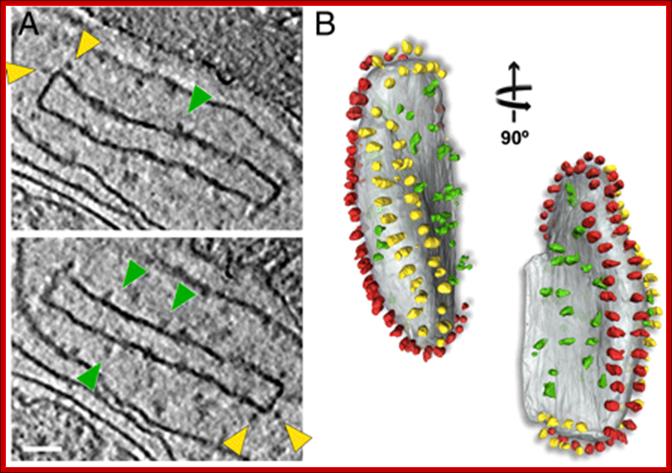
Fig: Isolated crista vesicle from P. anserina. (A) Two tomographic slices of the same box-shaped cristae vesicle showing ATP synthase dimers at approximately 90° bends in the membrane (yellow arrowheads) and complex I densities (green arrowheads) in flat membrane regions of the vesicle. (B) Segmented surface representation. Two rows of ATP synthase dimers (yellow and red) in the membrane (gray) run along the 90° membrane ridges. Irregularly distributed particles of complex I or other respiratory chain complexes (green) are confined to flat membrane regions, Scale bar, 50 nm.

Mitochondrion; http://tgesbiology.weebly.com/
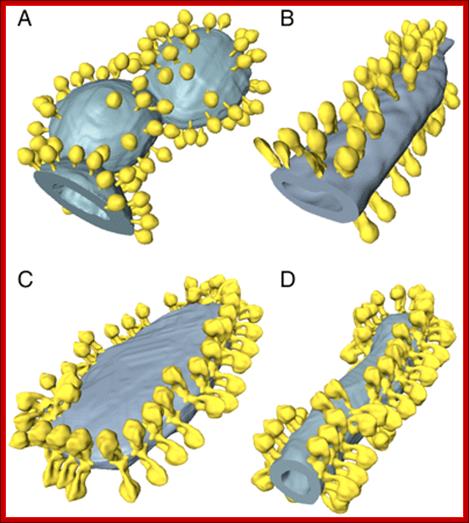
Fig: ATP synthase distribution in isolated mitochondrial membranes. Surface-rendered volumes of mitochondrial membranes from yeast strains lacking subunit g (A, B), and wild type (C, D). In the mutants, the ATP synthase complexes are monomeric, and randomly distributed over flat or gently curving membrane regions (A, B). By contrast ATP synthase from wild-type mitochondria form rows of dimers along the highly curved ridges of tubular (D) or disk-shaped (C) cristae vesicles. http://www.pnas.org/
Mitochondrial genome size varies from one species to the other species. In plant and fungi the mitochondrial genome is unusually large, when compared to that of Mammalian system. Ex. Yeast mitochondrial genome size if 84 kbp, in plants it is 150,000 to 200,000 bp. Chlamydomonas mitochondria contain 16,000bp long DNA and surprisingly linear? Human mitochondrial genome has ~16,590bp; it is the most compact, and smallest of all mitochondrial genomes. In spite of differences in size most of them actually code for 13 (14) polypeptides and 22 tRNAs. In certain cases tRNA coded for by nucleus is transported into mitochondria. There are 7-8 unidentified reading frames (URFs). The genome also codes for two rRNAs. Some plant mitochondria are exception to this.
Mitochondrial genome is a circular ds DNA. Each mitochondrion may contain multiple copies of the genome from 10-40 or more per mitochondrion. The number of mitochondria per cell varies from 100-1000. Both strands of the genome have coded information and they are used for transcribing to generate mRNAs and tRNAs.
Of the two strands, the one that codes for most of the genes, such as rRNA operon, 12 protein subunits and 14 tRNAs is called H-strand and the other strand that codes for one protein subunits, eight tRNAs and URFs is called L- strand.
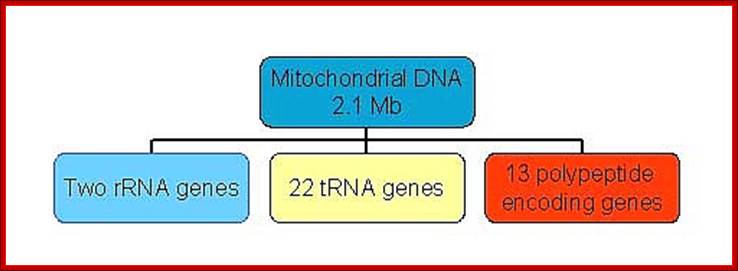
Mammalian (Hu) Mit DNA- encodes two rRNA gene, 22tRNA genes, 13 ORFs for polypeptides and 2-7URFs; L-strand codes for 8 tRNAs and H- strand codes for 12S and 16s rRNAs, 14 tRNAs and 13 polypeptide coding genes.
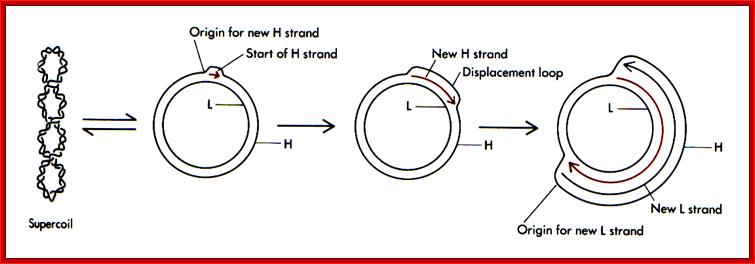
Replication of mitochondrial DNA uses different origins, one for H strand and another for L strand. They are located at different positions, Ori-H is at transcriptional initiation site called D-loop, and the Ori for L is 2/3 from the Ori-H. Interestingly the rRNA is transcribed 50-75 time more than the other transcripts; this might be due to termination of rRNA transcript more often than the others.
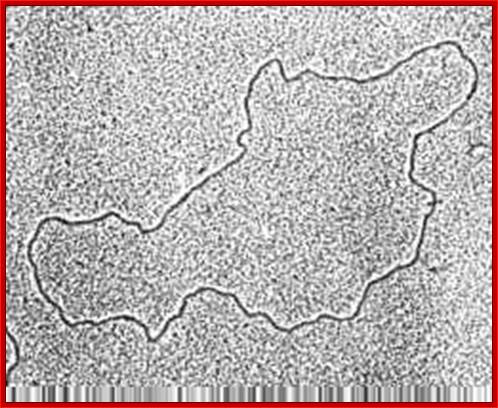
Mitochondrial genome-circular form (good TEM); http://bricker.tcnj.edu/; http://www.cytochemistry.net/
Gene in Organelles; http://bricker.tcnj.edu/
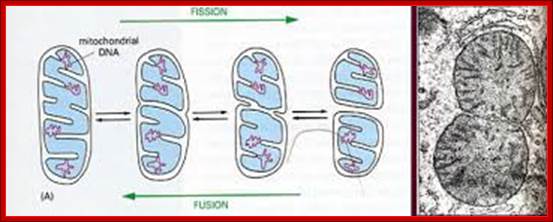
Replication of mitochondrial DNA is independent of cellular DNA replication which takes place once in its life cycle. But mit DNA replication takes place many times to keep the number per mitochondria for mitochondria themselves undergo division by fission.
The origin of Replication is located in what is called D-loop. Replication requires mitochondrial DNA polymerase (gamma DNAP) 140kDa, a viral like Polymerase, it lacks proof reading, hence more mutations. Replication requires mt transcriptional factors mtTF-A (Nuclear coded), but imported, mtRNA polymerase, mt SSB and an helicase. Origin requires the opening of the ds mit DNA. This is facilitated by the binding of mit TFA upstream of HSP and the mitRNA polymerase which uses its nine base promoter 5’TATAAGTTA3’ and transcribes. The transcript is processed to produce a short primer by an endo-ribonuclease RNase-MRP (mitochondrial processing). This primer at conserved block (CSBs) I-III, is used for extending the transcript and displacing the H strand. The displaced ssDNA is supported by SSBs. As mit DNAP progresses beyond 2/3ds of the mtDNA, it opens up the origin for L-strand replication. It extends one complete round. In this L region a promoter is used to produce a primer and then it is extended full length. The D-loop provides space for transcriptional initiation from either sides.
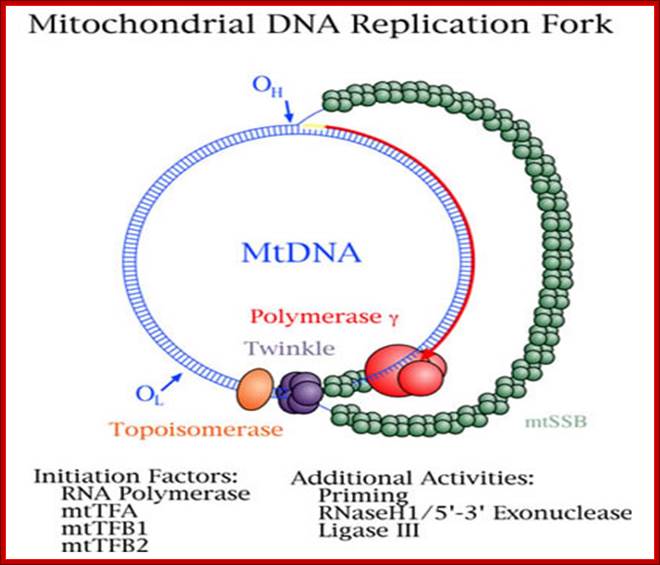
Mitochondrial Diseases. William C.;Copeland, Ph.D; http://www.niehs.nih.gov/
Mitochondrial Genome Transcription:
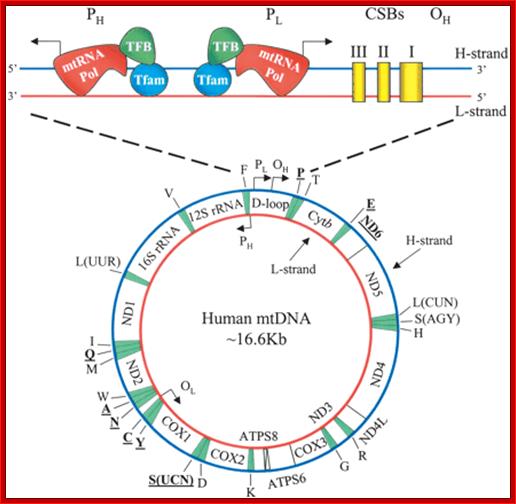
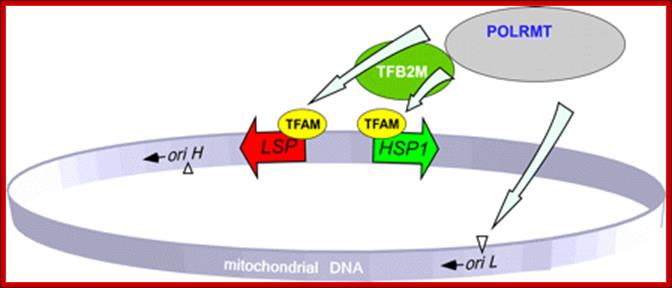
Transcriptional regulatory Circuits; Human mitochondrial DNA (mtDNA).16569bp; The genomic organization and structural features of human mtDNA are depicted in a circular genomic map. The D-loop regulatory region is expanded and shown above. Protein coding and rRNA genes are interspersed with 22 tRNA genes (denoted by the single-letter amino acid code). The D-loop regulatory region contains the L- and H-strand promoters (PL and PH, respectively) along with the origin of H-strand replication (OH). mtDNA transcription complexes containing mitochondrial RNA polymerase, Tfam, and TFB are depicted in the expanded D-loop along with the conserved sequence blocks (CSB I, II, and III). The origin of L-strand replication (OL) is displaced by approximately two-thirds of the genome within a cluster of five tRNA genes. Protein-coding genes include cytochrome oxidase (COX) subunits 1, 2, and 3; NADH dehydrogenase (ND) subunits 1, 2, 3, 4, 4L, 5, and 6; ATP synthase (ATPS) subunits 6 and 8; and cytochrome b(Cytb). ND6 and the eight tRNA genes encoded on the L-strand are in bold type and underlined; all other genes are encoded on the H-strand; Daniel P. Kelly et al;http://genesdev.cshlp.org/
There are two promoter elements for transcriptional initiation one for H-strand and the other for L-strand, called HSP and LSP respectively; each of them is 15 bp long. Replication of mitochondrial ori for H strand is located in between F and T genes and Ori for L is between N and C genes.
Human mitochondrial genome transcription little varies from others. Map of human mtDNA is circular double-stranded. The genome encodes 13 proteins, 22 tRNAs and two rRNAs; they are mapped (The author of this web site has done work on mit genome tranascriptional mapping in 1982 J. Biochemistry. The two strands of the mtDNA are termed heavy and light. Heavy-strand transcription is initiated from two sites, HSP1 and HSP2. The HSP1 transcript terminates at the 3′ end of the 16S rRNA, in a region bound by the mTERF protein. A second binding site for mTERF (mTERF*) has recently been identified and is important for stimulation of transcription from HSP1. The HSP2 transcript produces a polycistronic molecule that corresponds to almost the entire heavy strand, including 12 mRNA molecules and tRNAs (blue). Transcription from the light-strand promoter (LSP) produces the ND6 mRNA molecule (yellow) and primers for initiation of DNA synthesis at the heavy-strand origin of DNA replication (OH). Non-coding regions are indicated in green.

http://ars.els.cdn.com/content
The D-loop is the site at which transcription starts, one called HSP1 and the other HSP2. Transcription from LSP is in opposite direction producing a long transcript and processing releases tRNAs and one mRNA.

