Organization of Few More Operons:
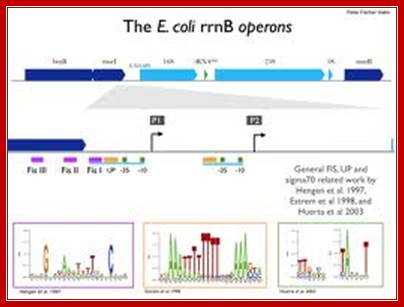
RrnB P1 promoter:
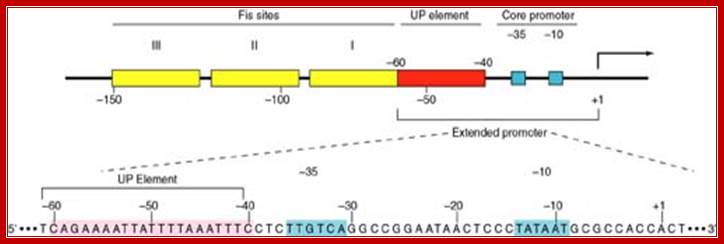
Core promoter
I-150--I-----I---110---II--60----I--50----I--35---I--10----I+à--
The core promoter region has an upstream element at -60 to –50. The upstream segment from –150 to –110 acts as an enhancer element. First, RNAP complex binds to –150 to –110 and not to the core. The core promoter element runs up to –60, so it is termed as extended promoter.
http://www.cbs.dtu.dk/
TCAGAAATTATTTTAAATTTCCTCTTGCAGGGCCGAATAA--CTCCC –10 TATAATGCGCCA CC A+ CT
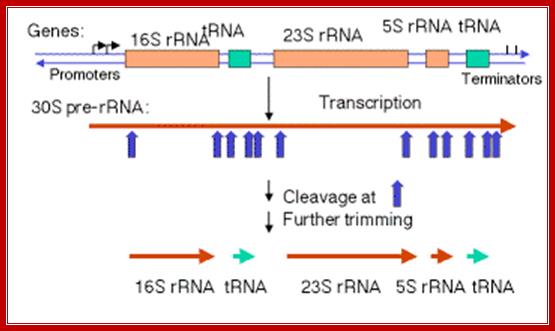
Rrn Operon:
+ > +>
---------I--------I----I------I----------------------------------------------
P1 P2 16s tRNA1 23s-5s tRNA2
P1 is located 300 bp upstream of the start of 16s RNA. P1 and P2 are 110bp apart. First 15 bp regions differ in different rRNA genes. The P1 and P2 are used differentially according to situations of the cell.
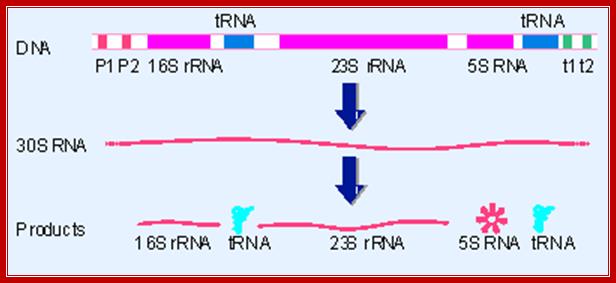
The rrn operons contain genes for both rRNA and tRNA. The exact lengths of the transcripts depend on which promoters (P) and terminators (t) are used. Each RNA product must be released from the transcript by cuts on either side. http://flylib.com/
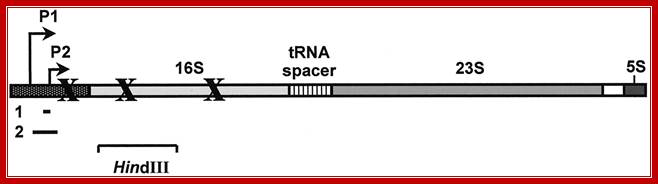

A schematic drawing of a typical rrn operon of E. coli. This schematic drawing is made to illustrate the relative position of the different rRNAs within all seven rrn operons of E. coli but does not take into account the specific characteristics of each one of these operons. 16 S, 23 S, tRNA spacer, and 5 S represent regions encoding the corresponding stable RNAs. Xs indicate the relative positions of the transcriptional blockage sites identified in this study. Lines 1 and 2 indicate the position and size of the oligonucleotide and the probes, respectively, used in the primer extension experiments and in the Northern blot analysis respectively. HindIII indicates the position and size of the HindIII fragment from the rrnB operon used in this study. P1 and P2 are the first and second promoters, respectively, of a rrnoperon of E. coli. The drawing is not to scale. www.apresentacao-sequenciamento-dna; https://bio.libretexts.org ; https://bio.libretexts.org
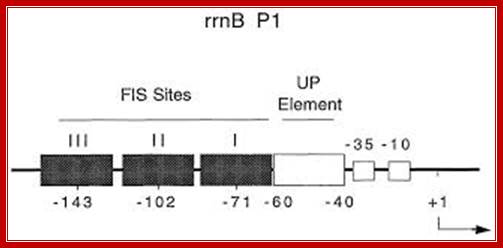
Molecular anatomy of a transcription activation patech:RNAP interacts with E.coli rrrBP1 prmoter; Anton J. Bokal, Wilma Ross, et al; http://emboj.embopress.org/
rrnB promoter elements; Mol Biol. http://ec2-107-21-65-169.compute-1.amazonaws.com
rRNA endonuclease site for processing; http://www.personal.psu.edu/
Aro-H codes for one of the three enzymes that catalysis the initial reactions in a common pathway that leads to the synthesis of an aromatic amino acid.
O +--->
Trp EDBC --------------------------I---------------------------I------
-12 to +10
O +---->
Trp-R--------------------------I---------------------------I-------
-12 to +10
Trp-R controls three unlinked set of genes. It also has autogenous regulation.