Regulation of Gene Expression during Sporulation:
When external conditions become unfavorable, bacteria like B. subtilis, one of the gram-positive bacteria, reluctantly switches to Sporulation mode as ultimate resort for survival. Bacterial endospores were first described by Cohn and Koch in B.subtilis and B. anthracis respectively. Many other systems such as Clostridium, Sporosaracina, B.anthracis, B.thuringienesis, B.cereus, Clostridium titani, C. botulinum, C. perfrigens and few others do generate spores.
Spores thus produced can survive up to 100 to thousands of years or more; there are records? Some have survived for one million years. Nearly 7500 years old spores of Actinomycetes (they are not true spores) have been revived in the lab. They withstand temperature from –80 to + 80, UV and Gamma radiations. They can over come the action of many deadly chemicals, detergents digestive enzymes and survive extreme pH conditions. Some of the present bacteria are becoming superbugs; resistant to many drugs, ex. one of the strains of E.coli in Europe (2011) was found to be resistant to most of the antibiotics; this has caused panic all over the world. WHO claims India has one such superbug; yet to be confirmed. But many of the sporulation bacteria are important in industries and medicine.
Center for Bacterial Cell Biology; http://www.ncl.ac.uk/
Normal cell; http://www.ncl.ac.uk/
Sporulating bacteria with demarcation of fore spore; http://www.ncl.ac.uk/
Dark bodies are spores; stained by malachite green (Schaffer-Fulton)
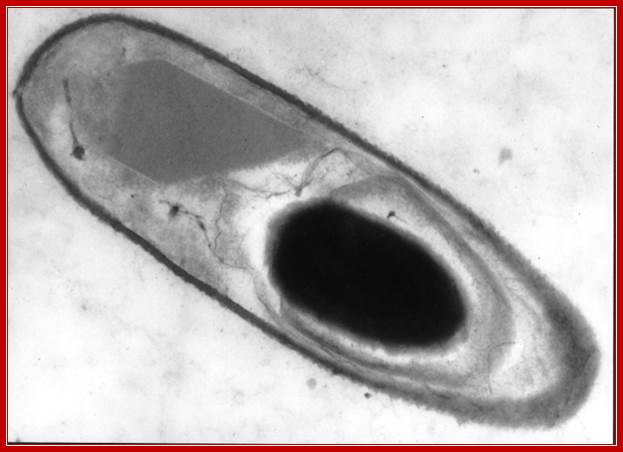
TEM of B.thuringienesis with an endospore; Vincent Sanchis; http://www.komunich.de/
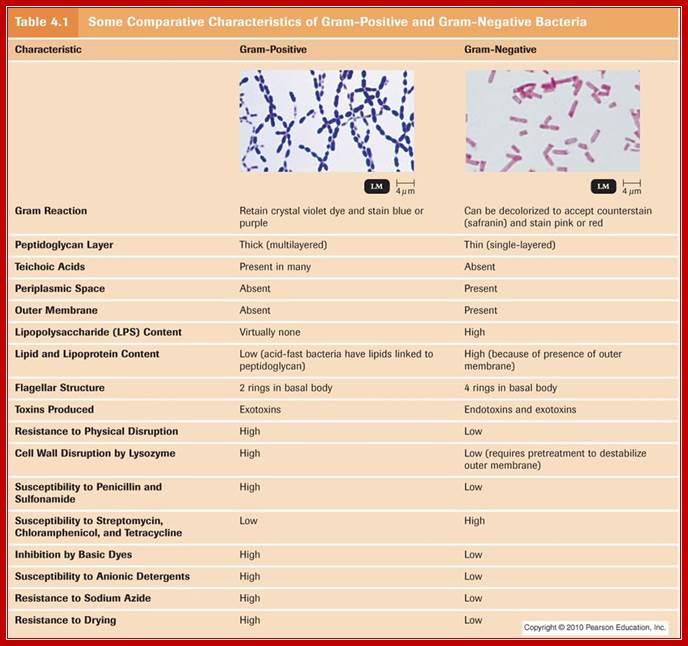
B.subtilis has been studied extensively for its sporulation strategy. It is a soil bacterium, gram positive and non pathogenic. Its genome size is ~4,215,814 (4.2 X 10^6) bp, contain ~4100 protein coding genes. The said number of genes is organized into hundreds of operons; sporulation operons are estimated to be 24 encoded with 121 genes. SpoOA is a regulon controls hundred of genes organized into 24 operons and many monocistronic genes. Nearly forty of them are positively regulated and 81 of them under negative regulation. This estimate was discerned by chromatin Immunoprecipitation (Chip) plus chip-on chip, transcriptional profiling and gel analysis of expressed proteins and DNA micro analysis (Virginie Molle et al). Cell wall structural features of gram positive and gram negative bacteria are given below.

http://drsachattha.blogspot.com/http://www.pangxiang.com/
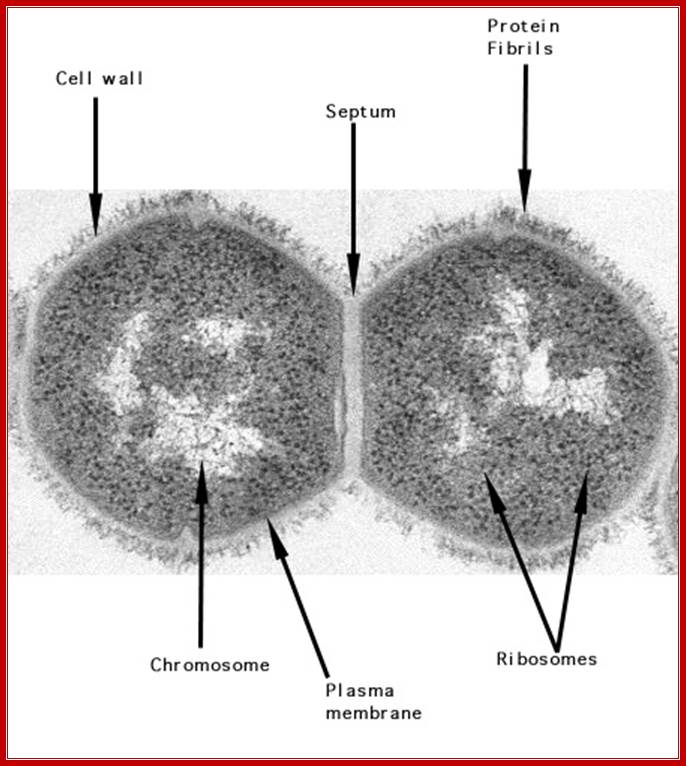
Gram positive cell with peptidoglycan coat and an internal nucleoid DNA; http://textbookofbacteriology.net/

B.subtilis cell surface (gram positive) ; http://cmgm.stanford.edu/