ABCs of mitochondrial transcription, Eric A.Shoubridge, Two forms of a transcription factor, similar in sequence to an rRNA modifying enzyme, have been identified as the missing components of the transcription initiation machinery for human mitochondrial DNA (mtDNA). Transcription from mtDNA promoters can now be reconstituted in vitro with three recombinant proteins: mitochondrial transcription factors A and B and the core mitochondrial RNA polymerase. www. nature.com
1. Figure: Human Mitochondrial Transcription Revisited; Schematics of the basal transcription initiation machinery in human mitochondria. Human mitochondrial genome contains two promoters located in the opposing DNA strands. The HSP1 promoter is responsible for synthesis of most mitochondrial genes and is activated by POLRMT-TFAM-TFB2M complex. During replication, when DNA region near oriL becomes single-stranded and forms stem-loop structure, transcription by POLRMT generates short RNA primers. This initiation event is TFAM1 and TFB2M-independent. Transcription from the LSP promoter generates primers for replication at oriH as well as the rest of the tRNAs and mRNA. This initiation event, similar to HSP1, requires cooperative action of POLRMT, TFAM, and TFB2M for efficient transcription and replication; Dmitry Temiakov et al; http://www.jbc.org/
The diagram below shows plant mitochondrial Promoter elements;

Promoter elements called HSP and LSP and the transcription ensues in opposite direction. There are no TATAAT sequences as found bacteria; they are the consensus sequence for Dicots and Monocots reapectively; http://edoc.hu-berlin.de/

Model of mtDNA transcription initiation machinery and promoters: www.slideplayer.com; www.myshared.ru
Mitochondrial transcription is bidirectional and starts in the D-loop region where the promoters HSP1, HSP2 and LSP are located. Transcription initiation requires the cooperation of TFAM, TFB2M and the RNA polymerase POLRMT. TFAM protein preferentially binds the mtDNA upstream of the promoters. Transcription initiated in the HSP1 promoter is terminated at the tRNALeu(UUR), transcribing only for the tRNAVal, the tRNAPhe and the 2 ribosomal RNA (12S and 16S). However transcription initiated from the HSP2 promoter transcribes the full-length mtDNA codes for 12 proteins and 14 tRNAs. On the other hand the other strand codes for eight tRNAs and one protein subunit and 7URFs. MTERF family members, MTERF1, MTERF2 and MTERF3 bind to the promoter region and modulate mtDNA transcription. MTERF1 also binds to the tRNALeu(UUR) inducing transcription termination. Transcripts originated from LSP promoter proceed through the entire mtDNA molecule or could be terminated prematurely to prime mtDNA replication. OH indicates the origin of replication on the heavy strand.
TFB2 is a Transient Component of the Catalytic Site of the Human Mitochondrial RNA Polymerase;
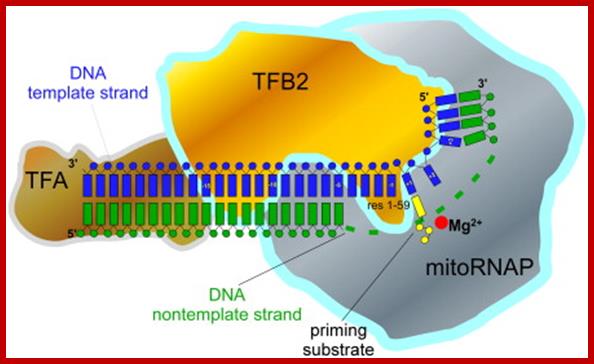
Transcription in human mitochondria is carried out by a single-subunit, T7-like RNA polymerase assisted by several auxiliary factors. We demonstrate that an essential initiation factor, TFB2, forms a network of interactions with DNA near the transcription start site and facilitates promoter melting but may not be essential for promoter recognition; Marina Sologub1, , Dmitry Litonin, 3, Michael Anikin1, Arkady Mustaev2 and Dmitry Temiakov.
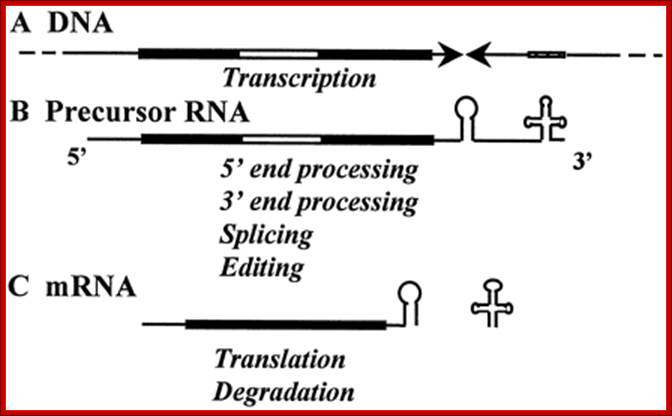
Once the transcription is initiated, it goes through the entire length and ends where it has started i.e. in the D-loop. Thus it produces full-length transcripts. The transcript is processed into individual rRNAs, tRNAs and mRNAs. Each of these RNAs is further processed to functional rRNAs, tRNA and mRNAs respectively. In some species like Lieshmania and others, mitochondrial mRNAs are edited and in some cases they are spliced.
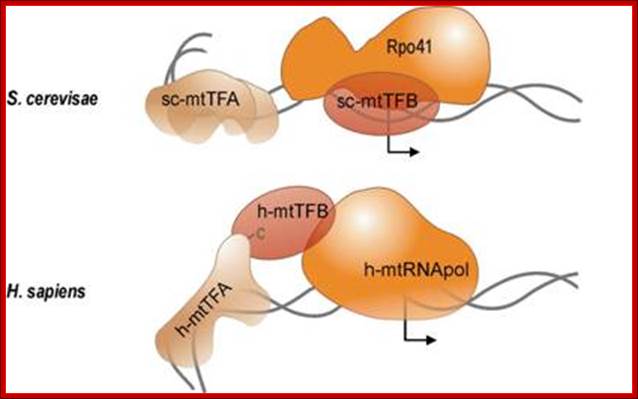
Sc mtTFA and h-mtTFB are transcriptional factors for S.c and Human respectively. The Rpo41and Sc-mtTF-B and h-mtRNA pol are responsible for mitochondrial Genome transcription. The diagram shows the RNAP bound to their promoter elements and they are in the processes of transcriptional initiation. http://edoc.hu-berlin.de/

All mRNAs are poly-adenylated at 3’ends with 50 or so A-ntds. So far no one has isolated full-length transcripts, which means they are processed as they are transcribed.
Some Sno RNAs have been found in certain mitochondria, they are implicated in mitochondrial RNA processing.
The light chain transcription on L strand is also initiated very near to the H strand, but transcribes the opposite strand and in opposite direction (that is anti-clock wise). This also generates a long transcript; it is processed to their respective RNAs.
Mitochondrial L and H promoters:
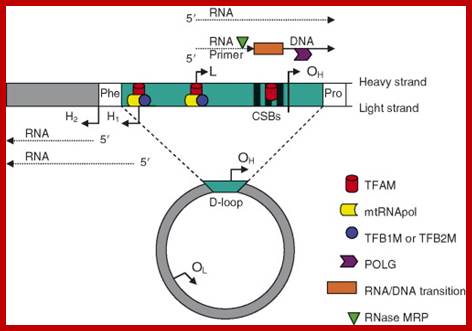
http://www.sciencedirect.com
LSP HSP
-I---------+<I----I- - - -- -- - - - - - - - - - - - I-----I+>----------I-----
mt TF 1 mt TF
TACCGCCAAAACATA AAACCCCAAAGACAC
Human mitochondria contain more than 151 different polypeptides, and it has a very high protein-to-phospholipid ratio (more than 3:1 by weight, which is about 1 protein for 15 phospholipids). A published human mitochondrial DNA sequence revealed 16,569 base pairs encoding 37 total genes: 22 tRNA, 2 rRNA, and 13 peptide genes. It also contains 7 URFs. The 13 mitochondrial peptides in humans are integrated into the inner mitochondrial membrane, along with proteins encoded by nuclear genes.
Mt RNAPs: Mitochondrial RNAP consists of single polypeptide chain (RPO41), the molecular weight of S.cerviciae is 145kD and another factor is called initiation factor MTF1. The enzyme shows significant similarities with that of T7 and Sp6 bacteriophage RNAPs
Promoters in a plant Arabidopsis thaliana: Kristina Kühn, Andreas Weihe and Thomas Börner.
Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Mitochondrial genes in the plant Arabidopsis thaliana are transcribed by two phage-type RNA polymerases encoded in the nucleus. Here, 30 transcription initiation sites of 12 mitochondrial genes and gene clusters have been determined using 5′-RACE and ribonuclease protection analysis of primary transcripts labeled in vitro by guanylyltransferase. A total of 9 out of 12 genes were found to possess multiple promoters, revealing for the first time that multiple promoters are a common feature of mitochondrial genes in a dicotyledonous plant.
Nearly half the identified transcription initiation sites displayed immediately upstream a CRTA core sequence, which was mostly seen within the previously described CRTAAGAGA promoter motif or a novel CGTATATAA promoter element. Plant mitochondrial genomes considerably vary in size but contain a fairly stable number of 50–60 genes. In contrast to metazoan mitochondria, which encode fewer, closely spaced genes and initiate transcription of the mitochondrial DNA (mtDNA) at a single unidirectional or bidirectional promoter on each strand.
In mitochondrial promoters of dicots, this core motif is embedded in an extended consensus of 9 nt, CRTAAGAGA, with the initiating nucleotide at the penultimate position. The primary transcripts carry triphosphates at their 5’end.
Identification of transcription initiation sites by 5’-RACE; Analysis of mitochondrial promoter elementsd in Arabidopsis thaliana
To learn about promoter specificities of the mitochondrial transcription machinery in Arabidopsis, mitochondrial transcription initiation sites were experimentally determined using a 5’-RACE technique first described by Bensing et al. (1996) (Figure 7), which since has been applied to define primary transcript 5’ termini in different groups of bacteria (Argaman, et al., 2001;Vogel, et al., 2003) and in plastids (Miyagi, et al., 1998). In bacteria as in mitochondria and plastids, primary transcript 5’ ends carry triphosphates while processed transcripts have monophosphates at their 5’ ends. Only the latter are a substrate to RNA ligase, and are in the experimental procedure selectively ligated to an RNA oligonucleotide, to which a forward primer will anneal in a subsequent 5’-RACE step. Primary 5’ termini may be ligated only after removal of a 5’ pyrophosphate through tobacco acid pyrophosphatase (TAP). Consequently, 5’-RACE will yield products from TAP-treated RNA for both primary and processed transcripts, whereas without exposure to TAP, products resulting from primary transcript termini will be significantly reduced or absent. Comparison of 5’-RACE products obtained from TAP-treated and untreated RNA (lanes +T and –T in Figure 8 and Figure 11) would thus identify primary transcripts; http://edoc.hu-berlin.de
Human mtDNA contains two major promoters for transcription, the heavy (H)-strand promoter (HSP) and the light (L)-strand promoter (LSP) (1). The HSP has been localized to -16 to +7 of the transcriptional start site (1, 2), and the LSP, to -28 to +16 of the transcriptional start site.
HSP—CCAAACCCCAAAGAC; LSP—ATACCGCCAAAAGAT;
Purple colored letter indicate the transcription start points.
Summary of nucleotide sequences around experimentally defined transcription initiation sites in Arabidopsis mitochondria. Two sequence logos are shown that were generated using Web Logo [http://weblogo.berkeley.edu/logo.cgi, (45,46)] from an alignment of 20 promoter sequences activating transcription initiation at an adenine nucleotide (upper sequence logo) and from an alignment of 11 sequences supporting initiation at a guanine nucleotide (lower sequence logo). Position +1 corresponds to the transcriptional start.
Chloroplast promoters are similar to bacterial promoters having –10 and –35 sequences from the start position. Plastid RNAPs are coded by nuclear genomes, and translated products are transported into plastids. Plastid-Cyanobacteria since they become symbionts lost their genetic autonomy and depend on nuclear contribution; it shows how one become dependent on the other though once one has all the genetic autonomy.
Mammalian mitochondrial fission and fusion:
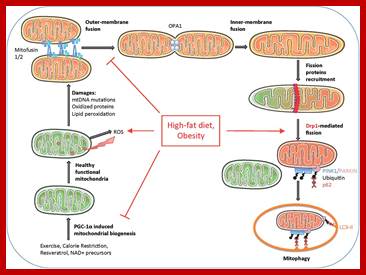
Mitochondrial dynamics, quality control and miRNA regulation in skeletal muscle: implications for obesity and related metabolic disease; ;Dennis Dahlmans et al; http://www.clinsci.org
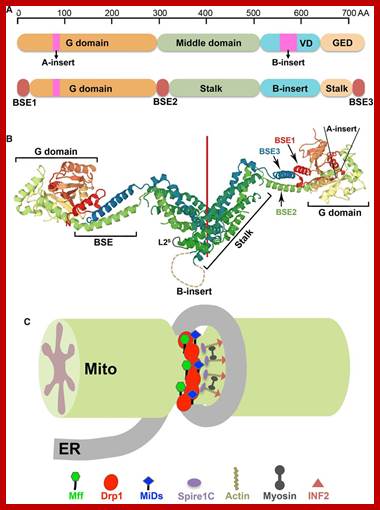
http://greatcourse.cnu.edu.cn/

