GAL and Histidine Genes Expression:
Yeast, an unicellular, eukaryotic system offers an excellent model for understanding various aspects of molecular mechanisms of gene expression and development.
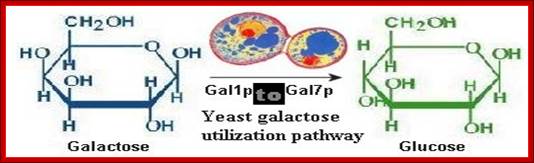
When yeast is grown on Galactose as an alternative carbohydrate source, instead of glucose, a set of genes are expressed to utilize Galactose and earlier glucose utilizing genes slow down or switched off.


www.m-direct.com.my;Disulfide bonds foemmation;http://www.crc.dk/Yeast cell; Subcellular localization of S. cerevisiae ABC transporters. http://mmbr.asm.org
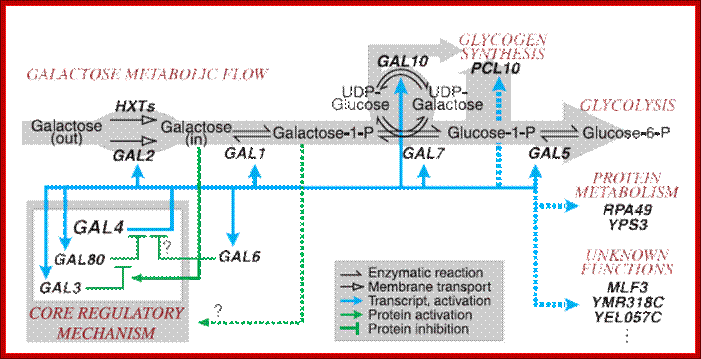
A cluster of GAL genes are required for the growth of yeast on galactose and they comprise structural (GAL1, GAL10, GAL2 and GAL7) and regulatory genes (GAL4, GAL80 and GAL3). A genome-wide analysis of promoters bound by Gal4 and induced by galactose found that in addition to the previously identified GAL genes, MTH1, PCL10 and FUR4 also belong to the GAL regulon (Ren et al, 2000). These genes are not required for galactose metabolism, but rather for global adaptation to growth on galactose. For example, the induction of MTH1, a repressor of glucose transporter gene expression results in the inhibition of glucose transport. Approximately 300 potential Gal4-binding sites have been identified in the yeast genome
As shown in the Figure above, galactose utilization consists of a biochemical pathway that converts galactose into glucose-6-phosphate and a regulatory mechanism that controls whether the pathway is on or off. This process has been reviewed extensively (2, 3) and involves at least three types of proteins. A transporter gene (GAL2) encodes a permease that transports galactose into the cell; several other hexose transporters (HXTs) may also have this ability (4). A group of enzymatic genes encodes the proteins required for conversion of intracellular galactose, including galactokinase (GAL1), uridylyltransferase (GAL7), epimerase (GAL10), and phosphoglucomutase (GAL5/PGM2). The regulatory genes GAL3, GAL4, and GAL80 exert tight transcriptional control over the transporter, the enzymes, and to a certain extent, each other. GAL4p is a DNA-binding factor that can strongly activate transcription, but in the absence of galactose, GAL80p binds GAL4p and inhibits its activity. When galactose is present in the cell, it causes GAL3p to associate with GAL80p. This association causes GAL80p to release its repression of GAL4p, so that the transporter and enzymes are expressed at a high level. Carsten Friis; http://www.cbs.dtu.dk/
![Canonical yeast galactose utilization pathway (adapted from[26]).Gray arrows indicate cellular processes likely impacted by GAL gene deletions. Abbreviations: intracellular galactose (galIN), galactose-1-phosphate (gal1P), glucose-1-phosphate (glu1P), uridine diphosphate (UDP), UDP-glucose (UDPglu), UDP-galactose (UDPgal).](Gene_Expression_II10-GAL_and_Histidine_Gene_Expression_files/image005.jpg)