B.subtilis endospore; http://www.normalesup.org/
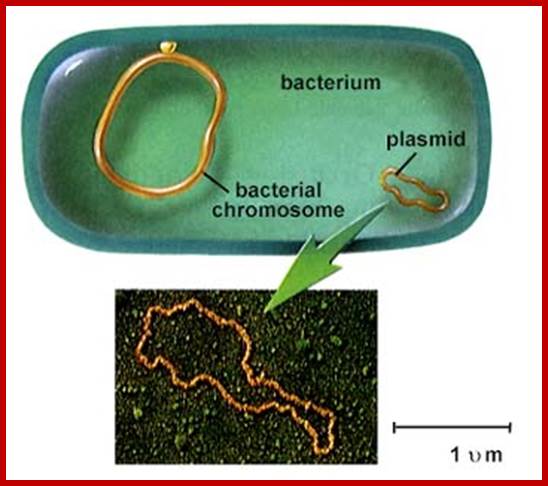
Top fig; http://www.nature.com/Didier Wion and Joseph Casadesus; Bacterial chromosome and a plasmid; Bottom fig. biological Diversity; bacteria and Archaea; http://www.biologie.uni-hamburg.de/
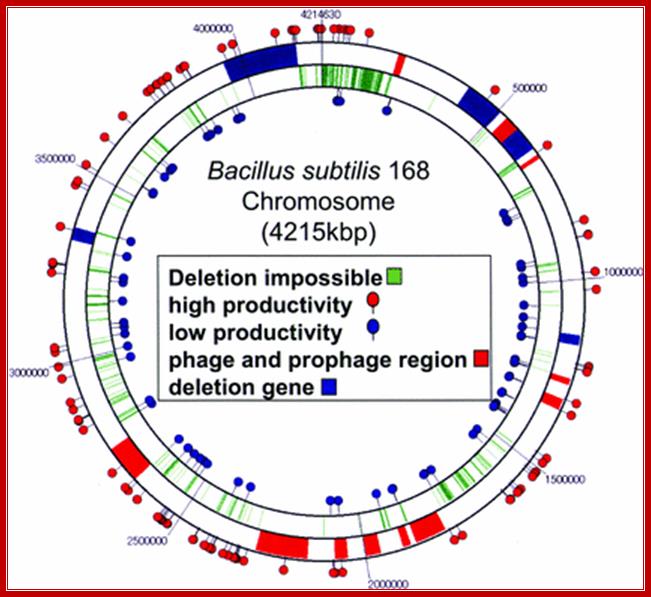
B.subtilis168 - genome map; http://www.babonline.org/
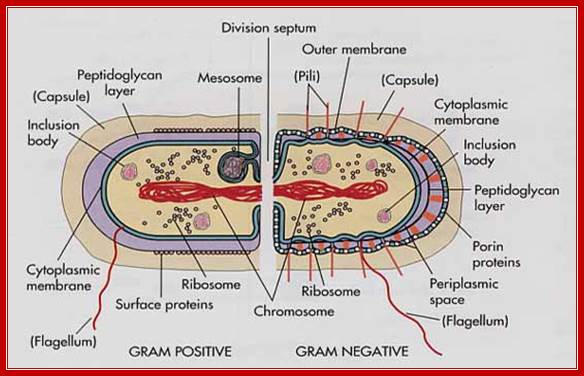
Cell wall features (general) of both gram (+) and (-) cells; http://micro.digitalproteus.com/
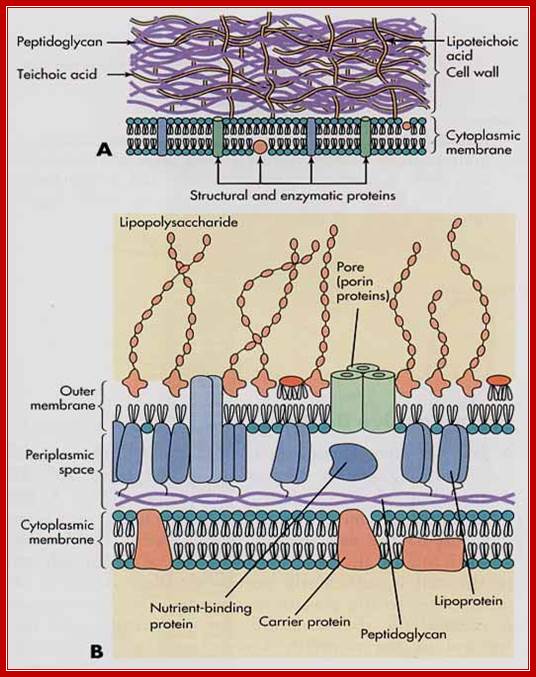
Figure A is gram positive bacterial wall and the figure B is of gram negative; http://micro.digitalproteus.com/
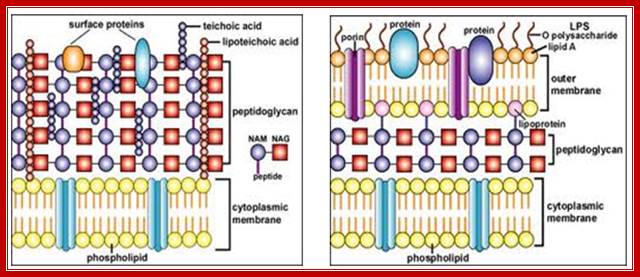
Gram positive and Negative Bacterial Cell Wall. http://nptel.ac.in/
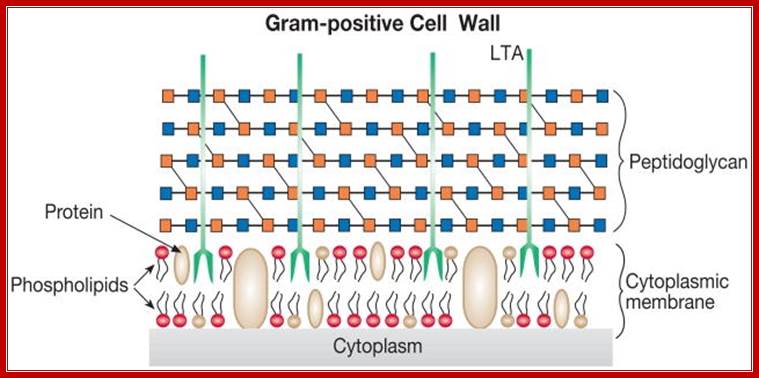
Gram positive wall; http://amit1b.wordpress.com/
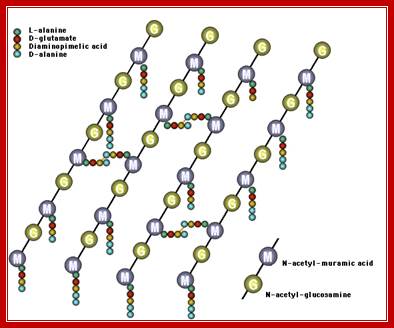
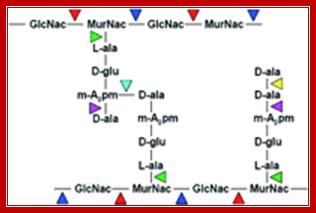
Peptidoglycan linkages; Five a.a chain of one poly G-M strand cross linked to another G-M strand by cross linking between d-alanine (5th and last) with 4th DAP of the other parallel chain; http://pathmicro.med.sc.edu/
Typical bacterial features
|
Structure |
Chemical composition |
Function |
|
Flagella |
Proteins |
Swimming /movement |
|
Pili |
Proteins |
Conjugation |
|
Sex pili |
Proteins |
Female, transfers DNA |
|
Common pili |
Filamentous protein |
Protect from phagocytosis |
|
Capsule-slime |
Polysaccharides with proteins |
Protect against descication, reserve nutrients, |
|
Cell wall |
|
|
|
Gram(+) |
Several layers of peptidoglycans |
Provides rigidity, prevents plasmolysis |
|
Gram (-) |
Two membranes on either side of a peptidoglycans layer, with periplasmic space in between. |
Prevents osmolysis, proteins have various functions |
|
Plasma membrane |
Teichoic acid and Phospholipids and proteins |
Contains receptors embedded and transmembrane receptors, transporters, contain sensors, |
|
Ribosomes |
RNA and proteins |
Protein sysnthesis |
|
Cell inclusions |
Lipids, proteins |
Reserve food? |
|
Chromosomes |
DNA with proteins |
Gnetic material-threads |
|
Plasmids |
DNA circular |
Extra chromosomal DNA |
Some comparison between gram positive and negative bacteria:
|
Property |
Gram(+) |
Gram(-) |
|
Wall thickness |
20-80nm thick |
10nm |
|
Membrane layers |
1 |
2 |
|
Peptidoglycan |
>50% |
10-20% |
|
Teichoic acid |
Present |
absent |
|
Mem-Lipid/lipoprotein |
0-3% |
58% |
|
Protein |
0 |
9% |
|
Lipopolysaccharides |
0 |
13% |
|
Sensitive to penicillin |
yes |
no |
|
Sensitivity to lysozyme |
yes |
no |
Gram (+) - Cytoplasmic inclusions
|
Inclusions
|
Composition |
Function |
|
Glycogen |
Polyglucose |
Reserve food in most bacteria |
|
Polybetahydroxybutyric acid(PHB) |
Poly hydroxybutyrate |
Reserve food |
|
Volutin threads |
Polyphosphates linear or cyclic polymers |