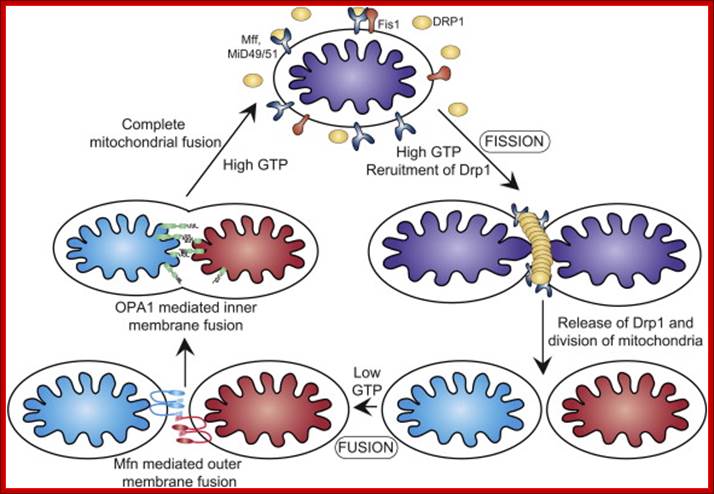
Mitochondrial division occurs when the cytosolic Drp1 is recruited to the mitochondrial outer membrane resulting in organelle constriction. Fis1, Mitochondrial fission factor (Mff) and Mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51) have been proposed to act in recruitment and assembly of Drp1 at the outer mitochondrial membrane. In the presence of GTP, Drp1 forms concentric rings around the scission site. GTP hydrolysis causes constriction of the Drp1 rings and facilitates scission of the mitochondrion. Other proteins involved in mitochondrial fission include Endophilin B1 that is proposed to modulate membrane curvature, and Lipin 1b that converts phosphatidic acid to diacylglycerol (DAG). Intermembrane space MTP18 and outer membrane GDAP1 also have proposed roles in mitochondrial fission. The core protein factors for mitochondrial fission and fusion are dynamin proteins that possess membrane-remodeling properties.


http://journal.frontiersin.org
Hakjoo Lee, Yisang Yoon;http://www.biochemsoctrans.org
The dynamics of mitochondrial fission and fusion. The localization, as well as some interaction and modification of the principal proteins involved in the two processes are shown. Once dephosphorylated, DRP1 is recruited to the outer membrane by FIS1 or by another, unknown, component. The oligomerization of DRP1 is followed by constriction of the membrane and mitochondrial fission. The pro-fusion proteins (MFNs on the outer membrane and OPA1 on the inner membrane) oligomerize to induce fusion of the membranes. Other additional components of the machinery are shown. BAX, BCL2-associated X protein; BNIP3, BCL2/E1B 19 kDa-interacting protein 3; CAMK1a, calcium/calmodulin-dependent protein kinase 1a; DRP1, dynamin-related protein 1; FIS1, fission protein 1; GDAP1, ganglioside-induced differentiation-associated protein 1; l-OPA1, long form of OPA1; MFN, mitofusin; MIB, mitofusin-binding protein; MTP18, mitochondrial protein 18 kDa; OPA1, optic atrophy 1; PKA, protein kinase A; PLD, phospholipase D; s-OPA1: short form of OPA1. https://www.researchgate.net
Mitochondria division: Strategy using mass spectrometry or tandem mass spectrometry (MS/MS) to analyze proteolytically digested proteins. Peptide mass fingerprinting (PMF) of digested peptide fragments using matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) is the preferred method for an initial protein identification after separation by 2-DE due to its high throughput and cost efficiency.
Yale Univ.
Mitochondrial dynamics: Steady state mitochondrial morphology requires a balance of fission and fusion events. Organelle division is mediated by Drp1 which forms high molecular weight oligomers on the mitochondrial surface. Once Drp1 is released fission is complete.
Mitochondrial fusion is a two-step process that requires outer and inner membrane fusion. Outer membrane fusion is facilitated by mitofusin tethering of adjacent membranes. In high GTP environments, OPA1 isoforms allow inner membrane fusion.
Mitochondrial fusion is promoted by homotypic/heterotypic interactions of the Mitofusin 1 and Mitofusin 2 dynamin-related GTPases at the outer mitochondrial membrane (OMM) of adjacent mitochondria and by Opa-1, also a dynamin-related GTPase, at the inner mitochondrial membrane (IMM). Inhibition or loss of any one of these proteins impedes mitochondrial fusion leading to increased mitochondrial fragmentation and is associated with clinical neuropathy in Charcot–Marie–Tooth disease and Autosomal Dominant Optic Atrophy, highlighting the critical role played by mitochondrial fusion in cellular homeostasis, particularly in the nervous system.
Mitochondrial fission requires the recruitment of a different dynamin-related GTPase, Drp1 to the OMM where it forms ring-like oligomers that pinch off mitochondria into smaller fragmented mitochondria. Fission is important ahead of mitosis to ensure even distribution of mitochondria to daughter cells but also occurs when cells undergo mitophagy or apoptosis. Recruitment of cytosolic Drp1 to the mitochondria during fission is a regulated process involving post-translational modification of Drp1 and its interaction with putative receptors at the OMM such as Mitochondrial Fission Factor (Mff), Fis1, MiD49, MiD51, and possibly other proteins with which Drp1 interacts. Recent work has also highlighted a role for the endoplasmic reticulum (ER) that is intimately associated with mitochondria, in determining the sites at which fission will occur . The constriction of mitochondria at points of contact with the ER are set up prior to recruitment of Drp1 to mitochondria and independent of Mff. Intriguingly, the mitochondrial fusion protein Mfn2 also plays a role in tethering mitochondria to the ER, in a manner required for proper calcium uptake by the mitochondria from the ER. Screens in yeast have identified additional putative molecular regulators of mitochondrial tethering to the cell cortex and the ER in ways that regulate mitochondrial positioning in cells and inheritance by daughter cells but whether similar mechanisms operate in mammalian cells is unclear; http://journal.frontiersin.org
Some of the well-known
Disease caused by malfunctioning of Mitochondria:
Diabetes mellitus and deafness (DAD), Leber's hereditary optic neuropathy (LHON), Wolff-Parkinson-White syndrome, multiple sclerosis-type disease affects 1 in 50,000 people in Finland, Leigh syndrome, Neuropathy, ataxia, retinitis pigmentosa, and ptosis (NARP), Dementia, Myoneurogenic gastrointestinal encephalopathy (MNGIE), Myoclonic Epilepsy with Ragged Red Fibers (MERRF), Mitochondrial myopathy, encephalomyopathy, lactic acidosis, stroke-like symptoms (MELAS)mtDNA depletion mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) (WIKIPEPDIA);
Symptoms include poor growth, loss of muscle coordination, muscle weakness, visual problems, hearing problems, learning disabilities, heart disease, liver disease, kidney disease, gastrointestinal disorders, respiratory disorders, neurological problems, autonomic dysfunction and dementia.
2. Plastids:
Introduction:
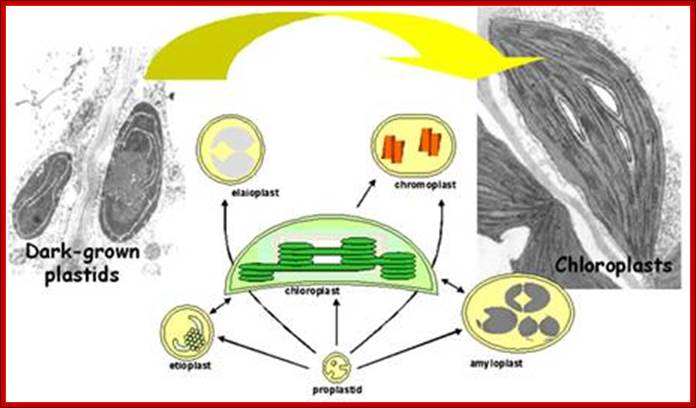
Diversity of plastid forms and their interconversions:

· Plastids exist in different forms, and the identity and abundance of each are controlled by developmental and environmental cues. Different types interconvert (see the arrows) following reorganization of the organellar proteome, Paul Jarvis
& Enrique López-Juez; http://www.nature.com
Plastids are sine qua non plant organelles found in all plants including algae. But cyanobacteria has all the structural features of chloroplast, in fact plastids are endsymbiotics of cyanobacteria. They perhaps originated about 450 million years ago. Cyanobacteria do not contain their own plastids for their cells themselves act as plastids. It is these unicellular bacteria entered eukaryote cells and become symbionts. These organelle harvest solar energy combining with CO2 fix it in the form of (CH2O)n, which is used in various forms for plants animals and microorganisms for their growth and reproduction. Plants such as algae some contain a single plastid, some contain two or more. Higher plant cells possess 60-100 plastids per cell.

Justin Graykin; https://justinegraykin.wordpress.com
Development of plastids in early embryonic cells start as colorless vesicle like proplastids and develop into different forms such as elaioplasts, proteinoplasts, amyloplasts, chromoplasts and chloroplasts. Plastid is dependent on cell nucleus and cell is dependent on plastids. Both are in communication with each other. Plastids are once free living cyanobacteria and many billions of years ago by endosymbiosis, become permanent residents of plant cells. They do show certain bacterial characteristic features. Animal cell with acquisition of mitochondria and then acquiring plastids become autonomous as photosynthesizing organisms called Plants. They also developed other structural features specific for plants. During adaptation to endosymbiontic form, more than 90% of bacterial genome was transferred to the nucleus of the host cell, but retained few genes.

Developmental Stages; http://www.ivy-rose.co.uk/Biology/Cells/Plant-Cell-Structure.php
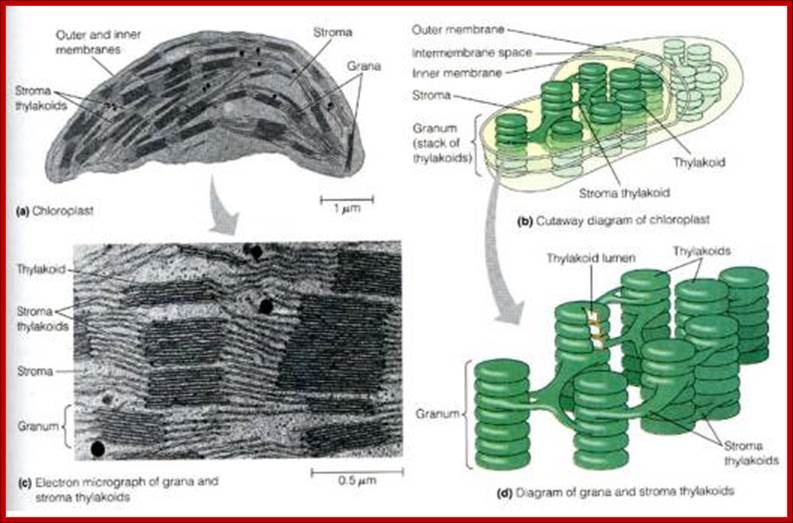
Plastids, like mitochondria are bound by outer and inner membranes, whose structure and functions are different. The two membranes at some points are bound to each other through attachment or transporter complexes, where one can find protein transporters. The inner membrane by invagination and invasion developed into circular membrane discs, which are organized into stacks; they are called thylakoids. The thylakoids are stacked one above the other, but intercalated with inter granal membranes. Such stacks of thylakoids are called Grana. There can be 100ds of such grana in a single plastid. They are interconnected by interregnal membranes. Each of these membranes are specialized structures with proteins and unique pigments. These are responsible for harvesting solar energy and fix CO2 in to carbohydrates. Chloroplasts are classified into different types based on their structure and function- Chloroplasts, Etioplasts, Chromoplasts, Leucoplasts (monoterpenes), Gerontoplasts, Amyloplasts, Elaioplasts (store fats), Proteinoplasts (store proteins) and Tannosomes (produce tannins).
Dr Enrique Lopez Juez; http://pure.rhul.ac.uk/
http://greatcourse.cnu.edu.cn/
- Outer membrane, 2. Intermembrane space,3. Inner membrane (1+2+3: envelope), 4.Stroma (aqueous fluid), 5.’ Thylakoid lumen (inside of thylakoid) ‘6. Thylakoid membrane, 7.Granum (stack of thylakoids), 8.Thylakoid (lamella), 9. Starch, 10. Ribosome, 11. Plastidial DNA,12.Plastoglobule (drop of lipids). http://en.wikipedia.org/
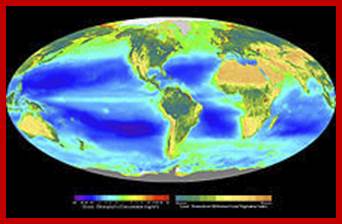
Composite image showing the global distribution of photosynthesis, including both oceanic phytoplankton and terrestrial vegetation. Dark red and blue-green indicate regions of high photosynthetic activity in ocean and land respectively; http://en.wikipedia.org/wiki/Photosynthesis
How the metabolism of Glycolipids is coordinated in plant cells. Authors, EriC Marechal et al have studie coordination of lipid synthesis within the plastid envelope and at whole cell level. They have analyzed the role of phosphatidic acid in regulating the synthesis of galactolipids.
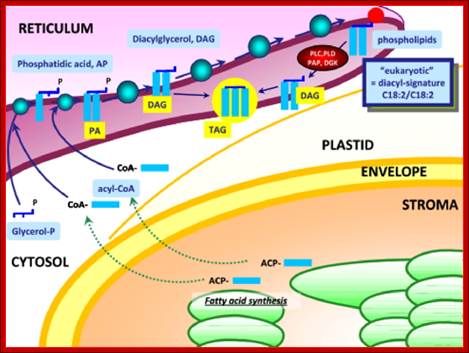
Synthesis of Phospholipids and oils takes place in ER; www.typicalfruits.de
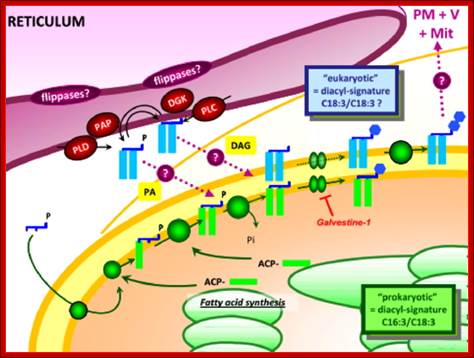
Synthesis of Galactolipids is localized in the envelop that limits plastids. Endomembranes and as if envelop membranes are in dialogue with each other in transferring lipids, http://www-dsv.cea.fr/en/institutes/institute-of-life-sciences.
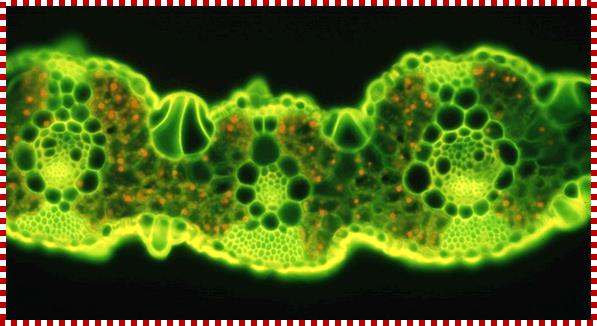
TS of a grass leaf; Mesophyll cells contain C3 plastids AND Bundle sheath cells contain C4 plastids;The families of Flowering Plants L. Watson and M. J. Dallwitz;http://delta-intkey.com/
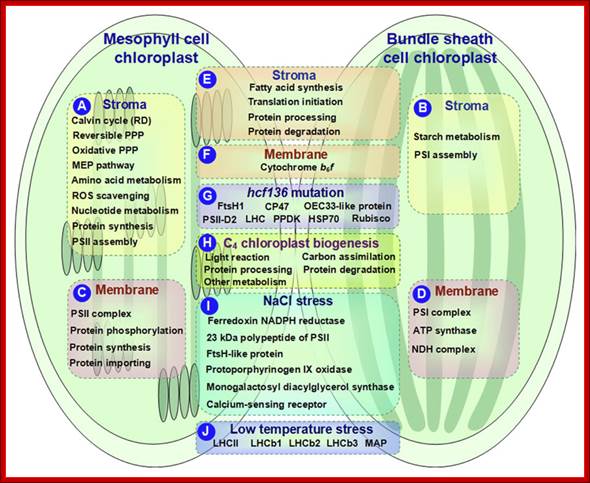
Schematic presentation of cell-specific or stress-responsive pathways and proteins in maize chloroplasts revealed from proteomics studies. (A,B) Preferential metabolic pathways in the stroma of M and BS, respectively; (C,D) Preferential metabolic pathways and protein complexes in the chloroplast membrane of M and BS, respectively; (E,F)Metabolic pathways and protein complexes equally distributed in the chloroplast stroma/membrane of M and BS; (G) Differentially expressed proteins in chloroplasts of maize hcf136 mutant in comparison to wild-type; (H)Dynamics of metabolic pathways during maize chloroplast biogenesis; (I) Salt-responsive proteins in maize chloroplasts; (J) Low temperature-responsive proteins in maize chloroplasts. ATP, adenosine triphosphate; HSP, heat shock protein; LHC, light harvesting complex; MAP, minor antenna proteins; MEP, methylerythritol phosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; NDH, NAD(P)H dehydrogenase; OEC, oxygen evolving center; PPDK, pyruvate orthophosphate dikinase; PPP, pentose phosphate pathway; PSI, photosystem I; PSII, photosystem II; RD, reductive phase; ROS, reactive oxygen species; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase; Qi Zhao et al;. http://journal.frontiersin.org/