Authors below; We investigated a thoroughly characterized network, the yeast galactose utilization pathway. This network, which is depicted in Figure 2, involves three regulatory genes (GAL3, GAL4 and GAL80) and five structural genes (GAL1, GAL2, GAL6, GAL7 and GAL10) that enable yeast to detect and metabolize galactose. Quantitative traits were measured for a library of single and double GAL gene deletion strains in a genetic background expressing yeast enhanced green fluorescent protein (yEGFP) from the promoter of the GAL10 gene (see Materials and Methods). Growth rates during early log-phase (fitness) and reporter expression were determined in rich media containing raffinose under inducing (+galactose; “ON”) and non-inducing (-galactose; “OFF”) conditions (see Materials and Methods). Phenix H et al; http://openi.nlm.nih.gov/
Galactose is transported by the gene product Gal 2p. Once inside, Galactose is phosphorylated by a kinase to Galactose1-phosphate; and the gene is called Gal 1. Galactose1p is then added on to UDP by Gal-1-phosphate Uridyl transferse; the gene is Gal 7p. And then UDP Galactose is converted to UDP-Glucose1p by UDP-glucose4-epimerase; the gene is Gal 10. Then Glucose1p is converted to Glucose 6p, that is then drawn into glycolytic pathway; Phenix H, Morin K, Batenchuk C, et al; http://openi.nlm.nih.gov/
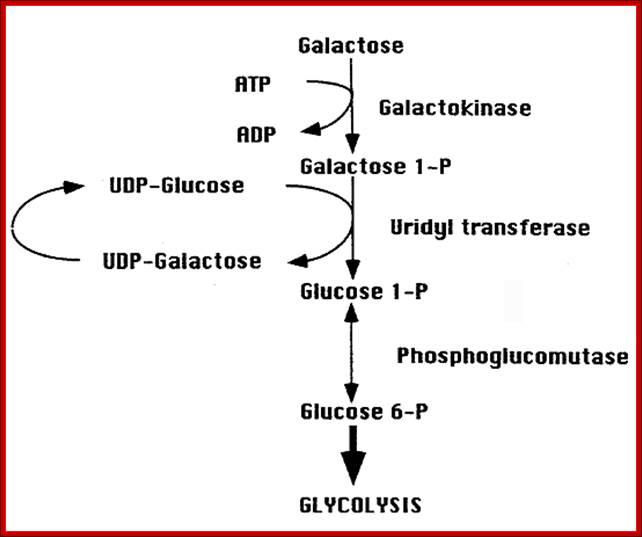
First step- transport of Galactose across the cell membrane; second step- Galactose to Gal1-P, next Gal1-P to Uridyltransferase that produces UDP-Galactose, then to Glucose1-p;Glucose1P is transformed into Glucose6-P, it is then subjected Glycolysis and then to Kreb’s cycle; Namrata Chhabra; http://www.namrata.co/
GAL genes and their functions:
Galactose transport ( Gal 2),
Galactose to galactose1-p (GAL 1 or Gal K)
Galactose1-p to UDP-Galactose= (Gal 10 or GAL T)
UDP Galactose is converted to UDP Glucose by epimerase (GAL7 or GAL E)
UDP Glucose is converted to Glucose1P by GAL5-p.


www.slideplayer.com; http://slideplayer.com
· The above mentioned genes are located very near to each other on chromosome 2. Regulator genes such as GAL4, GAL80 and GAL3 are located on chromosome XVI, XIII and IV respectively. The regulatory gene Gal4 product regulates more than 22 or more galactose related genes, hence it is called Regulon.
|
GAL-7 P |
GAL-10 P |
P GAL-1 |
|
<---------------------I------I |
<----------------------I-----I |
I-----I----------------------> |
<---GAL7---------------<I--P--I<-------GAL10------------<I--P-I-I--P- >----GAL1----à
GAL1 promoter elements:
----UAS1--UAS2--UAS3--UAS4----URS------TATA---InR---DPE
UAS = Upstream activator sequences,
URS = Upstream regulator / repressor sequences,
Mig = Multicopy inhibitor of Galactose 1 gene,
TUP = Thymidine uptake repressor Protein,
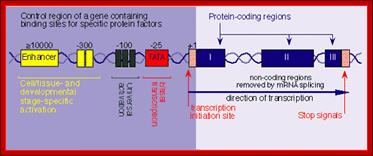
Each of these genes has their own promoter elements in the upstream of TATA, at –350; they are UAS (upstream activator sequence) and URS (at -80) (upstream regulatory sequences). Each of them is nearly 30 bp long.
The distance between TATA and the start (InR) can be 30 to 40 bp long. Each of the upstream elements consists of two inverted repeats of CGG -N11- CCG (consensus sequence). But the GAL 4 has CYCCRSNWWWWW
[UAS]--------------- [URS] -------------TATA…AAA…InR+>
-350 -80
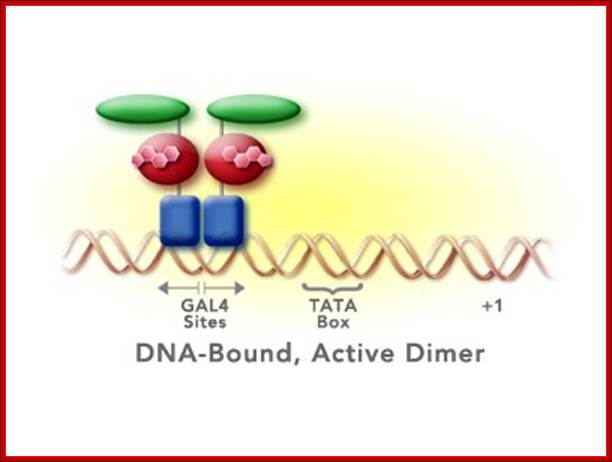
The GAL-4 protein is constitutively synthesized, binds to UAS sequences as a dimer but remains in inactive state; because a protein called GAL-80 is bound to GAL-4 and makes it inactive. In this state it acts as a repressor.
To Bind or Not to Bind; undergratuate course at Davidson College;http://www.bio.davidson.edu/Courses/Molbio
Biomoleculaes Gallery;http://www.abren.net/image/dna-protein.