Reserve phosphate-in corynebacteria |
|
Sulphur globules |
Elemental sulfur |
Reserve electron, energy source, purple and green sulfur bacteria |
|
Gas vesicles |
Protein hulls |
Buoyancy, in Cyanobacteria |
|
Parasporal bodies |
proteins |
Toxic to some- in bacillus species |
|
Magnetosomes |
Magnetite-Fe3O4 |
Orienting magnetic lines, some aquatic bacteria |
|
Carboxysomes |
Enzymes for carbon fixation |
CO2 fixation, cyanobacteria |
|
Chlorososomes |
Bacterio-chlorophyll, lipid and proteins |
Light harvesting pigments-bacteria |
|
|
|
|
|
|
|
|
Comparison between B.subtilis vegetative cells and endospores:
|
Property |
Vegetative |
Endospores |
|
Wall coats |
Murein cell wall-polymers |
Thick spore coat, cortex and peptidoglycans |
|
Appearance |
Non-refractile |
Refractive |
|
Calcium-DAP |
Absent |
Present in the core |
|
Water content |
High |
Very low |
|
Enzyme activity |
High |
Absent when dry |
|
Synthetic activity |
yes |
Zero when dried |
|
Heat resistance |
Low, susceptible |
High resistance |
|
Resistance to chemicals |
sensitive |
resistant |
|
Lysozyme |
sensitive |
resistant |
|
Dyes and drugs |
sensitive |
resistant |
|
Macromolecular synthesis |
Present |
Absent |
|
Spore proteins |
absent |
present |
|
Enzyme activity |
high |
Low or nil |
|
pH |
high |
low |
https://www.pinterest.com
|
Similarities between Eubacteria, Archaea and Eukaryotes |
|||
|
|
Eubacteria |
Archaea |
Eukaryotes |
|
Nucleus |
No |
No |
Yes: membrane-bound |
|
Nucleosomes/histones |
No |
Yes |
Yes |
|
Operons/polycistronic mRNAs |
Yes |
Yes |
No |
|
Introns |
No |
No |
Yes |
|
TATA Box binding protein |
No |
Yes |
Yes |
|
Organelles |
No |
No |
Yes: mitochondria, lysosomes, endoplasmic reticulum etc. |
|
Chromosomes |
One Circular |
One Circular |
More than one |
|
RNA polymerase |
One (simple) |
More than one (complex) |
More than one (complex) |
|
Protein initiator amino acid |
N-formyl methionine |
Methionine |
Methionine |
|
Protein synthesis sensitivity to diphtheria toxin |
Insensitive |
Sensitive |
Sensitive |
|
Peptidoglycan |
Yes |
No |
No |
|
Protein synthesis |
of Archaea are more similar to those of eukaryotes than eubacteria |
||
Induction of Sporulation:
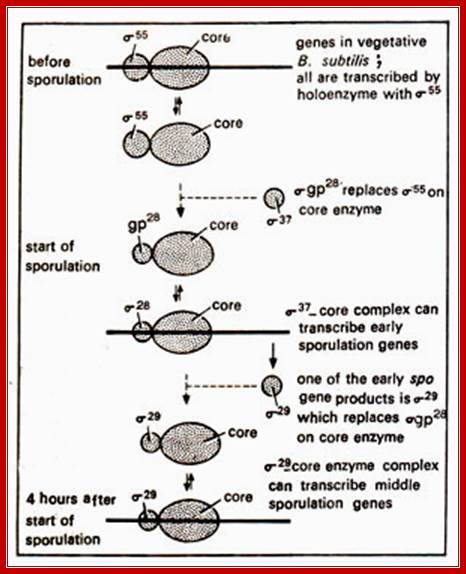
Environmental factors especially, stress such as starvation induces differential gene expression which triggers differential gene expression and unequal partition. Once the small prespore and the mother cells are demarcated, along with a number of genes are differentially expressed in stage specific manner to produce an endospore. In all these events, the switching on and off of several genes takes place; regulated by a cascade of sigma factors. Each of the sigma factors operates in switching on a cluster of genes in temporal fashion, some of them can be considered as Regulons.