In C3 plants Photosynthesis and carbon fixation takes place in the same plastid; http://hyperphysics.phy-astr.gsu.edu/
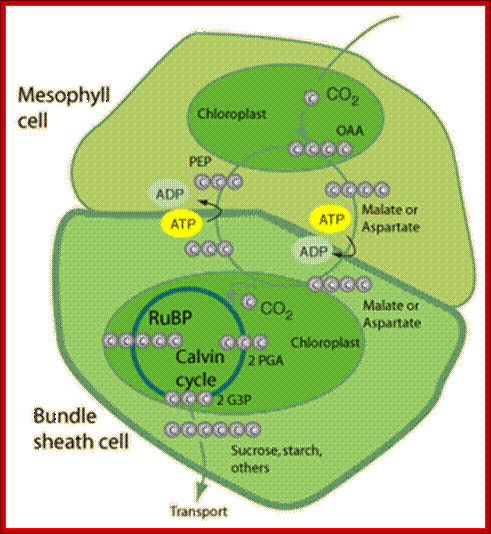
In C4 plants C3 carbon fixation takes place in mesophyll cells and C4 carbon acids are transferred to bundle sheath cells, where Calvin’r cycle occurs in chloroplasts and protects from photorespiration; http://hyperphysics.phy-astr.gsu.edu/
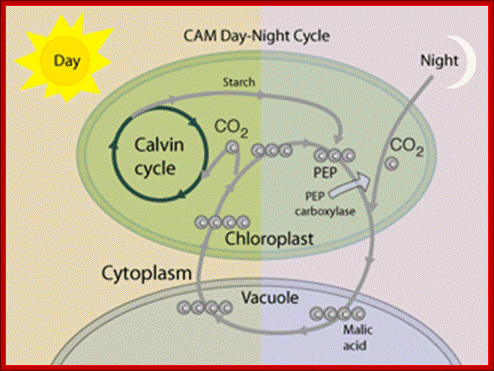
In CAM plants photosynthesis and initial carbon fixation occurs at night and 4-carbonic acid is stored in cell’s Vacuole. During day Calvins’ cycle operates in the same chloroplast. http://hyperphysics.phy-astr.gsu.edu/
Plastid DNA (cpDNA/ctDNA):
Plastids contain ds circular genomic DNA of 150Kbp to 350kbp size. The numbers of such genomes per plastid can 100 to 280 or more copies. The copy number increase during growth and differentiation. Occasionally in certain strains of Chlamydomonas spa19 two forms of spa (-) and spa(+) exist. Word spa is the suppression of polyadenylation. Proteins from stained spots were identified by matrix-assisted laser-desorption ionization (MALDI)-tandem time-of-flight (TOF/TOF) analysis using the MASCOT database search algorithm for unambiguous protein identification (see “Materials and Methods”). This method has identified about 369 proteins and etioplast contain 237.
The plastid genome is encoded with for more than ~150 genes. Among them Light harvesting complexes are important. The genome code for rRNAs, tRNAs and many proteins required for light harvesting. Besides there a large number of imported protein that are involved in the development of plastids and function of plastids. Plastid DNA exists as complex of proteins and associated with inner membrane and exists as ‘plastid Nucleoids’.
Plant chloroplast DNA molecules are about 120 - 180 kb in size. Higher plant cpDNA codes for about ~120 genes, including rRNAs, 30 tRNAs, several ribosomal proteins, RNA polymerase subunits, several respiration-related proteins, plus other 40 proteins. cpDNA has introns. However the primary transcriptome of Barley chloroplasts contain numerous noncoding RNAs coded by plastid RNAPs.
Microsatellite locations superimposed on chloroplast genetic maps of (a) rice, (b) tobacco, (c) black pine and (d) liverwort. A/T mononucleotide repeats are represented by purple, asterisked and numbered lines outside each circle. In liverwort, for clarity, this type of repeat is illustrated in the same way within the inverted repeats, but by short purple bars within the circle for the remainder of the genome. Dinucleotide repeats in liverwort are represented by blue, asterisked and numbered lines, and a G/C mononucleotide repeat in black pine is represented by a green, numbered and asterisked line. Exact nucleotide locations of microsatellites are available from the authors. Genes shown on the outside of the circle are transcribed in a counterclockwise direction, and those shown on the inside are transcribed in a clockwise direction. LSC means large, single-copy region and SSC means small, single-copy region; IR, inverted repeat.

Revisewd Genetic map of M.polymorpha, Ira,IRb,SSC and LSC on inner circle indicate inverted repeat regions, the small single copy region and the large single copy region respectively.
The plastid genome is well illustrated with inverted repeats of rRNA genes, Gene content, organization and molecular evolution of plant organellar genomes and sex chromosomes — Insights from the case of the liverwort Marchantia polymorpha; http://openi.nlm.nih.gov/;Ohyama K, Takemura M, Oda K, Fukuzawa H, Kohchi T, Nakayama S, Ishizaki K, Fujisawa M, Yamato K - Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. (2009)
In angiosperms, chloroplast DNA mainly exhibits maternal inheritance and its gene flow is therefore principally via seeds. Hence, chloroplast (and mitochondrial) genomes may exhibit different patterns of genetic diversity compared to nuclear genomes. Chloroplast DNA can be used as a tool in plant population biology. The occurrence of mononucleotide repeats within the Chloroplast genome of seed plants, bryophytes and algae as a consequence polymorphism within these regions may be used to study both intraspecific and interspecific variability. Chloroplast polymorphism in plant biology is of considerable practical value in monitoring gene flow, population differentiation and cytoplasmic diversity. In some species the plastid genome exists in linear form of 120-150kbp.
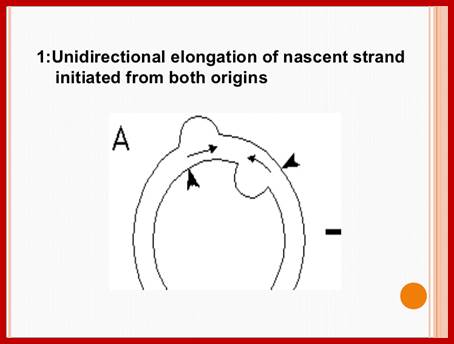
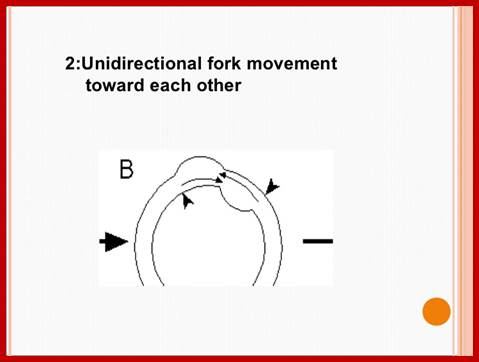
Plastid genomic DNA replication origin is located at 3’ end of 23srRNA gene in Rice. In the early 1970s, electron microscopy analyses of replicating chloroplast intermediates from pea and corn drew a model of replication. This model was based on two displacement loops (D-loops) separated by some distance on the genome, where the displacement of the two D-loops occur on opposite strands of the parental DNA molecule and subsequently, move towards each other. As a result of this mechanism, half of each displaced parental strand (from either origin until the centre of two origins) is left single-stranded on both sides of the pair of inverted repeats. This discovery of Cairn's replication mechanism in pea and corn chloroplast genomes was followed by a series of studies independently confirming this model for various plant species (Euglena gracilis; single D-loop]; Nicotiana tabacum; Chlamydomonas reinhardtii; Oenothera; Zea mays. The rolling circle mechanism could be initiated after one round of Cairns type of replication, so as to generate multiple copies of the chloroplast genome even though replication is initiated only once (pea and corn,). Electron microscopy analyses of certain in vitro tobacco chloroplast replication intermediates also revealed Y-arc patterns, indicative of rolling circle replication; Neeraja M Krishnan and Basuthkar J Rao.
Plastid genomes of peridinin-containing dinoflagellates are unique in that its genes are found on multiple circular DNA molecules known as ‘minicircles’ of ~2–3 kb in size, carrying from one to three genes. The non-coding regions (NCRs) of these minicircles share a conserved core region (250–500 bp) that are AT-rich and have several inverted or direct repeats. The migration pattern of the APBs in a 2D-gel electrophoresis show replication uses rolling circle mode, rather than the bubble-forming type. These minicircles contain one gene per one circular DNA, very unique.
Cp DNA replication:
Chloroplast DNA, whatever may be the size, most of them exist in supercoiled state. They use D-loop type of replication origin. Transcription start at this region and the RNA is used as primer for replication.
In molecular biology, a displacement loop or D-loop is a DNA structure where the two strands of a double-stranded DNA molecule are separated for a stretch and held apart by a third strand of DNA. The third strand has a base sequence which is complementary to one of the main strands and pairs with it, thus displacing the other main strand in the region. Within that region the structure is thus a form of triple-stranded DNA. A diagram in the paper introducing the term illustrated the D-loop with a shape resembling a capital "D", where the displaced strand formed the loop of the "D".
The mt DNA pol is called gamma DNAP of 105kD. Plastids too contain gamma like DNAps. The Pea DNAP is 70kD insensitive to dideoxynucleotide (d2NTP). For its activity, it requires 43kD protein; it binds to DNAP. Binding of 43kD protein to DNAP increases processive activity. It is a glycoprotein. The enzyme lack detectable 5’à3’exonuclease activity, but contain 3’à5’ exonuclease activity.
Replication plastid DNA shows the origin of H strand and L strand replication. It also exhibits two origins located at the end of 23splastid rRNA genes, both start open up , as the replication progresses the meet and merge and replication continues bidirectionally. http://bioinfosu.okstate.edu/,
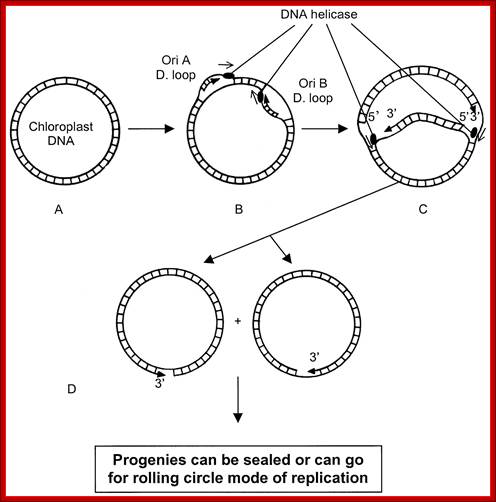
Plastid DNA replication steps; http://galleryhip.com/
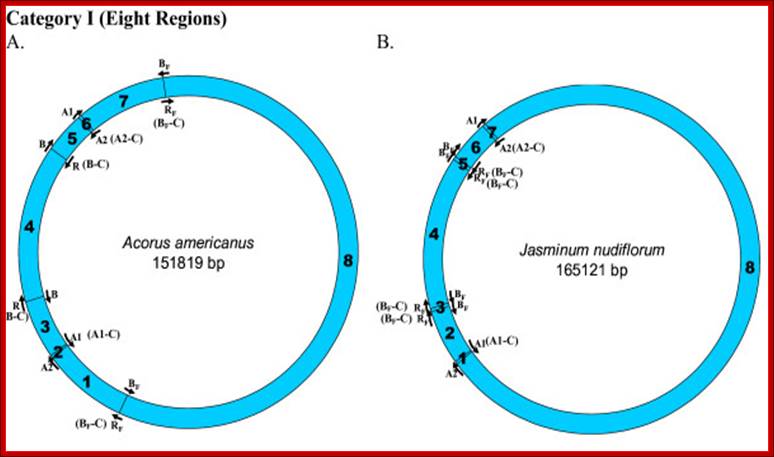
Locations of replication origins mapped on the tobacco chloroplast genome. The figure depicts the relative positions of known replication origins (A1, A2, B and R) and their complementary copies (origin type appended with '-C'), on the circular tobacco chloroplast genome, mapped using SimVector 4.22 http://www.premierbiosoft.com webcite. The six regions between each pair of replication origins are annotated respectively as 1 (between A2 and A1 [A1-C]), 2 (between A1 [A1-C] and R [B-C]/B), 3 (between R [B-C]/B and B/R [B-C]), 4 (between B/R [B-C] and A1), 5 (between A1 and A2) and 6 (between the A2s on either strands. Krishnan NM, Rao BJ - BMC Genomics (2009)https://openi.nlm.nih.gov
Mapping of replication origin-like sequence homologues in second category of chloroplast genomes: Representative genomes from the second category are depicted here. Categorization of chloroplast genomes were performed as described in Figure 2. www.Biomedcentral.com
https://www.slideshare.net
https://www.slideshare.net