· In this inactive form, the Gal-4 as dimer is bound to their respective promoter regions of each of the genes and all are rendered inactive.
The GAL-80 has two sites, one for binding to GAL-4 and the other to the inducer Gal3. The Gal3 binds to when Galactose is made available and then it binds to GAL80. Binding of the Gal3-Galactose to GAL-80, induces the conformational change in the protein such that the GAL-80 dissociates from the GAL4. Release of GAL80 from GAL4 induces the GAL4 to become active and now it acts as an activator. GAL4 is positive regulator of genes such as GAL1, GAL2, GAL7, GL10 and MEL1.
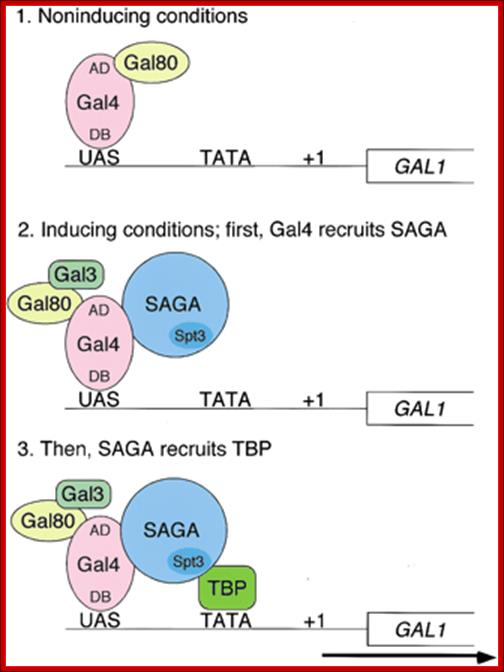
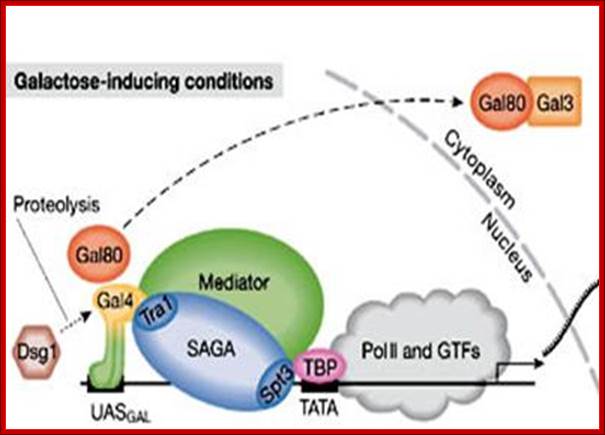
A model for SAGA functioning as a coactivator for Gal4 by facilitating TATA-binding-protein (TBP) binding to the TATA box of the GAL1 gene. (1) Under noninducing conditions, Gal4 is bound to the UASG via its DNA-binding domain (DB), and the Gal4 activation domain (AD) is blocked by Gal80. (2) After the addition of galactose, the Gal3 inducer is activated and alters the Gal80–Gal4 complex such that the Gal4 activation domain is no longer blocked by Gal80. This change allows the Gal4 activation domain to recruit SAGA to UASG. The presence of the Gal3 protein at the promoter is suggested by the formation of a Gal3–Gal4–Gal80 complex in vitro and in vivo (Chasman and Kornberg 1990; Leuther and Johnston 1992; Parthun and Jaehning 1992; Platt and Reece 1998; Sil et al. 1999). However, other data have suggested that Gal3 is cytoplasmically localized (Peng and Hopper 2000). (3) Once recruited to the promoter, SAGA, mainly via a Spt3-–TBP interaction, recruits TBP to the TATA box to allow transcription initiation; Genes and Development. Erica Larschan and Fred Winston,
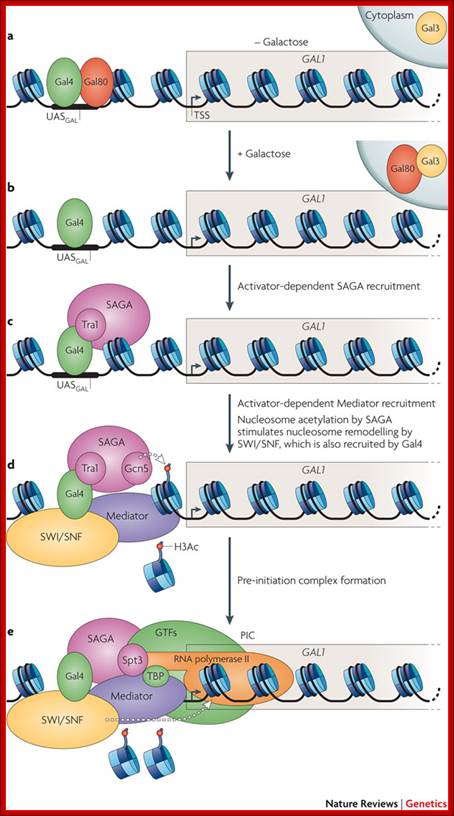
In yeast, in the absence of galactose, the acidic activator Gal4 is bound by its repressor Gal80 (a). Addition of galactose to the growth medium causes an inducer protein, Gal3, to bind and sequester Gal80 in the cytoplasm, releasing it from Gal4 (b). Gal4 binds target UASGAL (upstream activating sequence) sites in the promoters of Gal genes such as GAL1 and sequentially recruits co-activators, such as the acetyltransferase SAGA (c) and Mediator (d). Gal4 also recruits ATP-dependent nucleosome-remodelling complexes such as SWI/SNF that remove nucleosomes at the promoter and are stimulated by SAGA-catalysed histone acetylation. Together, SAGA and Mediator recruit RNA polymerase II and the general transcription factors (GTFs), leading to formation of the pre-initiation complex (PIC) (e). Nucleosome removal, catalysed by SWI/SNF, aids in the kinetics of Mediator and GTF recruitment, thereby facilitating rapid PIC formation and initiation of transcription at Gal genes. H3Ac, histone H3 acetylation; TBP, TATA-binding protein; TSS, transcription start site; Nature Reviews, Genetics; Vikki M. Weake & Jerry L. Workman