Environ factors like high temperature, draught, deficiency of nutrition, oxidative stress and others send signals across the signal transducers found in or on cell membranes; they are the key factors that induce sporulation. It is an altruistic process and it is the last resort for the bacterial survival. With the onset of hostile environmental changes, Bacillus subtilis does not initiates spore formation, but delays and it will be triggered as an ultimate mode of survival. It takes about 7-8 or even 10 hours to complete spore formation from vegetative cell.
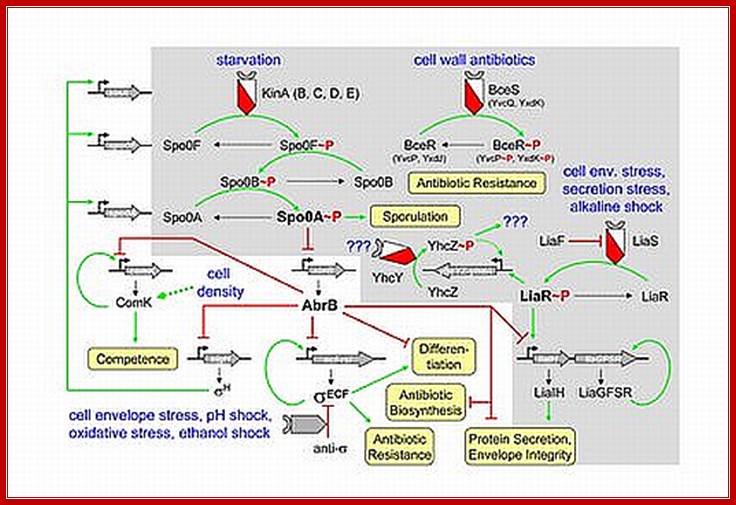
This diagram shows starvation and hostile cell wall antibiotics activate early signal transduction to induce molecular events and induce sporulation. http://syntheticmicrobe.bio.lmu.de/
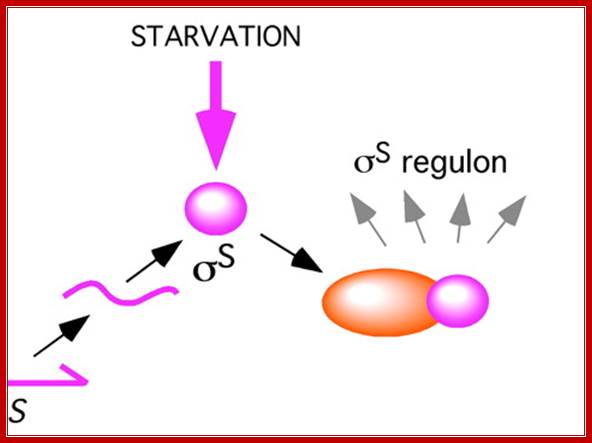
In some systems starvation leads to the activation of Sigma S which acts as a Regulon; means a single factor activates several operons. http://www.pnas.org.
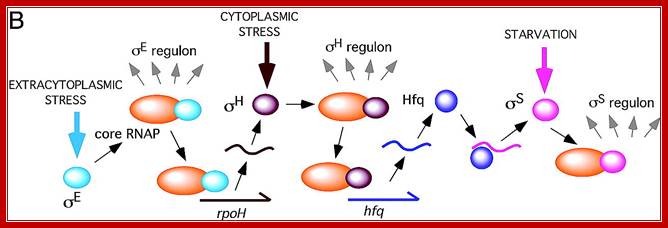
In the case of B.subtilis extra cytoplasmic stress factors transduce cytoplasmic factors which in turn trigger S regulon activity? Sig E( sig E regulon) activates rpo H (regulon) > which in turn activates Hfq > Hfq then activates sig S (sig S acts as regulon); A cascade of regulons such as regulon E >regulon H> regulon>Hfq > > S regulon operate in sporulation. http://www.pnas.org
The diagram below is an example of enteric bacteria shown how starvation regulates a whole set of genes activated S regulon. Extracellular stress activates sigE which activates specific genes using common RNAP; and sigE acts as a regulon in activating hundred of genes. Among them, one sig H acts as cytoplasmic response Regulon. One of the factors produced in response to sig H is Hfq which activates sig S which responds to starvation and acts as a regulon and activates a whole set of genes.
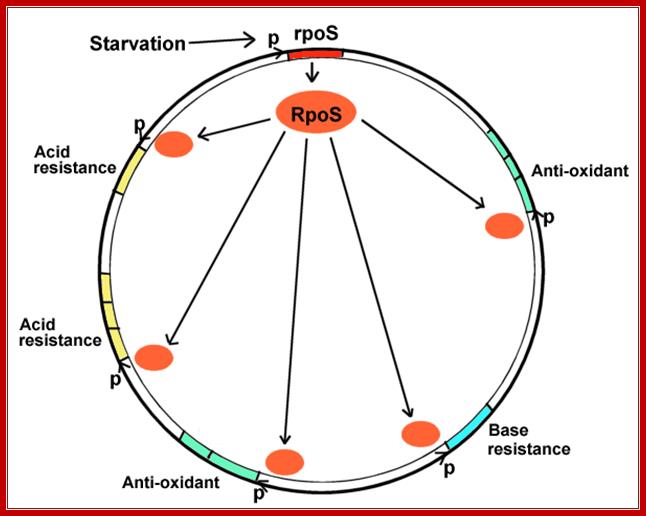
http://www.u.course.cl
The diagram shows stress in the form of starvation induces rpoS regulon of an enteric E.coli bacteria; the diagram is repeated in context. The σS (RpoS) and the σB (SigB) have been identified as general stress responsive alternative sigma factors in Gram-negative and in Gram-positive bacteria, respectively. The σS was identified in both E. coli and in S. Typhimurium as a Group II sigma factor that activates expression of numerous genes required to maintain cell viability as the cell leaves exponential growth conditions and moves into stationary phase. Mol.Biol
Sigma factors are classified into two structurally unrelated families, the σ70 and the σ54 families. The σ70 family includes primary sigma factors (e.g., Bacillus subtilis σA), sig70 = sigA, as well as related alternative sigma factors; σ54 forms a distinct subfamily of sigma factors referred to as σN, sig54 = sigN, in almost all species for which these proteins have been characterized to date. Several examples of alternative sigma factors have been shown to contribute to virulence in at least one organism. For each sigma factor, when applicable, examples are drawn from multiple species.
When bacteria enter digestive tract; how do they adjust to body's defense mechanisms such as acid and antioxidant stress? When bacteria exit the intestine how do they cope with starvation?
- When bacteria use up their carbon sources, they express RpoS, the starvation sigma factor. (Review, what is a sigma factor?).
- RpoS joins RNA polymerase to initiate transcription of different environmental stress related genes; the genes protecting against all the different stresses that the bacteria might encounter before they enter a new human intestine. This phenomenon is known as cross-protection.