Source: mcmanuslab.ucsf.edu;’ https://image.slidesharecdn.com
Often the plastid genome, once the circular DNA go through replication, it switches to what is called rolling circle mode for the production more number of genomes in short time.
Plastid Genome Transcription:
Reasonably, when compared to mitochondria, plastids contain more than 120-150 genes?; in some nearly 200 genes. They are protein coding, tRNA and rRNA genes. But rRNA genes are located at sites on either sides of D-loop region as inverted repeats. Most of the tRNA genes are clustered and found in between protein or RNA coding regions.
Transcription is performed by two types of RNA polymerases; one plastid genome coded called PEP RNAP. The second type is nuclear coded called NEP. The PEP coded RNAP is more or less similar to that of bacteria coded for by plastid genes rpoA, rpoB, rpoC1, and rpoC2, but requires a sigma70 like proteins for identification of promoter elements and activating the polymerase. The molecular mass of this complex is ~900kDa. The beta subunit is essential for PEP transcriptional activity.
The NEP coded RNAP consists of single polypeptide of 110kD. It recognizes distinct types of promoters with sequence similarity to plant mitochondrial promoters. NEP promoters were found to be more active in early leaf development.
Fractionation studies have identified several candidate sigma factors in purified RNA polymerase preparations, it was only 4 years ago that the first sigma factor genes were cloned from two photosynthetic eukaryotes, both of which were from red algae. More recently this achievement has extended to the identification of families of sigma like factor genes from several species of vascular plants. Recent results suggest that accumulation of individual sigma like factories controlled by light, by plastid type and/or by a particular stage of chloroplast development. First sigma factor genes were cloned from two photosynthetic eukaryotes, both of which were red algae. More recently this achievement has extended to the identification of families of sigma like factor genes from several species of vascular plants. We propose that σ interaction with the −10 element in PrrnP1 is replaced in part by direct PEP-RUA (protein–DNA) interaction or by protein–protein interaction between the PEP and an RUA binding transcription factor, Jon Y. Suzuki, Priya Sriraman etal. The rrn operon in tobacco is transcribed by the multisubunit, plastid-encoded RNA polymerase (PEP) from a σ70-type promoter (PrrnP1). Some have P1 and P2 promoters.
The PEP transcribes rrn operon containing -35 and -10 promoter sequences. PrrnP1 contains the conserved −35 (TTGACG) and −10 (TATATT) promoter elements. The promoter also contain upstream of -35, GTGGGA and activator RUA (rRNA upstream activator). PEP is reported to increase its activity during chloroplast maturation.
PrrnP1:
-------------- TTGACA------ >--- TATATT - --+1
Most of the house keeping genes of plastids is transcribed by both enzymes. Genes of photosystems I and II are completely dependent on PEP transcription. Cyanobacteria, the chloroplast progenitors, have a plethora of noncoding RNAs at their disposal to regulate gene expression. However, only a few potentially regulatory RNAs have been reported to be transcribed from plastid genes. Therefore, chloroplasts may contain additional hitherto undetected genes for regulatory ncRNAs. (Nishimura et al., 2004; Georg et al., 2010; Hotto et al., 2010; Zghidi-Abouzid et al., 2011).
RBCL/S- promoters:
-18 GATCAG---- +1 >----- +63-AAGCTT--+1>--
RbcS-- LRE-TTTCAAAA---TTACT CATGG--+1>
RbcS is a multigene family consisting 2-22 genes.
Plastid genes in higher plants are mainly organized as operons, of which more than 60 have been described in the tobacco (Nicotiana tabacum) chloroplast genome.
Plastids mRNA protein coding transcripts are polycistronic and their 3’ end has inverted repeats. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. It is concluded that intercistronic processing is not required for translation of these RNAs, although certain processing steps may enhance translational efficiency. The first characterization of transcriptional, posttranscriptional, and translational processes of heterologous operons expressed via the tobacco (Nicotiana tabacum) chloroplast genome is reported; Tania Quesada-Vargas etal.
Plastid mRNA stability is mainly influenced by the presence of 5′ untranslated regions, or UTRs (Eibl et al., 1999; Zou et al., 2003), nucleus-encoded factors, posttranscriptional and translational patterns have been the target of several studies that showed that intercistronic processing enhanced translation of chloroplast operons, including the maize (Zea mays) psbB and pet clusters, which comprise genes from the PSII reaction center and the cytochrome b6/f complex, respectively (Barkan,1988).
In Chlamydomonas, nearly all chloroplast genes appear to be transcribed as monocistronic mRNAs, with translation being an essential regulatory step of gene expression
Other mechanisms, such as editing, which can produce alternate start codons, have been linked to alternative processing and to a complete, different translation pattern.
A protein barrier mechanism was proposed based on analysis of the pentatricopeptide repeat (PPR) protein PPR10: PPR10 binds two intercistronic regions and impedes 5′- and 3′-exonucleases, resulting in processed RNAs with PPR10 bound at the 5′- or 3′-end. Chloroplast mRNA populations are characterized by overlapping transcripts derived by processing from polycistronic precursors. The mechanisms and functional significance of these processing events are poorly understood. We describe a pentatricopeptide repeat (PPR) protein, PPR10, whose binding defines mRNA segments derived from two transcription units in maize chloroplasts. PPR10 interacts in vivo and in vitro with two intergenic RNA regions of similar sequence. The processed 5′ and 3′ RNA termini in these regions overlap by approximately 25 nucleotides
1. Many of the chloroplast mRNAs possess Shine-Delgarno (SD)-like sequences (typically GGAGG) in the 5′-untranslated regions, but the position is highly variable. The rbcL mRNA has a typical SD-like sequence; so also the mRNA of atpE. However, SD-like sequences in the rps12 mRNA and in the petB mRNA is located far from (–44 to –42) and not close to (–5 to –2) the initiation codon; these results indicate that functional properties; Tetsuro Hirose1 and Masahiro Sugiura
Post transcription processing of chloroplast mRNAs can be cis splicing, alternate splicing and 1st and 2ng group II intron mediated splicing. Even mRNAs are subjected editing.
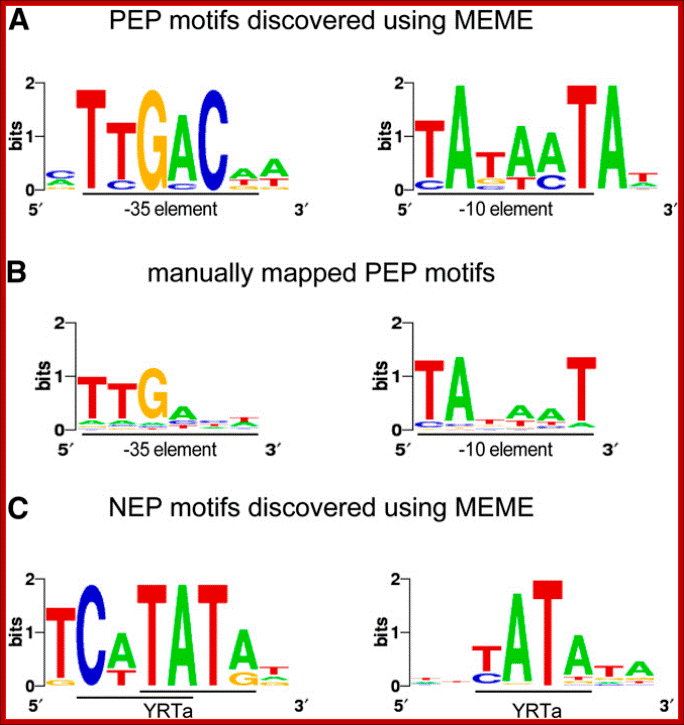
Sequence Logos of Promoter Motifs Detected in Green and White Plastids: Maximization for Motif Elicitation (MEME); Yusuke Yagi and Takash Shina; http://frontierin.org
In green plastids, MEME analysis discovered a -10 (right) and -35 (left) PEP consensus element upstream of 44 and 20 TSSs, respectively. The motifs were found to be significantly enriched in green pre-TSS sequences. (B) A manual search for the PEP promoter elements detected the -10 box (right) in 156 TSSs and the -35 box (left) in 109 of the TSSs with mapped 10 elements. (C) Two versions of the YRTa motif were discovered by MEME in white plastids. A TCaTATat motif (left) was found upstream of 22 of the white TSSs and Gene expression in plastids of higher plants is dependent on two different transcription machineries, a plastid-encoded bacterial-type RNA polymerase (PEP) and a nuclear- encoded phage-type RNA polymerase (NEP), which recognize distinct types of promoters, yATata (right) upstream of 151 (62%).
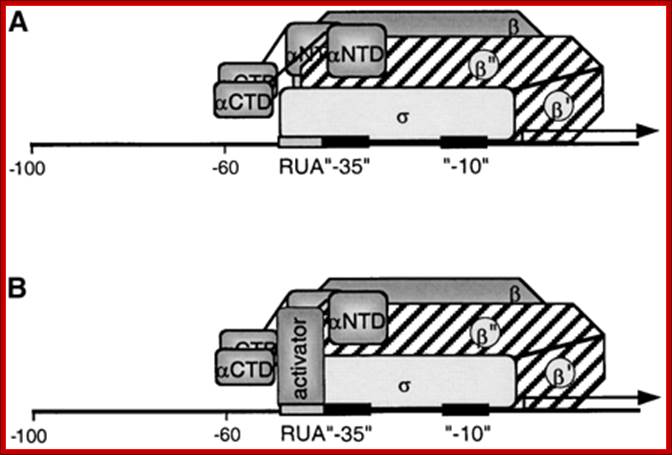
Model for the Interaction of the PEP with the PrrnP1 promoter: (A) Factor-independent activation of PrrnB1 transcription. Depicted is the σ interaction with the RUA −35 region. Note that the actual subunit involved in the interaction may be a different subunit. Identified are the α- (αCTD and αNTD), β-, β′-, β′′-, and σ-subunits and the RUA, −35, and −10 promoter elements. (B) Factor-dependent activation of PrrnP1 transcription by activator bound to RUA. https://www.researchgate.net/


The PEP coded RNAP complex. Model illustrating regulation by AtSIG6 phosphorylation at two different chloroplast promoters.(a) Transcription of the atpB/E operon is driven both from an NEP and a PEP promoter. The latter is recognized by the PEP/SIG6 complex, yet only if the sigma factor is phosphorylated at defined regulatory site(s).(b) In contrast, there is no apparent effect of SIG6 phosphorylation state on transcription at the psbA PEP promoter. The more complex architecture of this promoter (Figure S4 could be part of a scenario in which SIG6 is capable of binding at various phosphorylation sites, perhaps even further complicated by functional overlap with other sigma factors.Colour code: green, promoters; red, PEP; orange, AtSIG6.
As depicted in the model shown in Figure 6(a) for atpB transcription, AtSIG6 is able to confer productive binding to, and initiation from, the PEP promoter only in its phosphorylated state. The most critical phosphorylation sites seem to be those at S94/95 and S174, as inferred from the data indicating that mutational changes at these sites lead to dramatic alterations in phenotype (Figure 4) and plastid gene expression at the atpB PEP promoter in vivo (Figure 5). Interestingly, none of the analysed mutant lines show any appreciable effect on psbA gene expression in vivo (Figure 5). Furthermore, EMSA control experiments using the psbA promoter (Figure S5) did not reveal differences in binding activity of the phosphorylated versus unphosphorylated forms of AtSIG6. Together, this could mean that binding and initiation at the psbA promoter (Figure 6b) by AtSIG6 may be regulated differently compared with the atpB promoter; https://www.researchgate.net