 T
T
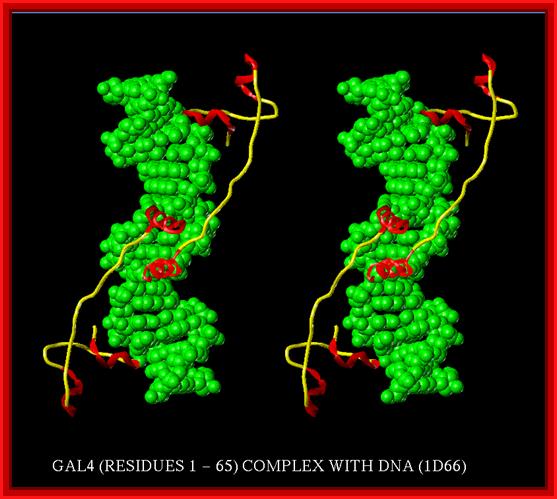
The dimerized GAL4 DNA binding domain of an active dimer will bind to one of the 17 bp GAL4 binding sites in the promoter of the transgene. Other molecules of the active dimer may bind to adjacent GAL4 binding sites
· The protein GAL-4 is 881 amino acids long (~92kDa). It is Zn2-Cys4 homodimer protein. It has several domains, one for DNA binding, second for dimrization, third for GAL-80 binding and two more domains for activating. The protein binds to UAS sequences as dimer and remains bound to their respective promoters for any number of generations.

Modular structure of GAL4 protein; http://www.personal.psu.edu/
1: DNA binding domain 1-74 a.a (zinc finger structure) and nuclear localization.
2:Dimerization domain.
3: Transcription activation (acidic region) and Gal80 interaction domain at the end of protein.
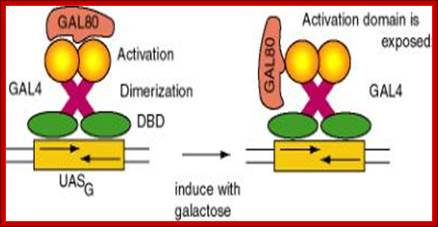
Negative Regulation and induction of GAL4; http://www.personal.psu.edu/

https://www.youtube.com
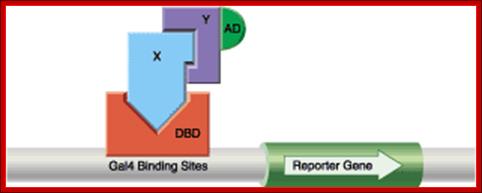
The two-hybrid method: One protein (X) is fused to the Gal4 DNA-binding domain (DBD) and a second protein (Y) is fused to the Gal4 activation domain (AD). If X and Y interact, the activation domain is brought into the proximity of a reporter gene regulated by Gal4 binding sites, resulting in expression of the reporter protein.
From N-terminal end 1 to 74 a.a it has a DNA binding domain with Helix-loop Helix structure, having 6 cys and 2 Zinc cores. Another from 74 to 147, the sequences generates a secondary structure that facilitates dimerization; the dimerized region has Leucine zipper motifs. The linker is found between 40 and 49 aa. In between 851 to 881aa, the protein has a site for the binding of GAL-80 protein; it covers GAL4 activator domain. Two activation domains are found, one at 148 to 196 and the other at 768 to 881.
The activated GAL4 is 200
to 400 times efficient in activating gene expression, this feature is typical
an enhancer, but this element is called upstream activator element.
· The GAL-4 free from GAL-80 acts like a positive regulator where GAL binding to it Activator-enhancer sequence represses gene expression.
· Yeast’s UAS elements can be equated to that of Enhancers.
Gal4 product also regulates 25 other genes related to Galactose metabolism such as Gal3, Gal80, Gal4, Gal2, and few other glyco related genes.