- The stress genes can be used for conditions as unrelated as acid or base resistance.
- The stress genes activated may or may not be part of multi-gene operons. They may face in opposite directions, from many different promoters, at all different loci around the genomic map.
Bacteria are the best genetic system to study genetic regulons. However, stress genes discovered in bacteria have been shown to have homologs in eukaryotic systems. For example, heat shock genes have been found in all organisms, including humans. Heat shock genes in humans also show cross-protection; for example, one heat shock protein interacts with the progesterone receptor and the contraceptive drug RU486.
In most of the cases for example environ stress, osmo-regulatory stress such as salt or H2O2 oxidative stress are perceived at cellular surface, which harbors a whole lot of sensory receptors. In general histidine kinase acts as an important sensor which triggers downstream events.
Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp PCC 6803:
Kay Marin * , † , ‡, Iwane Suzuki † , §,Katsushi Yamaguchi †, Kathrin Ribbeck *, Hiroshi Yamamoto †,Yu Kanesaki †, Martin Hagemann *, and Norio Murata † , § , ¶
Perception of salt stress in the cyanobacterium synechocystis sp. strain PCC 6803 is sensed by histidine kinase system. A library of strains with mutations in all 43 histidine kinases was screened by DNA microarray analysis of genome wide gene expression under salt stress.
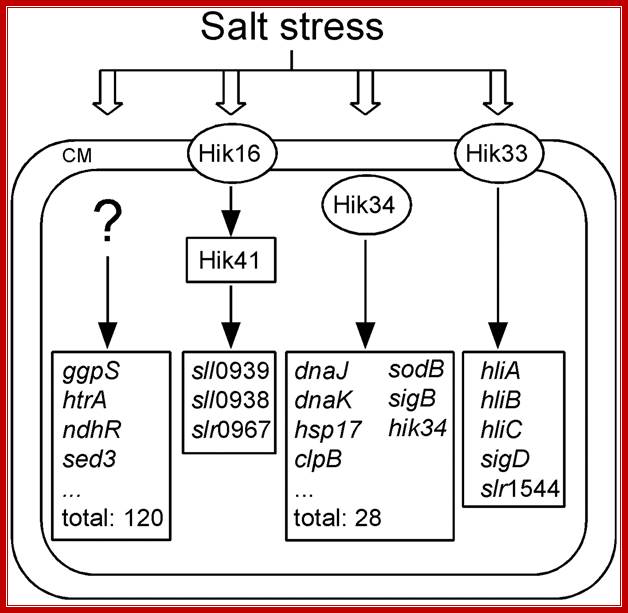
Salt stress results in the activation of cell surface receptors which in turn activates several groups of Histidine kinase HiK genes; http://www.pnas.org/

Stress Response and Signal Transduction inCorynebacterium glutamicum;;C. glutamicum harbors 13 different two component sensor systems and one of them (MtrBA) is directly related to transcription regulation of target genes involved in the response to hyper-osmotic stress. Two component systems are in general comprised of a membrane bound sensor histidine kinase and a soluble response regulator protein located in the cytoplasm. A physical stimulus, related to a particular external stress factor, is perceived by the sensor kinase and transduced to an intracellular signal transduction cascade via (auto) phosphorylation of the sensor kinase, (trans phosphorylation of the response regulator, and regulatory interaction with specific binding sites near target genes at the DNA. Prof. Dr. Reinhard Krämer, Dr. S. Nicklisch, K. Börngen, S. Maksimov, E. Glees, U. Meyer; http://www.kraemerlab.uni-koeln.de/
Bacterial Signal Transduction:
Two-Component Signal Transduction (TCS): Intramembrane-sensing histidine kinases:
The mechanism of stimulus perception in bacterial two-component systems: Two-component signal transducing systems are ubiquitously distributed communication interfaces in bacteria. They consist of a histidine kinase that senses a specific environmental stimulus, and a cognate response regulator that mediates the cellular response, mostly through differential expression of target genes. Histidine kinases are typically transmembrane proteins harboring at least two domains: an input (or sensor) domain and a cytoplasmic transmitter (or kinase) domain. They can be identified and classified by virtue of their conserved cytoplasmic kinase domain. In contrast, the sensor domains are highly variable, reflecting the plethora of different signals and modes of sensing. In order to gain insight into the mechanism of stimulus perception by bacterial histidine kinases, we surveyed sensor domain architecture and topology within the bacterial membrane, functional aspects related to this topology, and sequence and phylogenetic conservation. Based on these criteria, three groups of histidine kinases can be differentiated: (I) Periplasmic-sensing histidine kinases detect their stimulus (often small solutes) through an extracellular input domain. (II) Histidine kinases that detect stimuli (usually membrane-associated, such as ionic strength, osmolarity, turgor, functional state of the cell envelope) via their membrane-spanning segments and – sometimes – additional short extracellular loops. (III) Cytoplasmic-sensing histidine kinases (either membrane-anchored or soluble) detect cellular or diffusible signals reporting the metabolic or developmental state of the cell (Mascher et al. 2006, Microb Mol Biol Rev 70:910-38, see Fig. 4 for details). [Project: Diana Wolf]
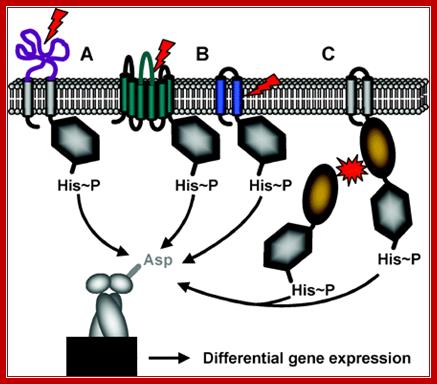
Schematic representation of the three different mechanisms of signal perception: (A) Periplasmic-sensing HK, (B) HK with sensing mechanisms linked to the transmembrane regions (signal perception can occur either with the membrane-spanning helices alone, or by combination of the transmembrane regions and short extracellular loops), (C) Cytoplasmic-sensing HK (either soluble or membrane-anchored proteins). The signal is represented by red arrows or red stars. The parts of the proteins involved in signal perception are highlighted in color. [Anna Staron], [ Sina Jordan, Beatrice Gutt] (Mascher 2006, FEMS Microbiol Lett 264:133-44; Thorsten Mascher et al; http://mmbr.asm.org/