Recent advances in the study of chloroplast gene expression and its evolution; Chloroplasts are semiautonomous organelles which possess their own genome and gene expression system. However, extant chloroplasts contain only limited coding information, and are dependent on a large number of nucleus-encoded proteins. During plant evolution, chloroplasts have lost most of the prokaryotic DNA-binding proteins and transcription regulators that were present in the original endosymbiont. Thus, chloroplasts have a unique hybrid transcription system composed of the remaining prokaryotic components, such as a prokaryotic RNA polymerase as well as nucleus-encoded eukaryotic components. Recent proteomic and transcriptomic analyses have provided insights into chloroplast transcription systems and their evolution. Here, we review chloroplast-specific transcription systems, focusing on the multiple RNA polymerases, eukaryotic transcription regulators in chloroplasts, chloroplast promoters, and the dynamics of chloroplast nucleoids. Note- There are Two Basic Chloroplast Transcription Machineries with Different Evolutionary Origin; Yusuke Yagi and Takashi Shiina2;http://journal.frontiersin.org
Nearly 89 genes of 113 plastid genes are a part of ~20 operons, while 24 are transcribed as monocistronic transcripts. Plastid genes in higher plants are mainly organized as operons, of which more than 60 have been described in the tobacco (Nicotiana tabacum) chloroplast genome- they are more or less related. Chlamydomonas contain monocistronic transcripts, in maize, tobacco, dicistronic to tricistronic mRNAs. Of the chloroplast genome from rice (Oryza sativa L.) 134,524bp, contains four rRNA genes, 30 tRNA genes, and over 100 genes that encode proteins. Most of the genes on the chloroplast DNA are organized as clusters and are co-transcribed as long primary transcripts. We identified 16 polycistronic transcripts from the rice chloroplast genome.
Coping with cryptic and defective transcripts in plant mitochondria; Sarah Holec, Heike Lange and Jean Canaday, Dominique Gagliardi
Plant mitochondria are particularly prone to the production of both defective and cryptic transcripts as a result of the complex organization and mode of expression of their genome. Cryptic transcripts are generated from intergenic regions due to a relaxed control of transcription. Certain intergenic regions are transcribed at higher rates than genuine genes and therefore, cryptic transcripts are abundantly produced in plant mitochondria. In addition, primary transcripts from genuine genes must go through complex post-transcriptional processes such as C to U editing and cis or trans splicing of group II introns. These post-transcriptional processes are rather inefficient and as a result, defective transcripts are constantly produced in plant mitochondria. In this review, we will describe the nature of cryptic and defective transcripts as well as their fate in plant mitochondria. Although RNA surveillance is crucial to establishing the final transcriptome by degrading cryptic transcripts, plant mitochondria are able to tolerate a surprising high level of defective transcripts.
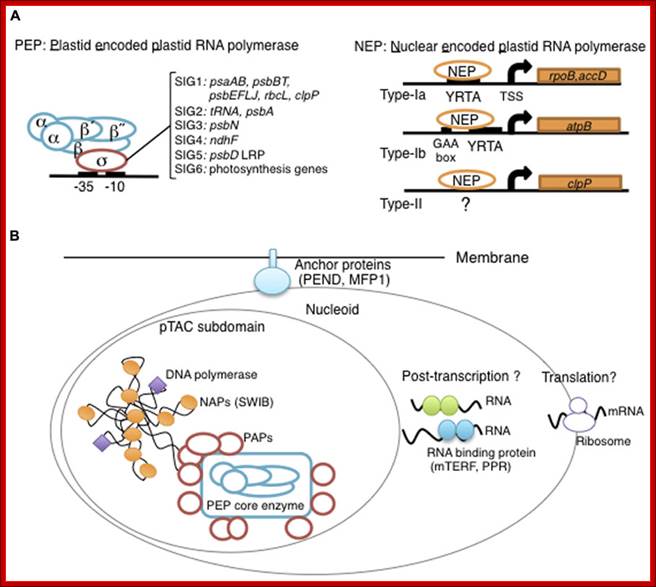
Overview of chloroplast transcription. (A) Basic transcriptional machinery in higher plants. Higher plants have two distinct types of chloroplast RNA polymerase: plastid-encoded plastid RNA polymerase (PEP; left panel) and nucleus-encoded plastid RNA polymerase (NEP; right panel). PEP is a bacterial-type multi-subunit RNA polymerase composed of the core enzymatic subunits α, β, β′, β″ (blue) and a sigma subunit (red) that is responsible for promoter recognition. Plastid sigma factors are divided into six subgroups, SIG1–SIG6, and selectively recognize bacterial-type promoters in the plastid. NEP (right panel) is a monomeric enzyme that resembles mitochondrial T7-type RNA polymerases. NEP is involved in the transcription of housekeeping genes such as rpo genes for PEP core subunits, and ribosomal protein-coding genes. Positioned upstream of genes transcribed by NEP are three distinct types of promoter structures (Type-Ia, Type-Ib, and Type-II). (B)The chloroplast nucleoid subdomain and its components. Chloroplast nucleoids are attached to the membrane (envelope or thylakoid) by anchor proteins (PEND and MFP1). The plastid transcription active chromosome (pTAC) is one of the nucleoid subdomains, which contains the transcription factory. Chloroplast genomic DNA is packed by chloroplast-specific nucleoid-associated proteins (NAPs; orange circle). The mature chloroplast contains a large PEP complex with several PEP associate proteins (PAPs; red circles). Recent proteome analysis suggested that chloroplast nucleoids contain additional subdomains, which regulate post-transcriptional RNA maturation and translation; Yusuke Yagi1 and Takashi Shiina http://journal.frontiersin.org/
Plant mitochondria lack quality-control of defective mRNAs. Two exons are shown as green rectangles and a promoter as a bent arrow in a diagram of a part of the mitochondrial genome. Fully processed RNAs are in green, non-processed in red. Editing status is indicated by the corresponding letter (C or U). After transcription, primary transcripts undergo C to U editing, splicing and extremity processing. These processes are inefficient and give rise to a heterogeneous population of mRNAs: some are non-spliced, mis-processed or not fully edited. Defective as well as fully processed transcripts are translated. In most cases, only proteins produced from fully processed mRNAs accumulate (green ovals) while proteins produced from non-processed mRNAs are degraded (red ovals with blue crosses). Protein polymorphism due to partial editing has been detected in at least one case and is shown as a green oval with a red spot.
Plastids contain more than ~1000 proteins, most are nuclear coded then imported. The PEP RNAP is the major enzyme accounting for 88% TSS. A large number of NEP promoters become activated in the absence of PEP, as white plastids yielded a larger number of TSS than green plastids, of which the majority was not found in green plastids. Numerous ncRNAs transcripts were detected from intergenic regions as well as antisense transcripts to ∼35% of genes in green plastids. This represents a rich resource for investigating potential regulatory ncRNAs activity in plastids. Many of the plastid mRNAs have introns, which spliced either cis or trans-splicing.

(a) The ndhB gene in chloroplast DNA, consisting of two exons (1 and 2) separated by an intron. Nucleotide positions are marked below. (b) The ndhB gene in chloroplast mRNA showing the location of the AtropaNad2 primers. (c) Location of AtropaNad2a primer (bold type, underlined) in exons 1 and 2 (large case letters). Part of intron sequence shown in small case letters.

Trans splicing in Chlamydomonas.
RNA Editing:
Many a times the mRNA produced by nuclear genome and organelle genome contains wrong nucleotides, which change the codon information. Such changes can be one or two nucleotides in a mRNA. In some hundreds of such changes are found. This is due to faulty sequence in the genome. So organisms have designed methods to correct them by producing the required proteins and even complementary RNAs called Guide RNAs, which are used for correcting the mistakes generated by nuclear genome.
RNA editing in higher plant organelles results in the conversion of specific cytidine residues to Uridine residues in RNA. The recognition of a specific target C site by the editing machinery involves trans-acting factors that bind to the RNA upstream of the C to be edited. Several pentatricopeptide repeat (PPR) proteins from the PLS subfamily that are essential for the editing of particular RNA transcripts.

Genomics aims at unraveling the blueprint of life; however, DNA sequence alone does not always reveal the proteins and structural RNAs encoded by the genome. The reason is that genetic information is often encrypted. Recognizing the logic of encryption, and understanding how living cells decode hidden information—at the level of DNA, RNA or protein—is challenging. RNA‐level decryption includes topical RNA editing and more ‘macroscopic’ transcript rearrangements. The latter events involve the four types of introns recognized to date, notably spliceosomal, group I, group II, and archaeal/tRNA splicing. Intricate variants, such as alternative splicing and trans‐splicing, have been reported for each intron type, but the biological significance has not always been confirmed. Novel RNA‐level unscrambling processes were recently discovered in mitochondria of dinoflagellates and diplonemids, and potentially euglenids. These processes seem not to rely on known introns, and the corresponding molecular mechanisms remain to be elucidated. WIREs RNA 2012, 3:213–228. doi: 10.1002/wrna.1106 Sandrine Moreira, Sophie Breton, Gertraud Burger.
Coping with cryptic and defective transcripts in plant mitochondria; Sarah Holec, Heike Lange, Jean Canaday, Dominique Gagliardi
Plant mitochondria are particularly prone to the production of both defective and cryptic transcripts as a result of the complex organization and mode of expression of their genome. Cryptic transcripts are generated from intergenic regions due to a relaxed control of transcription. Certain intergenic regions are transcribed at higher rates than genuine genes and therefore, cryptic transcripts are abundantly produced in plant mitochondria. In addition, primary transcripts from genuine genes must go through complex post-transcriptional processes such as C to U editing and cis or trans splicing of group II introns. These post-transcriptional processes are rather inefficient and as a result, defective transcripts are constantly produced in plant mitochondria. In this review, we will describe the nature of cryptic and defective transcripts as well as their fate in plant mitochondria. Although RNA surveillance is crucial to establishing the final transcriptome by degrading cryptic transcripts, plant mitochondria are able to tolerate a surprising high level of defective transcripts.
In Arabidopsis thaliana (OTP80, OTP81, OTP82, OTP84, OTP85, and OTP86) are involved in editing several sites. In tobacco chloroplast transcripts 34 ntds are efficiently edited to U. No common consensus region is present around all editing sites; however, sites can be grouped in clusters that share short common sequences.

Plastid vectors to test in vivo RNA editing. (A) The pMR210 minigene vector with the maize rpoB site I editing segment (EF, NcoI–XbaI fragment) (Reed and Hanson, 1997). (B) The pSC2 kan fusion vector with the psbL editing segment (NcoI–NheI fragment) (Chaudhuri and Maliga, 1996). (C) The pRB51 3′‐UTR vector to accept editing segments as XbaI–BamHI fragments (Bock et al., 1996). The left and right plastid targeting sequences are in bold. The 5′‐UTR and 3′‐UTR of mRNA above the edited genes are depicted as a stem‐loop structure; the position of the edited nucleotide is also marked. Shown are the spectinomycin resistance (aadA) and kanamycin resistance (kan) genes; the plastid rrn16, trnV, psbE, psbF, psbL, psbJ, and 3′‐rps12 genes; EF, editing fragment; Prrn, promoter of the plastid ribosomal RNA operon; Trps16, 3′‐UTR of the plastid rps16 gene; PpsbA, promoter of the plastid psbA gene; TpsbA, 3′‐UTR of the plastidpsbA gene. Arrows mark transcription initiation sites.
Pentatricopeptide repeat proteins: a socket set for organelle gene expression; Christian Schmitz-Linneweber1, Ian Small2
Pentatricopeptide repeat (PPR) proteins are RNA-binding proteins that are particularly prevalent in terrestrial plants. Although the PPR protein family was only recognized eight years ago, it is already clear that these proteins have a range of essential functions in post-transcriptional processes (including RNA editing, RNA splicing, RNA cleavage and translation) within mitochondria and chloroplasts. Several PPR proteins have been shown to act as fertility restorer genes in commercially important cytoplasmic male sterility systems. Here, we discuss several recent papers that cover their evolutionary history and molecular mode of action. We use these new data to propose hypotheses for their physiological roles that could explain why PPR proteins are so numerous in terrestrial plants.

Molecular functions of chloroplast PPR proteins. This figure lists PPR protein functions that are well supported by genetic and biochemical experiments, including binding data. (1) Translation. PPR proteins participate in translation initiation by binding to specific sequence elements in the 5′UTR of mRNAs. (2) Editing. Binding of PPR proteins to short cis-elements immediately upstream of RNA editing sites is required for C-to-U processing. (3) Splicing. Sequence elements in group II introns of highly convoluted unspliced precursor RNAs are entry sites for PPRs. This interaction is essential for splicing. (4) RNA stability. PPR proteins have a role in RNA cleavage and stability. Binding of PPR proteins could recruit endonucleases and thus lead to RNA cleavage. Conversely, binding could prevent endonucleolytic cleavage by blocking access of RNases, thus stabilizing the RNA. Possibly, binding occurs in the vicinity of secondary structure elements, such as hairpins, that are known targets for endonucleases. Ultimately, PPR proteins guarantee production of a correctly processed, mature RNA that, in the case of mRNAs, is then subject to translation.
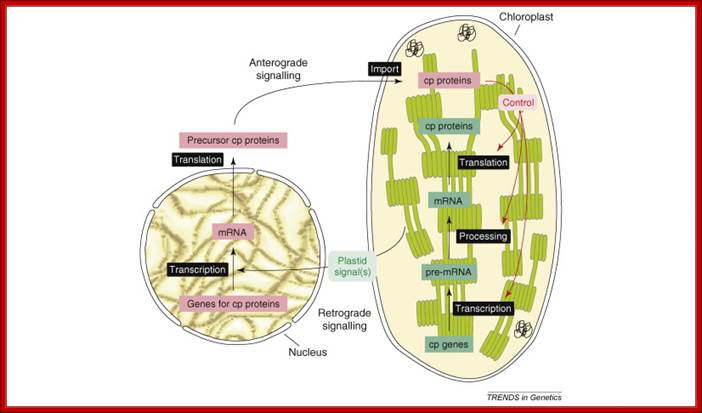
Concept of anterograde and retrograde signaling: The anterograde (nucleus-to-organelle) control of plastid properties including OGE occurs at several levels. Modulation of the level of transcripts of nuclear genes for chloroplast (cp) proteins controls the abundance of many cp proteins. The post-translational import of proteins into chloroplasts is facilitated by the Tic–Toc complex. Isoforms of this translocon exist with different substrate specificity, possibly enabling the accumulation of tissue-specific plastid proteomes. OGE, including transcription, transcript editing, maturation and processing, and the translation of plastid-encoded proteins, is in large parts mediated by nuclear-encoded factors. Post-translational events, such as the assembly of the multi-protein complexes of the thylakoid membrane, also require nuclear-encoded assembly factors. Organelle development, for instance organelle division, is tightly controlled by nuclear-encoded proteins. Current data support the conclusion that the regulation of OGE occurs mainly at the post-transcriptional level. Retrograde signaling describes organelle-to-nucleus signaling, in this case, chloroplast to nucleus. Plastid signals are thought to regulate the transcription of nuclear genes for cp proteins in response to the metabolic and developmental state of the organelle.