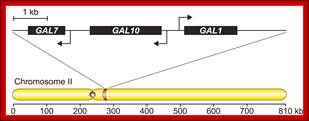
The GAL1-GAL10-GAL7 gene cluster. The GAL cluster consists of three genes (GAL1, GAL10, and GAL7), chromosomes7(XI); encoding enzymes that catalyze four sequential steps in galactose assimilation, that are clustered in a 7 kb region of ChromosomeII;
Dept. of Biological Sciences, Lehigh University; http://glanglab.com/research.html

GAL1 promoter and upstream elements

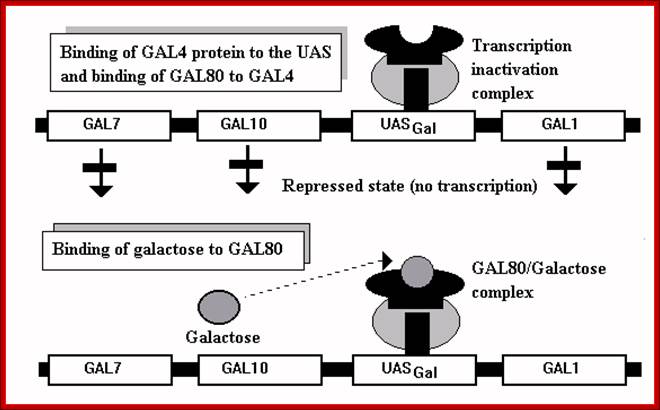
GAL binding to UAS, and GAL80 binding to GAL4; thus block transcription of GAL1 and other related genes. But the binding of Galactose to Gal3 and this complex binding to GAL4 induces protein conformation, this leads to the release of GAL80-GAL3-Galactose. The GAL4 free from the repressor undergoes conformational change and it acidic active site interacts with GTC and activates transcription.
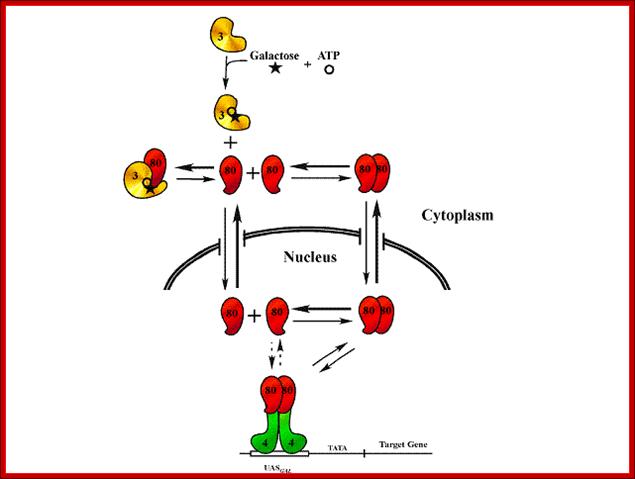
GAL80 shuttle model:GAL-gene switch are the Gal4, Gal80 and Gal3 proteins that constitute a molecular mechanism for galactose-responsive transcriptional activation (galactoseinduction) of the GAL genes. The key observations that our Gal80 Shuttle model takes into consideration are as follows: 1)Gal3 resides exclusively in the cytoplasm of the cell; 2) Gal80 shuttles rapidly between the cytoplasmic and nuclear compartments of the cell; 3) Gal80 exists as a monomer,dimer and perhaps a tetramer; 4) a Gal80 dimer is the unit of Gal80 that binds to and inhibits a dimer of Gal4 that is located at the UASgal sites within the nucleus; 5) a Gal80monomer is the unit of Gal80 that binds to Gal3; 6) Gal80 binds to Gal3 exclusively in the cytoplasm and in the presence of galactose; and 7) in response to galactose the binding of Gal80 to UASgal-associated Gal4 decreases. Jim Hopper;Dept Molecular Genetics; https://molgen.osu.edu/
[UAS]----------- [URS] ---------------TATA…AAA…InR+>
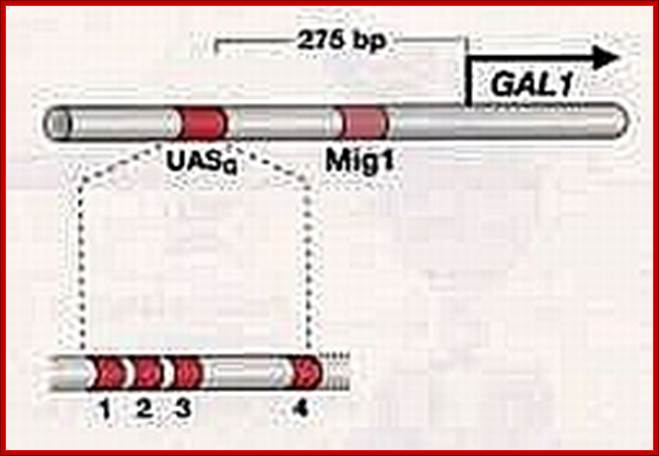
Upstream UAS is ~ 118bp long, consists of 17bp long GAL4 binding sites; www.en.ppt-online.org
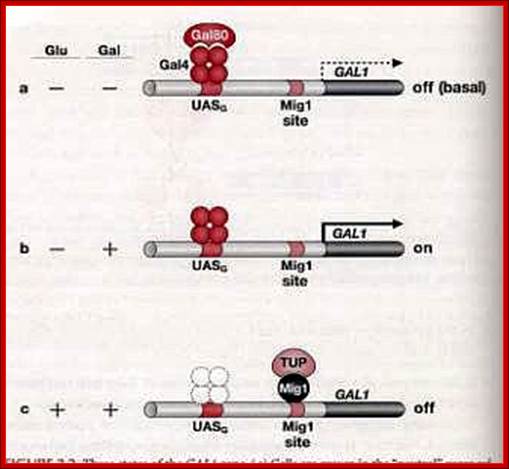
In the presence of glucose the GAL1 gene is blocked by Mig1 which is bound by TUP. www.en.ppt-online.org
In the presence of glucose the GAL genes are rendered inactive not just by the binding of GAL4-GAL80 but also the binding of Mig1 (Multi-copy inhibitor of galactose gene). The Mig1 protein is a cytosolic protein, but in response to Glucose it moves into the nucleus in hypo-phosphorylated state and binds to URS region, a AT—GC--AT rich site found in between UAS and the TATA box. The Mig1 then recruits Tup protein (Thymidine utilizing protein), which recruits deacetylases and repress the gene. Deacetylation leads methylation and recruitment of repressor proteins. In the absence of glucose the Mig1 gets phosphorylated through cAMP mode and moves out into the cytoplasm. The Mig1 is very effective on GAL4 gene itself in the presence of glucose.