Intramembranous-sensing HKs (IM-HK) are characterized by their short input domain, consisting solely of two putative transmembrane helices. They lack an extra-cytoplasmic domain, indicative for a sensing process at or from within the membrane interface, see the above Fig. Most proteins sharing this domain architecture are found in Firm cutes bacteria. Two major groups can be differentiated based on sequence similarity and genomic context: (1) BceS-like IM-HK that are functionally and genetically linked to ABC transporters, and (2) LiaS-like IM-HK, as a part of three-component systems; Most IM-HK sense cell envelope stress, and identified target genes are often involved in maintaining cell envelope integrity, mediating antibiotic resistance, or detoxification processes. Therefore, IM-HK seem to constitute an important mechanism of cell envelope stress response in low G+C Gram-positive bacteria,
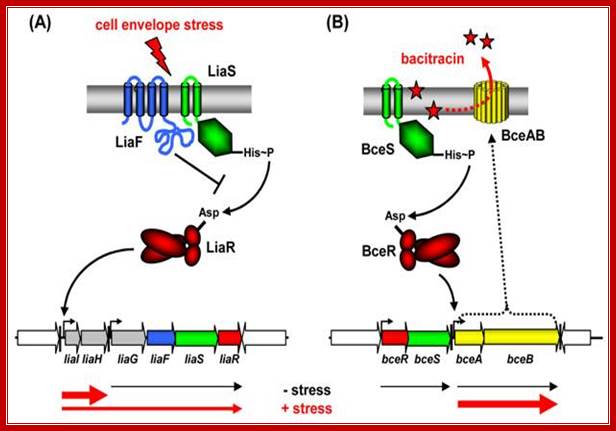
Stimulus perception by two component system (TCS): Cell envelope stress-sensing TCS in Bacillus subtilis: Sensors are shown in green, inhibitors in blue, transcriptional regulators in red. Arrows indicate activation, T-shaped lines repression. Transcripts with and without inducing conditions are indicated below by the red and black arrows, respectively. (A) LiaFSR three-component systems. LiaF functions as a strong inhibitor and is potentially involved in stimulus perception. (B) BceRS-BceAB-like TCS-ABC detoxification modules. http://syntheticmicrobe.bio.lmu.de/
Michael Hecker,1 Jan Pané-Farré,1 and Uwe Völker2
Institut für Mikrobiologie, 2Interfakultäres Institut für Genetik und Funktionelle Genomforschung, Ernst-Moritz-Arndt-Universität, Greifswald 17489, Germany;
One of the strongest and most noticeable responses of Bacillus subtilis cells to a range of stress and starvation stimuli is the dramatic induction of about 150 SigB-dependent general stress genes. The activity of SigB itself is tightly regulated by a complex signal transduction cascade with at least three main signaling pathways that respond to environmental stress, energy depletion, or low temperature. The SigB-dependent response is conserved in related gram-positive bacteria but is missing in strictly anaerobic or in some facultative anaerobic gram-positive bacteria. It covers functions from nonspecific and multiple stress resistance to the control of virulence in pathogenic bacteria. A comprehensive understanding of this crucial stress response is essential not only for bacterial physiology but also for applied microbiology, including pathogenicity and pathogen
Signal Transduction; TCS and ECF sigma factors; http://syntheticmicrobe.bio.lmu.de/
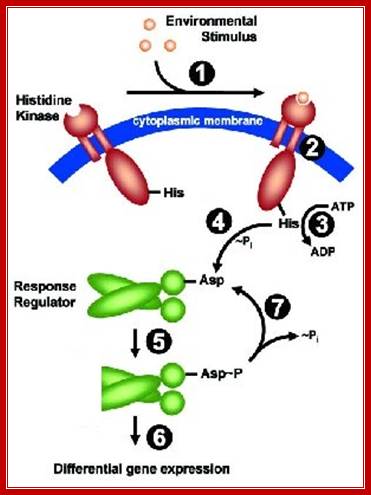
The cell envelope stress response network in Bacillus subtilis, activated HK phosphorylates Asp of response regulator. This in turn binds to specific gene promoter elements and activates the genes (shown below).
Two-Component Systems (TCS) interacting with DNA: http://syntheticmicrobe.bio.lmu.de/
Extra Cytoplasmic Factors (ECF), [PD Dr. Thomas Wiegert L]:
Our major research project is based upon results we obtained from global transcriptional analysis of alkaline stressed B. subtilis cells. Using DNA-macro-arrays, we defined the B. subtilis alkali stress stimulon and could show that a major part of genes induced at least four-fold after alkaline shock is member of the Sigma-W regulon. The alternative sigma factor Sigma-W belongs to the group of ECF sigma factors which control genes of extra-cytoplasmic function, and alkaline stress was the first stimulus described to induce the Sigma-W regulon, allowing us to identify additional Sigma-W-controlled genes and to define the promoter consensus sequence in more detail refers to Weigert (below). Later it was proposed that Sigma-W constitutes an antibiosis regulon, reacting on defects in cell wall biosynthesis, suggesting that an elevated extracellular pH imposes envelope stress to B. subtilis cells, (Wiegert et al., 2001, (Helmann, 2002)
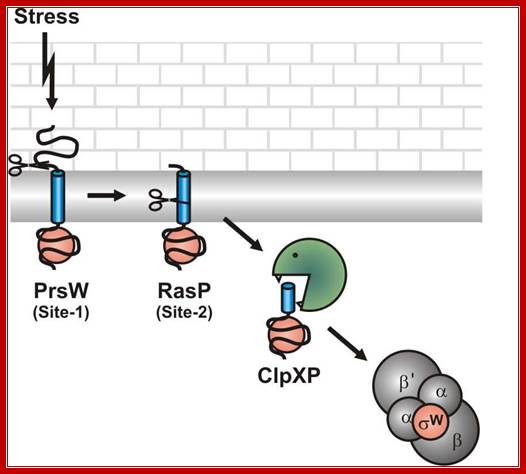
Model for the stress induced proteolytic cleavage of the B. subtilis Sigma-W anti-sigma factor RsiW. http://www.genetik.uni-bayreuth.de/
Alkaline shock imposes a kind of cell wall stress that activates PrsW to remove most of the extra-cytoplasmic part of RsiW (Site-1). The truncated RsiW created in this site-1 proteolytic step is substrate for RasP in site-2 proteolysis, where the transmembrane part RsiW is clipped (site2).The ClpXP protease completely degrades truncated RsiW and as a consequence Sigma-W is released to interact with the RNA polymerase core enzyme and Sigma-W-controlled promoters are transcribed.
Extra cytoplasmic factors (ECF) and sigma factors seem to be of crucial importance for prokaryotes in the adaptation to changing environmental conditions. For Streptomyces coelicolor and a Bacteroides strain living in the human gut more than 50 ECF sigma factors have been discovered. The function and regulation of ECF sigma factors is mostly unknown, some publications denote a role in cell wall stress response, antibiosis and pathogenesis. Our data on the Sigma-W / RsiW system of sigma factor /anti-sigma factor point to a general mechanism to modulate the activity of ECF anti-sigma factors in bacteria through regulated intramembrane proteolysis.