This figure covers four major chloroplast posttranscriptional processes:
RNA processing, editing, splicing, and turnover: RNA processing includes the generation of transcript 5′ and 3′ termini, as well as the cleavage of polycistronic transcripts. Editing converts specific C residues to U and often changes the amino acid that is specified by the edited codon. Chloroplasts feature introns of groups I and II, which undergo protein-facilitated cis- or trans-splicing in vivo. Each of these RNA-based processes involves proteins of the pentatricopeptide motif-containing family, which does not occur in prokaryotes. Plant-specific RNA-binding proteins may underpin the adaptation of the chloroplast to the eukaryotic context.
RNA Decay:
The most thoroughly studied pathway for RNA decay in chloroplasts involves the 3′→5′ exonuclease polynucleotide phosphorylase,
As in bacteria, chloroplast polynucleotide phosphorylase activity is stimulated by 3′ polyadenylation of its RNA substrate, and it is blocked by stable 3′ RNA structures. Recent results show that chloroplasts also have a protein-based mechanism for stabilizing 3′ termini that has no apparent analog in bacteria: a bound PPR protein can block 3′ exonucleases in vivo and in vitro.
Polyadenylation accelerates degradation of chloroplast mRNA.
J Kudla, R Hayes, and W Gruissem. Jost Seibler⁎, Frieder Schwenk⁎,
PCR analysis of the psbA and psaA-psaB-rpsl4 operons revealed other polyadenylated endonucleolytic cleavage products, indicating that poly(A) addition appears to be an integral modification during chloroplast mRNA degradation. Polyadenylation promotes efficient degradation of the cleaved petD RNAs by a 3'-5' exoribonucleases. Furthermore, polyadenylation also plays an important roles.
A, RNA metabolism in the chloroplast: Schematic representation of a monocistronic transcription unit in the chloroplast. UTRs are marked with a thin black line; sequences that code for amino acids with a thick black line; and introns with a thick white line. The inverted repeats characterizing the 3′-UTR are symbolized by two arrowheads. A tRNA gene usually presenting 3′ to the inverted repeats is shown as a dashed line. B, The precursor RNA transcribed from this transcription unit undergoes 5′- and 3′-end processing to generate the 5′ and 3′ ends of the mRNA, respectively. C, The 3′ end is located several nucleotides 3′ of the stem-loop structure formed by the inverted-repeats sequence. The introns are removed by splicing, and the tRNA gene is processed by RNaseP. The mRNA is then translated and later degraded.
Unique Mitochondria in Plasmodium falciform:
Plasmodium species during the intraerythocytic phase of their life cycle are known to possess a single mitochondrion. The mitochondrion among plasmodia spp. is heterogeneous with respect to size, shape, behavior upon subcellular fractionation, extent of internal structures.
Apicoplast and Mitochondrial connection:
The apicoplast and the mitochondrion in P. falciparum are always seen to lie in close contact with each other. Hence, transport of metabolites across these organelles was hypothesized62.
The mitochondrial genome in the human malaria parasite Plasmodium falciparum is most unusual. Over half the genome is composed of the genes for three classic mitochondrial proteins: cytochrome oxidase subunits I and III and apocytochrome b. The remainder encodes numerous small RNAs, ranging in size from 23 to 190 nt.
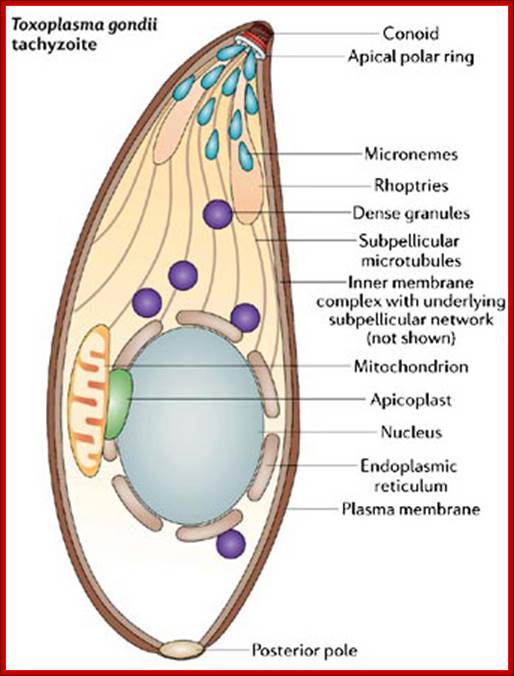
In the 1970s, scientists discovered a novel organelle in apicomplexan parasites, which was named the apicoplast. Like mitochondria and chloroplasts, apicoplasts contain their own DNA. When scientists analyzed apicoplast DNA, they were surprised to learn that apicoplasts shared sequence similarities with plastids (organelles found in the cells of photosynthetic organisms like algae and plants). Could the apicoplast be a vestigial chloroplast? How could apicomplexans and plants be related? What follows is the amazing story of how apicoplasts were discovered and how they have become a promising target for drug development.
The Apicoplast: An Organelle with a Green Past; Regulation of apicomplexan actin-based motility; Laura Vargas Parada,http://www.nature.com/
The identification of 14 additional small mitochondrial transcripts from P. falciparum and the assignment of 27 small RNAs (12 SSU RNAs totaling 804 nt, 15 LSU RNAs totaling 1233 nt) to specific regions of rRNA are supported by multiple lines of evidence. The regions now represented are highly similar to those of the small but contiguous mitochondrial rRNAs of Caenorhabditis elegans. The P. falciparum rRNA fragments cluster on the interfaces of the two ribosomal subunits in the three-dimensional structure of the ribosome.
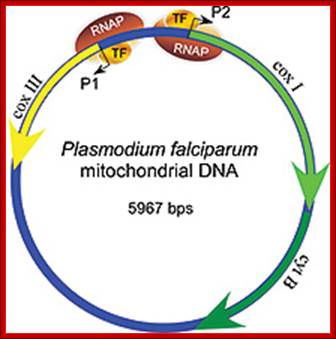
Malarial parasite contains only one mitochondria contains smallest possible genome. It encodes just for three proteins for electron transport. They are transcribed by mtRNAP with at least one TF involving two promoters P1 and P2, as shown in the figure.
Apicoplast
Plasmodium falciparum, and most other members of the phylum Apicomplexa, contain an organelle termed an apicoplast. The apicoplast is an essential plastid, homologous to a chloroplast, although the apicoplast itself lacks any photosynthetic function. Evolutionarily it is thought to have been derived through secondary endosymbiosis. As humans do not harbor apicoplasts, this organelle and its constituents are seen as a possible target for antimalarial drugs.
The malaria parasite carries a plastid called the apicoplast that has been the subject of intense study in the last 15 years. Having originated from red-algal plastids, the apicoplast has lost its ability to photosynthesize, but carries out other essential functions such as type-II fatty acid synthesis, biosynthesis of hame and isoprenoids synthesis.
It contains a 35-kb genome, which encodes for 30 proteins. The genome of this organelle has now been sequenced for several species. It appears to be conserved and to encode ~30 genes in all species examined.
The plastid genome replicates at the late trophozoite stage of the parasite intraerythocytic cycle. It proceeds predominantly via a D-loop/bi-directional ori mechanism with replication ori localized within the inverted repeat region. The process of replication involves a nuclear-encoded DnaJ homolog that binds to the ori site. The apicoplast exists in close proximity to the mitochondrion of Plasmodium and attains an intricate branched structure in the trophozoite stage. It is maternally inherited. It has 4 membranes-secondary symbionts, first involving a cyanobacterium- like prokaryote; the second involving an endosymbiotic alga and a eukaryotic host.
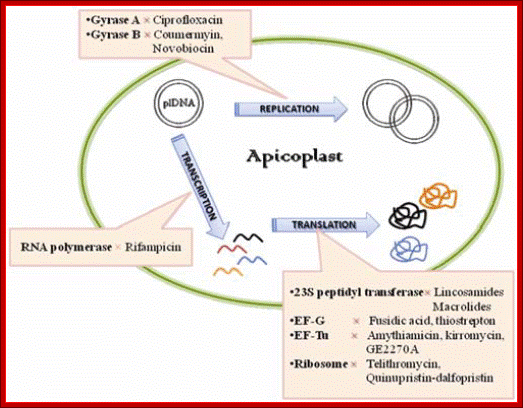
Unique Mitochondria in Plasmodium falciform:
Plasmodium species during the intraerythocytic phase of their life cycle are known to possess a single mitochondrion. The mitochondrion among plasmodia spp. is heterogeneous with respect to size, shape, behavior upon subcellular fractionation, extent of internal structures.
Apicoplast and Mitochondrial connection:
The apicoplast and the mitochondrion in P. falciparum are always seen to lie in close contact with each other. Hence, transport of metabolites across these organelles was hypothesized62.
The mitochondrial genome in the human malaria parasite Plasmodium falciparum is most unusual. Over half the genome is composed of the genes for three classic mitochondrial proteins: cytochrome oxidase subunits I and III and apocytochrome b. The remainder encodes numerous small RNAs, ranging in size from 23 to 190 nt.
The identification of 14 additional small mitochondrial transcripts from P. falciparum and the assignment of 27 small RNAs (12 SSU RNAs totaling 804 nt, 15 LSU RNAs totaling 1233 nt) to specific regions of rRNA are supported by multiple lines of evidence. The regions now represented are highly similar to those of the small but contiguous mitochondrial rRNAs of Caenorhabditis elegans. The P. falciparum rRNA fragments cluster on the interfaces of the two ribosomal subunits in the three-dimensional structure of the ribosome.

Malarial parasite contains only one mitochondria contains smallest possible genome. It encodes just for three proteins for electron transport. They are transcribed by mtRNAP with at least one TF involving two promoters P1 and P2, as shown in the figure.
Apicoplast
Plasmodium falciparum, and most other members of the phylum Apicomplexa, contain an organelle termed an apicoplast. The apicoplast is an essential plastid, homologous to a chloroplast, although the apicoplast itself lacks any photosynthetic function. Evolutionarily it is thought to have been derived through secondary endosymbiosis. As humans do not harbor apicoplasts, this organelle and its constituents are seen as a possible target for antimalarial drugs.
The malaria parasite carries a plastid called the apicoplast that has been the subject of intense study in the last 15 years. Having originated from red-algal plastids, the apicoplast has lost its ability to photosynthesize, but carries out other essential functions such as type-II fatty acid synthesis, biosynthesis of heme and isoprenoids synthesis.
It contains a 35-kb genome, which encodes for 30 proteins. The genome of this organelle has now been sequenced for several species. It appears to be conserved and to encode ~30 genes in all species examined.
The plastid genome replicates at the late trophozoite stage of the parasite intraerythocytic cycle. It proceeds predominantly via a D-loop/bi-directional ori mechanism with replication ori localized within the inverted repeat region. The process of replication involves a nuclear-encoded DnaJ homolog that binds to the ori site. The apicoplast exists in close proximity to the mitochondrion of Plasmodium and attains an intricate branched structure in the trophozoite stage. It is maternally inherited. It has 4 membranes-secondary symbionts, first involving a cyanobacterium- like prokaryote; the second involving an endosymbiotic alga and a eukaryotic host.
Plastid Division:
Chloroplast division by the division complex: A, Dividing chloroplasts in the moss P. patens (top panel) and localization of the FtsZ protein at the chloroplast division site in Arabidopsis (bottom panel). The green fluorescence shows the localization of FtsZ, and red shows the auto fluorescence of chlorophyll. Bars = 10 μm. B, Diagram showing the chloroplast division process (top panel) and localization of components of the division complex (bottom panel).
During division, protein-based ring structures form inside and outside the chloroplast’s double membrane. These rings tighten and pinch the organelle into two. The structure of the inner ring is based on FtsZ protein and the outer on a member of the dynamin protein family (FtsZ protein, which is of cyanobacterial origin.
Sequence of events in chloroplast division (from top left to bottom right). An FtsZ ring forms inside the chloroplast double membrane at the division site. A plastid division (PD) ring forms outside the chloroplast double membrane at the division site. During division, dynamin patches are recruited to the PD ring. A continuous dynamin ring forms at a late stage of division. The FtsZ ring disassembles just before completion of division. When division is complete, the remnant of the PD ring and the dynamin ring disassembles outside of the chloroplast.
The role of these dynamin proteins turns out to be critical to how chloroplasts lost their independence and how they are regulated by the cell—hence, to the development and control of photosynthesis itself.
Mitochondria in protists and chloroplasts in photosynthetic eukaryotes have evolved organelle-targeted forms of FtsZ, the prokaryotic ancestor of tubulin. In fungi, animals and plants, mitochondria no longer utilize FtsZ for division; two related to the filament-forming dynamins are involved in mitodivision.
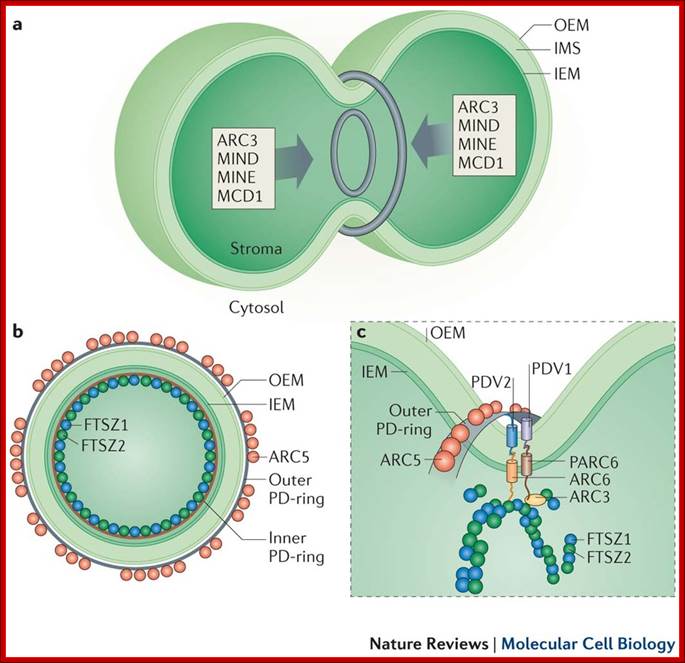
a | Plastids typically divide by binary fission. This process is mediated by concentric rings of a division machinery on both sides of the envelope (at the stromal and cytosolic surfaces of organelle). Correct positioning of the division machinery at the constriction zone is mediated by the MIN (MINICELL) system (grey arrows), which acts to control Z-ring formation. b | Multiple rings surround the organelle at the division site to enable constriction. On the stromal side, the Z-ring (comprising functionally distinct FTSZ1 (FILAMENTOUS TEMPERATURE SENSITIVE Z1) and FTSZ2 homologues) and the inner PD-ring (of uncertain composition) are present. On the cytosolic side, an outer PD-ring that is composed of polyglucan filaments and a discontinuous ring of dynamin-related ARC5 (ACCUMULATION AND REPLICATION OF CHLOROPLASTS 5) operate. c | Positional information from the stromal Z-ring is conveyed to the cytosolic components through ARC6 and PARC6 in the inner membrane and PDV1 (PLASTIC DIVISION 1) and PDV2 in the outer membrane through specific interactions in the intermembrane space (IMS). Correctly positioned PDV proteins enable the recruitment of ARC5 at the constriction zone. Z-ring formation is dynamically controlled by the action of PARC6 and ARC3. IEM, inner envelope membrane; MCD1, MULTIPLE CHLOROPLAST DIVISION SITE 1; OEM, outer envelope membrane. The images are adapted, with permission, from Ref. 107 © (2013) Elsevier and Ref. 156 © (2008) Elsevier.
Biogenesis and homeostasis of chloroplasts and other plastids;Paul Jarvis;& Enrique López-Juez; http://www.nature.com/