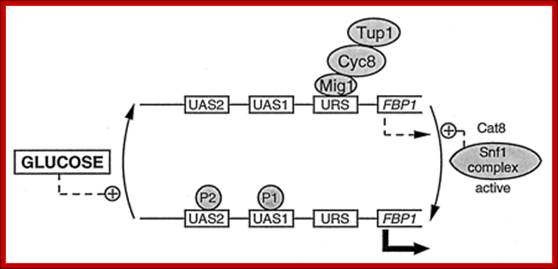
The URS called upstream regulator sequence consists of a GC rich region flanked by AT sequences.
When the cell is provided with glucose, it induces degradation of cAMP. In such situation Mig1protein in dephosphorylated state (multicopy inhibitor of Galactose-a gene repressor) that is normally found in the cytoplasm moves into the nucleus and binds to URS region. The Mig1 has a DNA binding Zif (zinc finger) motifs. This recruits Tup1p proteins (Thymine uptake repressor proteins) on to Mig.
But when Galactose is provided and glucose is withdrawn, cAMP is produced, which activate a kinase that phosphorylates Mig1p and the phosphorylated Mig1p moves out of the nucleus and remains in cytoplasm. So it removes repression at URS region.
The activated GAL4 bound in the upstream interacts with BTA, through DNA looping activates transcriptional complex. http://embor.embopress.org
Two hybrid systems is used to show two domains of GAL4 are involved in the regulation of GAL1 and other genes. One domain is DNA binding domain and the other domain is Activator domain.
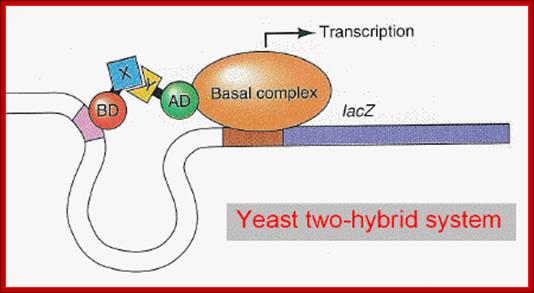
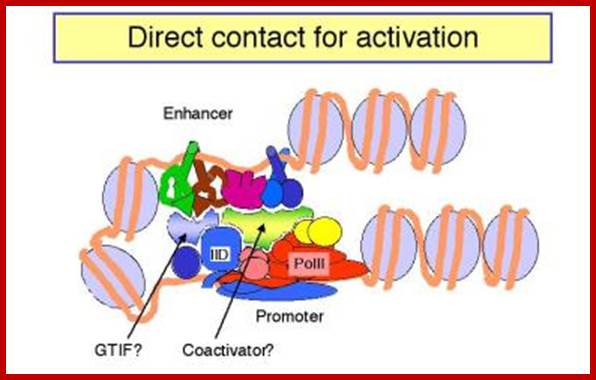
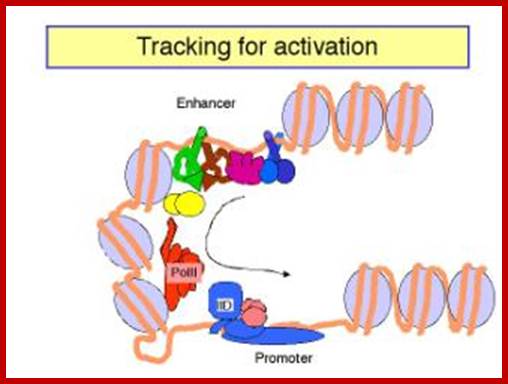
Activated GAL4 through its DNA binding domain (BD) binds to DNA sequences and through its Activator domain (AD) interacts with BTA- here the marker gene is Lac Z.; http://www.bx.psu.edu; http://www.bx.psu.edu
The nuclear protein GAL4 is a positive regulator of gene expression for the galactose-induced genes such as GAL1, GAL2, GAL7, GAL10, and MEL1. These genes encode enzymes which convert galactose to glucose. GAL4 recognizes a 17 base-pair long sequence in the upstream activating sequence (UASuas-g) of these genes, (5'-cggrnnrcynyncnccg-3'). GAL4 binds to the DNA as a homodimer.