Bacterial Signal Transduction: and Extracytoplasmic Function (ECF) σ Factors and Two-Component Regulatory Systems (TCS)
In order to respond to changes in environmental parameters, cells must be able to transmit the information from the cell surface (site of induction) to the cytoplasm (site of cellular response). The two transmembrane signal-transducing mechanisms most common in bacteria are two-component systems and extracytoplasmic function (ECF) s factors, Fig. below.
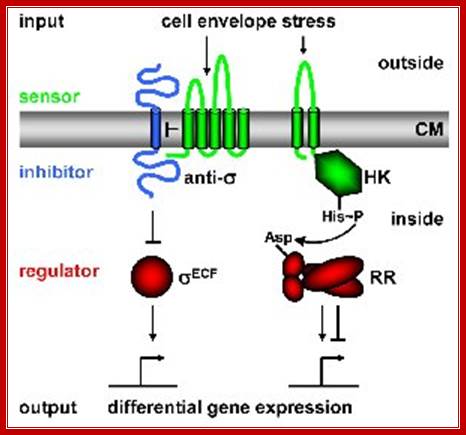
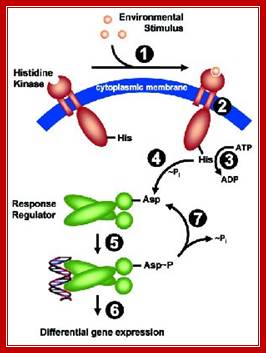
Regulatory principles orchestrating bacterial stress responses: ECF s factors (left) and TCS right; HK, histidine kinase; RR, response regulator). Sensors are shown in green, inhibitors in blue, transcriptional regulators in red. Arrows indicate activation, T-shaped lines repression. CM = cytoplasmic membrane. The HisàAsp phospho-transfer for TCS is indicated. See text for details; http://syntheticmicrobe.bio.lmu.de/.
Both systems consist of two proteins, a transmembrane sensor protein (histidine kinase and anti-s factor, respectively) that detects a specific stimulus (= input), and a cognate cytoplasmic effector protein (response regulator and s factor) that mediates the cellular response, usually differential expression of target genes. The two systems differ in the way the two proteins communicate and their output. Two-component systems are activated and communicate through a series of phosphotransfer reactions, i.e. phosphorylation and dephosphorylation of both histidine kinase and response regulator (Parkinson 1993, Cell 73:857-71; see Fig. below).
In contrast, ECF signal-transduction occurs through protein-protein interaction: In the absence of a signal the anti-s factor tightly binds the s factor, thereby keeping it in its inactive state. It is released in the presence of a stimulus and binds alternative promoter sequences upstream of its target genes (Helmann 2002, Adv Microb Physiol 46:47-110; see Fig). [Responsible scientists: Anna Staroń, Tina Wecke]
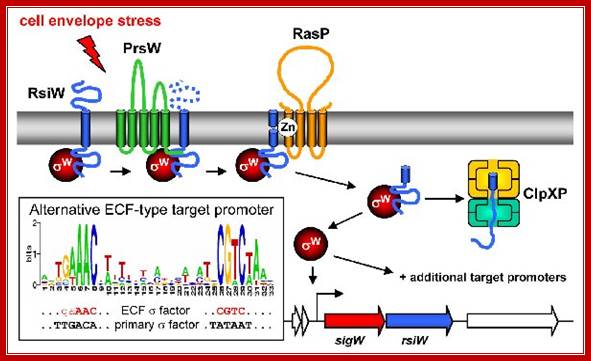 Mechanism of ECF σ- dependent signal transduction. The three-stepped
proteolytic cascade leading to anti-σ factor degradation and ECF σ
factor activation is illustrated for σW. Color-code as in Fig. 1. The
stimulus is represented by the red arrow. Additional proteins are shown in
yellow. The conserved alternative promoter sequence recognized by ECF σ
factors is illustrated in the inset. The autoregulatory feedback loop (binding
of the ECF σ factor to its own promoter) is indicated; http://syntheticmicrobe.bio.lmu.de/
Mechanism of ECF σ- dependent signal transduction. The three-stepped
proteolytic cascade leading to anti-σ factor degradation and ECF σ
factor activation is illustrated for σW. Color-code as in Fig. 1. The
stimulus is represented by the red arrow. Additional proteins are shown in
yellow. The conserved alternative promoter sequence recognized by ECF σ
factors is illustrated in the inset. The autoregulatory feedback loop (binding
of the ECF σ factor to its own promoter) is indicated; http://syntheticmicrobe.bio.lmu.de/
Mechanism of ECF s- dependent signal transduction:
The three-stepped proteolytic cascade leading to anti-s factor degradation and ECF s factor activation is illustrated for sW. Color-code as in Fig. above. The stimulus is represented by the red arrow. Additional proteins are shown in yellow. The conserved alternative promoter sequence recognized by ECF s factors is illustrated in the inset. The autoregulatory feedback loop (binding of the ECF s factor to its own promoter) is indicated.
The output of two-component systems is more flexible, allowing inhibition as well as induction of gene expression, while ECF s factors function as activators of expression only. The B. subtilis genome encodes 7 ECF s factors and 35 two-component systems to sense changes in and communicate with the environment. Many of these systems are still of unknown function.
Statistical Estimate of the Total Number of Operons Specific for B.subtilis Sporulation. D. HRANUELI,l P. J. PIGGOT,2 AND J. MANDELSTAM; Microbiology Unit, Department of Biochemistry, University of Oxford, Oxford OXI 3QU, United Kingdom
As an alternative to exhaustive mapping, an attempt has been made to obtain a rough estimate of the total number of sporulation operons involved; this is done by statistical analysis.
Sixteen sporulation mutants taken at random were characterized biochemically and morphologically. The mutations they contained were mapped to determine whether they fall into any one of 23 known operons or not. From the proportion that does so ('0A6), it is calculated that the most probable number of sporulation operons is 37 (68% confidence limits of 31 and 46). If allowance is made for the fact that two of the operons apparently contain mutagenic "hot spots" and the calculation is amended accordingly, the most probable numbers of operons becomes 42 (limits 33 and 59).
Molecular Dynamics of Response Regulators:
Spo0F belongs to a large class of proteins, the response regulators that participate in many different bacterial signal transduction pathways. Response regulators have diverse functions, though common to all of these proteins is a regulatory domain of ~120 residues that becomes phosphorylated at a conserved aspartate residue in a magnesium-dependent reaction with a histidine auto kinase. The overall fold of Spo0F consists of five a -helices surrounding five parallel b -strands, forming a hydrophobic sheet; a structure in common with other response regulators. The fold brings three aspartic acid residues into close proximity to form the binding pocket to accept the phosphoryl group. From structural studies enabled to determine in the orientation of secondary structure elements in the putative recognition surfaces and the relative charge distribution of residues surrounding the site of phosphorylation.