Biogenesis and homeostasis of chloroplasts and other plastids; Chloroplasts are the organelles that define plants, and they are responsible for photosynthesis as well as numerous other functions. They are the ancestral members of a family of organelles known as plastids. Plastids are remarkably dynamic, existing in strikingly different forms that interconvert in response to developmental or environmental cues. The genetic system of this organelle and its coordination with the nucleocytosolic system, the import and routing of nucleus-encoded proteins, as well as organellar division all contribute to the biogenesis and homeostasis of plastids. They are controlled by the ubiquitin–proteasome system, which is part of a network of regulatory mechanisms that integrate plastid development into broader programmes of cellular and organismal development. Comparison between bacterial and plastid divisions: Schematics of a dividing bacterium and a chloroplast are shown. Each complex division mechanism is simplified. Bacteria use an FtsZ ring and a peptidoglycan system, whereas plastids adopt inner and outer PD rings: a dynamin ring and a FtsZ ring. Peptidoglycan, which may or may not be present, is not shown in the plastids.
· Paul Jarvis & Enrique López-Juez- http://www.nature.com
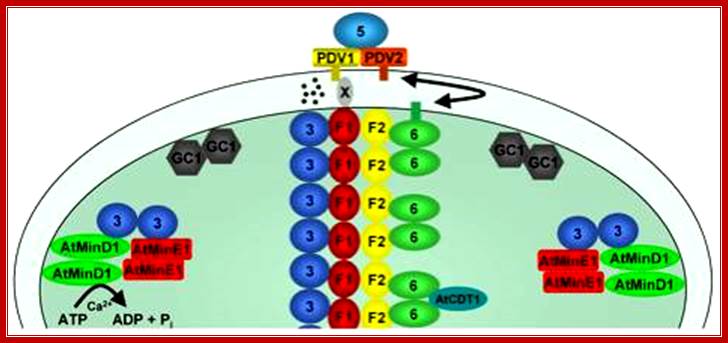
Stromal and cytosolic plastid division machineries. As the first step of stromal division machinery assembly AtFtsZ1-1 (F1) and AtFtsZ2-1 (F2) form a Z-ring at the centre of chloroplasts. ARC6 (6) and ARC3 (3) are recruited to the Z-ring through specific interactions with AtFtsZ2-1 and AtFtsZ1-1, respectively. AtCDT1 also interacts with ARC6, although the localisation of AtCDT1 is not known. The placement of the Z-ring requires the combined action of AtMinE1, AtMinD1 and possibly ARC3, which form a complex and can localise to plastid poles. GC1 localizes to the stromal side of the inner envelope membrane and forms dimers. PDV1 and PDV2 localise to ring-like structures on the cytosolic surface of the outer envelope membrane and recruit ARC5 to the division site to constitute the cytosolic division machinery. The inner and outer PD rings are not shown. Coordination and signalling between the two division machineries may occur through a direct interaction between known proteins (e.g. between ARC6 and PDV1), may require as yet unidentified intermembrane space proteins (x) or through the action of signalling components (black spots); Jodi Maple and Simon Geir Møller (2007 http://www.ux.uis.no/
Transplastomic plants:
Recently, chloroplasts have caught attention by developers of genetically modified plants. In most flowering plants, chloroplasts are not inherited from the male parent,[14][15] although in plants such as pines, chloroplasts are inherited from males.[16] Where chloroplasts are inherited only from the female, transgenes in these plastids cannot be disseminated by pollen. This makes plastid transformation a valuable tool for the creation and cultivation of genetically modified plants that are biologically contained, thus posing significantly lower environmental risks. This biological containment strategy is therefore suitable for establishing the coexistence of conventional and organic agriculture. While the reliability of this mechanism has not yet been studied for all relevant crop species, recent results in tobacco plants are promising, showing a failed containment rate of transplastomic plants at 3 in 1,000,000
Construction of marker-free transplastomic plants; Kerry A Lutz, Pal Maliga.
Because of its prokaryotic-type gene expression machinery, maternal inheritance and the opportunity to express proteins at a high level, the plastid genome (plastome or ptDNA) is an increasingly popular target for engineering. The ptDNA is present as up to 10 000 copies per cell, making selection for marker genes essential to obtain plants with uniformly transformed ptDNA. However, the marker gene is no longer desirable when homo-plastomic plants are obtained. Marker-free transplastomic plants can now be obtained with four recently developed protocols: homology-based excision via directly repeated sequences, e transcripts excision by phage site-specific recombinases, transient co-integration of the marker gene, and the co-transformation-segregation approach. Marker excision technology will benefit applications in agriculture and in molecular farming.

The transformation of the plastid genome, until recently restricted to tobacco, is now being extended to a rapidly growing list of crops. This perspective provides an overview of emerging trends of technology development in the field with a focus on vector design and marker excision systems. The new tools will facilitate engineering of the photosynthetic machinery and enable novel agricultural and industrial applications.
Homologous recombination yields transformed plastid genomes in higher plants. Recombination between the left (L) and right (R) targeting sequence in the vector and cognate sequences in the plastid genome (ptDNA) lead to the incorporation of the marker gene (mg) and the gene of interest (goi) in the ptDNA to yield the TP1 transplastome (TP1-ptDNA). Triangles flanking the marker gene symbolize recognition sequences of site-specific recombinases to be used for marker gene excision. http://pubs.rsc.org
![Figure 7. Marker gene excision from the plastid genome by Cre or Int site-specific recombinases [137]. A site-specific recombinase gene ( cre / int ) introduced into the nucleus by transformation, pollination or transient Agroinfiltration, encodes a plastid-targeted recombinase that excises selectable marker gene (SMG) from TP1-ptDNA after import into plastids. Excision of the marker gene by phage recombinases via the target sites (black triangles) yields marker-free TP2-ptDNA carrying only the gene of interest (GOI) and one recombinase recognition sequence [141, 142, 143].](Gene_Expression_II14-Mitochondrial_Gene_Expression_files/image086.jpg)
Marker gene excision by the phage Cre or Int site-specific recombinases: Site-specific recombinases (Cre/Int) expressed from nuclear genes (cre/int) excise marker genes (mg) from TP1-ptDNA after import into plastids. Excision of the marker gene by phage recombinases via the target sites (triangles) yields marker-free TP2-ptDNA carrying only the gene of interest (goi) and one recombinant copy of the recombinase recognition sequence. https://www.researchgate.net
Eukaryotic organelles:
Eukaryotes are one of the structurally complex cell type, and by definition are in part organized by smaller interior compartments, that are themselves enclosed by lipid membranes that resemble the outermost cell membrane. The larger organelles, such as the nucleus and vacuoles, are easily visible with the light microscope. They were among the first biological discoveries made after the invention of the microscope.
Not all eukaryotic cells have each of the organelles listed below. Exceptional organisms have cells which do not include some organelles that might otherwise be considered universal to eukaryotes (such as mitochondria). There are also occasional exceptions to the number of membranes surrounding organelles, listed in the tables below (e.g., some that are listed as double-membrane are sometimes found with single or triple membranes). In addition, the number of individual organelles of each type found in a given cell varies depending upon the function of that cell.
|
Major eukaryotic organelles |
||||
|
Organelle |
Main function |
Structure |
Organisms |
Notes |
|
photosynthesis, traps energy from sunlight |
double-membrane compartment |
plants, protists (rare kleptoplastic organisms) |
has some genes; theorized to be engulfed by the ancestral eukaryotic cell (endosymbiosis) |
|
|
translation and folding of new proteins (rough endoplasmic reticulum), expression of lipids (smooth endoplasmic reticulum) |
single-membrane compartment |
all eukaryotes |
rough endoplasmic reticulum is covered with ribosomes, has folds that are flat sacs; smooth endoplasmic reticulum has folds that are tubular |
|
|
sorting, packaging, processing and modification of proteins |
single-membrane compartment |
all eukaryotes |
cis-face (convex) nearest to rough endoplasmic reticulum; trans-face (concave) farthest from rough endoplasmic reticulum |
|
|
energy production from the oxidation of glucose substances and the release of adenosine triphosphate |
double-membrane compartment |
most eukaryotes |
has some DNA; theorized to be engulfed by an ancestral eukaryotic cell (endosymbiosis) |
|
|
storage,transportation, helps maintain homeostasis |
single-membrane compartment |
eukaryotes |
||
|
DNA maintenance, controls all activities of the cell, RNA transcription |
double-membrane compartment |
most eukaryotes |
contains bulk of genome |
|
Mitochondria and chloroplasts, which have double-membranes and their own DNA, are believed to have originated from incompletely consumed or invading prokaryotic organisms, which were adopted as a part of the invaded cell. This idea is supported in the Endosymbiotic theory.
|
Minor eukaryotic organelles and cell components |
|||
|
Organelle/Macromolecule |
Main function |
Structure |
Organisms |
|
helps spermatoza fuse with ovum |
single-membrane compartment |
many animals |
|
|
vesicle which sequesters cytoplasmic material and organelles for degradation |
double-membrane compartment |
all eukaryotic cells |
|
|
anchor for cytoskeleton, helps in cell division by forming spindle fibers |
Microtubule protein |
animals |
|
|
movement in or of external medium; "critical developmental signaling pathway". |
Microtubule protein |
animals, protists, few plants |
|
|
detects light, allowing phototaxis to take place |
green algae and other unicellular photosynthetic organisms such as euglenids |
||
|
carries out glycolysis |
single-membrane compartment |
Some protozoa, such as Trypanosomes. |
|
|
conversion of fat into sugars |
single-membrane compartment |
plants |
|
|
energy & hydrogen production |
double-membrane compartment |
a few unicellular eukaryotes |
|
|
breakdown of large molecules (e.g., proteins + polysaccharides) |
single-membrane compartment |
most eukaryotes |
|
|
pigment storage |
single-membrane compartment |
animals |
|
|
probably plays a role in Fe-S cluster assembly |
double-membrane compartment |
a few unicellular eukaryotes which lack mitochondria |
|
|
myocyte contraction |
bundled filaments |
animals |
|
|
ribosome production |
protein-DNA-RNA |
most eukaryotes |
|
|
not characterized |
not characterized |
fungi |
|
|
breakdown of metabolic hydrogen peroxide |
single-membrane compartment |
all eukaryotes |
|
|
degradation of unneeded or damaged proteins by proteolysis |
very large protein complex |
All eukaryotes, all archaea, some bacteria |
|
|
translation of RNA into proteins |
RNA-protein |
eukaryotes, prokaryotes |
|
|
material transport |
single-membrane compartment |
all eukaryotes |
|
Prokaryotic organelles:
Prokaryotes are not as structurally complex as eukaryotes, and were once thought not to have any internal structures enclosed by lipid membranes. In the past, they were often viewed as having little internal organization; but, slowly, details are emerging about prokaryotic internal structures. An early false turn was the idea developed in the 1970s that bacteria might contain membrane folds termed mesosomes, but these were later shown to be artifacts produced by the chemicals used to prepare the cells for electron microscopy.[32]
However, more recent research has revealed that at least some prokaryotes have micro compartments such as carboxysomes. These subcellular compartments are 100 - 200 nm in diameter and are enclosed by a shell of proteins.[1] Even more striking is the description of membrane-bound magnetosomes in bacteria,[33][34] as well as the nucleus-like structures of the Planctomycetes that are surrounded by lipid membranes.[35]
|
Prokaryotic organelles and cell components |
|||
|
Organelle/Macromolecule |
Main function |
Structure |
Organisms |
|
protein-shell compartment |
some bacteria |
||
|
light harvesting complex |
|||
|
movement in external medium |
protein filament |
some prokaryotes and eukaryotes |
|
|
magnetic orientation |
inorganic crystal, lipid membrane |
||
|
DNA maintenance, transcription to RNA |
DNA-protein |
prokaryotes |
|
|
DNA exchange |
circular DNA |
some bacteria |
|
|
translation of RNA into proteins |
RNA-protein |
eukaryotes, prokaryotes |
|
|
photosystem proteins and pigments. |
mostly cyanobacteria. |
||