GAL4 from the organism Saccharomyces cerevisiae contains such a domain, it binds two Cadmium (Cd) ions rather than Zinc ions. It is 43 amino acid residues long. It contains two right-handed alpha-helices shown here in red. The six conserved cysteine residues are shown in blue, and the two cadmium ions are shown in silver.
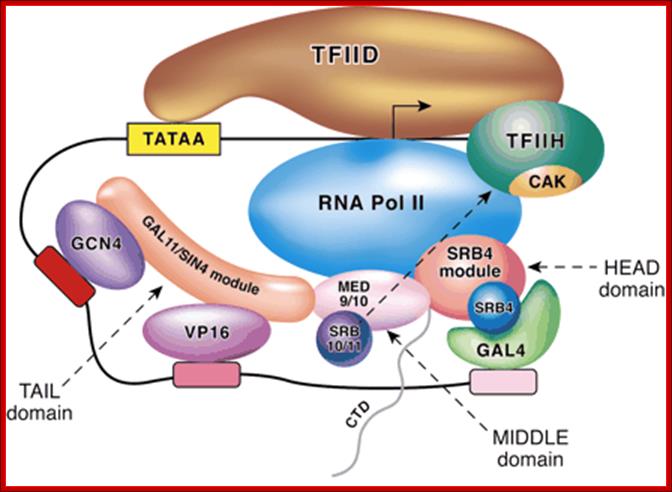
When GAL4 is activated it recruits the required components such as SRBs, VP16 and GC4 to the promoter region and activates RNAP II.
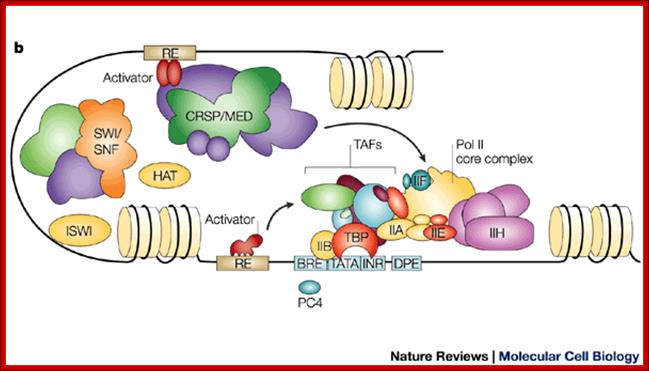
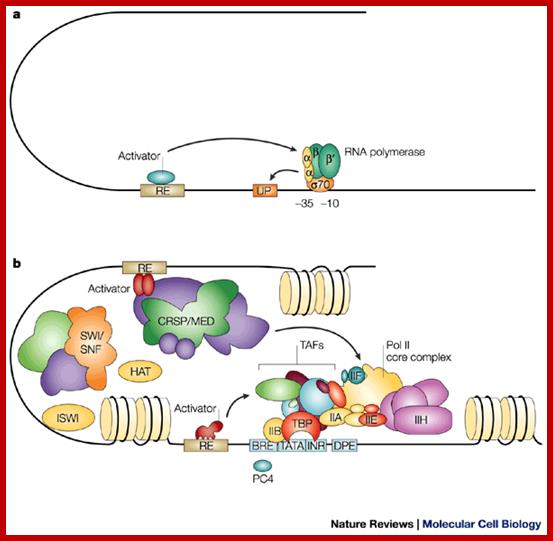
This is a general view, not of GAL4, showing various components of promoter elements and the assembly of all chromosomal remodeling components, transcriptional activation components join to activate transcription. www.flipper.diff.org
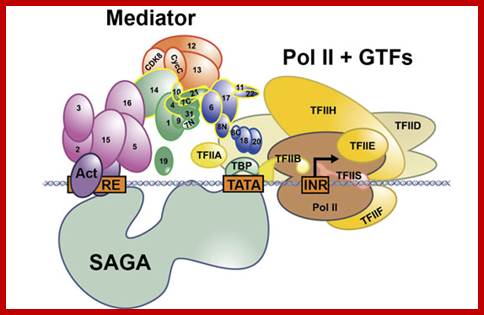
Schematic view of the Pol II preinitiation complex at the core promoter. Mediator bridges between activators (Act) bound to regulatory DNA elements (RE) and the basal transcription machinery (Pol II and the GTF). Pol II subsequently starts synthesizing RNA at the so-called Initiator DNA element (INR). Mediator modules are colored in blue (head), green (middle), magenta (tail) and orange (kinase). Individual Mediator subunits are shown.; https://www.elitenetzwerk.bayern.de
The activated Gal 4 is phosphorylated at serine-837. The Gal4 has an activator domain rich in acidic amino acids, in this case it is glutamic acid rich. They interact with various transcriptional components and recruit co activators such as GCN4 (5) (acetylase protein), mediator complex and transcriptional apparatus and initiate transcription. The binding of GAL-80 renders the GAL-4 inactive; in this state it cannot interact with PIC.
· This form of regulation is typical of Negative type and positive type; repressing by a repressor is negative and activating is positive type.
In this process, as both Gal 1 and Gal 10 genes on either side of regulator sequences, the transcriptional activators that bind bring both the genes’ together for using common transcriptional components by looping the Gal 1 and Gal 10, for they can initiate transcription from their respective Start point.
The activation is 200 to 400 times efficient, this feature is typical of an enhancer, but this element is called upstream activator element. The GAL-4 free from GAL-80 acts like a positive regulator. So yeast’s UAS elements can be equated to that of Enhancers.
In the case of Gal-1 gene the regulatory region is very elaborate. It consist of UAS at -350 and another regulatory region called URS at -215, then nearer to start point it has promoter elements such as TATA, InR and DPE elements.
Apart form the Gal4 as an activator, URS region also plays an important role in repressing GAL1 gene in spite of the activated Gal4. All the said genes that are regulated by Gal 4 have more or less the same consensus sequences at TATA box and UAS boxes.
GAL4 Gene Perse:
The GAL 4 gene lacks TATA box, so it is TATA- less but contains an UAS region( GATCCGAAGA), an UES (TATATAAA(T)4 (upstream essential sequence) and URS regions.
Under Galactose inducing conditions Gal80-Gal3-galctose are removed from the Gal4 repressor. On Galactose induction F-box protein called Grr1 is degraded in ubiquitinated mode otherwise it binds to Gal4 that inhibits its activation mode.
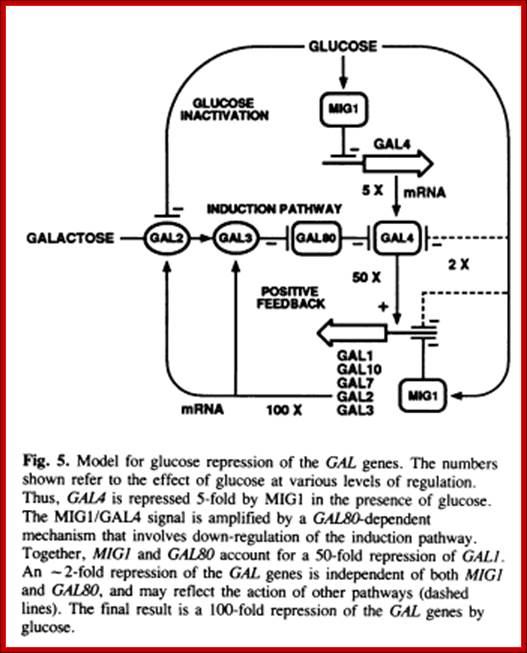
http://ppt-online.org
F-box contains a 50 a.a sequence that is targeted for protein degradation by ubiquitination and proteosome mediated process. Once the Gal4 becomes active it recruits SAGA complex and mediator complex and all the components required for transcriptional initiation i.e. BTA (basal Transcriptional Apparatus). The SAGA complex has histone acetylation proteins that are responsible for nucleosomal release and making the DNA free for the assembly of the activator proteins.
The Gal4 gene product acts on more than 25-300 galactoses related genes; it is a Regulon.
Histidine Gene Expression:
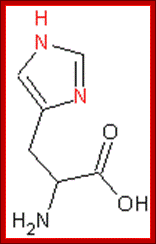
· Histidine gene 3 is expressed constitutively when amino acid level is normal.
· The promoter region has a two InR and start regions. One at normal position and the other at +12 positions. In the upstream region it has 17 bp A/T rich sequences. Such A/T rich blocks are found in many constitutively expressed genes.
There is correlation between the length of the A/T rich region and the level of gene expression. Increase in the length increases the level of gene expression. Such regions with A/T sequences fail to form nucleosomal structures.
· In His 3 genes there are two such A/T blocks, and they work in either orientation, but work as activator regions.
----IA: TI-----------IA: TI---------------TATA--------+1>-----+12>--->
[TGACTC] [TGACTC]
When conditions are normal, and amino acid levels are adequate, His-3 is expressed using +1 InR start point. Under starvation condition, a regulatory protein called GCN-4, General Control of Nitrogen genes, plays an important role. Under normal conditions, this protein, which is constitutively expressed, is in inactive state. But under amino acid starvation the GCN4 gets activated
· The activated protein now binds to TGACTC sequence and activates the expression of the gene using the InR start from +12 regions. The protein has leucine zipper motifs, consisting of 281 aa long chain and they form dimers.
· It has DNA binding domain at N-terminal region (1 to 60 aa) and it has an activator domain at 107 to 125 a.a positions. The activator domain contact RNA pol and activates the enzyme.