Initiation is triggered by environmental signal transduction across the cell membrane perhaps via histidine kinase or similar transduction events. Sensors are located in the membrane. The receptors look like multipass membrane proteins, sensing the environmental factors they activate a whole set of kinases such as Kinase A, kinase B, kinase C, kinase D and kinase E. Each of them is activated responding to different factors. Various mechanisms operate as depicted above in perception of stress signals and transduction of the same. Starvation activates a set of sigma factors as well as proteins. In this process new sig factors are produced which displace sig43 / sigA and use the same RNAP to transcribe different set of genes for sporulation process. In this process a cascade of gene expression takes place till the endospore is formed. This cascade of gene expression and gene inactivation is all sporulation specific.
Bug eats Bug:
With the onset of nutrient poor conditions, cannibal cells secrete two toxins that kill some of their ‘sensitive’ siblings, whilst the toxin-producing cells themselves are protected by simultaneous production of ‘immunity machinery’. The nutrients released by the killed cells then feed the community and delay the onset of sporulation.
An additional general strategy for eking out an existence in a potentially harsh environment is to encapsulate the population of cells in a viscous ‘slime’, biofilm that acts as a barrier to the external environment (which may include antibiotic molecules or immune cells). The biofilm is formed by a sub-population of cells that produce the extracellular matrix, so-called matrix producers. This amounts to a quorum sensing mechanism.
Daniel López, a postdoc working in the Kolter lab at the Harvard Medical School, has previously reported a ‘quorum-sensing’ peptide molecule called surfactin, produced by B. subtilis, which stimulates a cascade that results in the activation of spoOA and the triggering of biofilm matrix formation. Quorum sensing molecules are a means by which a cell population can monitor the density of cells in a given area, with high cell densities – and thus the potential for food competition – resulting in a high concentration of quorum sensing molecules. Such signals therefore trigger the population to take action.
Bug eats Bug; Jim Caryl; http://mentalindigestion.net/
The three pathways, sporulation, cannibalism and matrix production, are known to be controlled by a master regulator protein, spoOA; however the degree to which each pathway is controlled differs depending on the state of spoOA itself. The key has been to determine the environmental cues that differentially activate spoOA and how this in turn triggers one or more of the three pathways.
Grossman: Cell Signaling:
Grossman:
Cell-Cell signaling, quorum sensing, and competence development:
B. subtilis, like many organisms, has mechanisms for sensing and
responding to high cell density (quorum sensing). In B. subtilis, both the
initiation of sporulation and competence development (the natural ability to
bind and take up exogenous DNA) are regulated, in part, by high cell density.
We have purified and characterized two different extracellular peptide
signaling molecules that are produced by B. subtilis and accumulate in culture
supernatant as cells grow to high density. Both peptides stimulate
phosphorylation (activation) of the transcription factor, ComA. ComX pheromone
is a 10 amino acid peptide with a hydrophobic modification on a tryptophan
residue. The pheromone contains the C-terminal 10 amino acids from a 55 amino
acid precursor. The cellular response to the pheromone requires the
membrane-bound histidine protein kinase, ComP. ComP autophosphorylates and
donates phosphate to the transcription factor, ComA. The competence and
sporulation stimulating factor, CSF, is an unmodified pentapeptide. It
corresponds to the C-terminal 5 amino acids of a 40 amino acid precursor
(phrC). The cellular response to CSF requires transport back into the cell via
oligopeptide permease, a member of the large family of ATP-dependent
import/export systems found in both prokaryotes and eukaryotes. CSF stimulates
accumulation of ComA~P and competence gene expression by inhibiting activity of
the phosphatase, RapC. Thus, the two factors stimulate competence gene
expression by affecting activity of a kinase and an opposing phosphatase. Grossman; http://openwetware.org/
Flow of signal transduction:
Stimulus >>Cell surface Receptor >>Hitidine Kinase >>KinA, KinB (kinC and kinD and kinE) >>SpoOF>>SpoOB>> SpoOA
The flow of signal cascade through the phosphorelay to Spo0A is highly controlled at several levels. Although the primary signals transduced by the two kinases, KinA and KinB, responsible for Spo0F phosphorylation remain obscure, genetic studies have revealed a series of genes unique for the activation of each kinases. The activity of each of the kinases is regulated by complex signal-transduction pathways that respond to environmental, metabolic, and cell-cycle signals. It is now clear that all the signals that affect sporulation cannot be processed by the kinases alone. Access to the phosphorelay for additional signals is provided by two families of phosphatases, which dephosphorylates either Spo0F~P or and act to prevent sporulation.
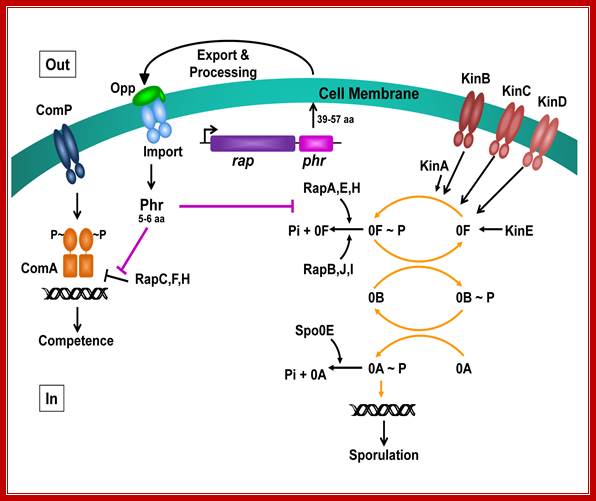
The phosphorelay for sporulation initiation in B. subtilis: In the phosphorelay, the activity of five sporulation kinases (KinA, B, C, D and E) is counteracted by six response regulator aspartyl phosphate phosphatases (RapA, B, C, Spo0E, YisI and YnzD). In a phosphorelay cascade the first to be SpoOF then SpoOB and ultimately SpoOA getsa activated http://www.plosbiology.org/

Stress signal activate a cascade of kinases and phosphotases; in this cascade the most important product is SpoOA. This activates the process of sporulation first forming prespore and fore spore and mother cell. Many events are aided by specific sig factors; which act as a part of regulalons, perhaps as Modulons. This process starts as stimulon by environ signals (quorum semsing) and end as modulon culminating in endospore formation. http://www.devbio.biology.gatech.edu/
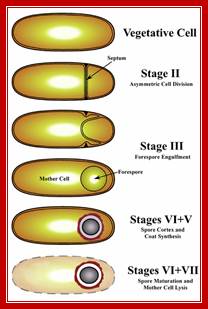
Triggered by nutrient limitation, B. subtilis undergoes a switch from binary fission to asymmetric division, in which a division septum is laid down close to one of the cell poles (Fig.1). Polar septation divides the developing cell into two unequal-sized cellular compartments called the mother cell (the larger cell) and the forespore that follow dissimilar pathways of differentiation
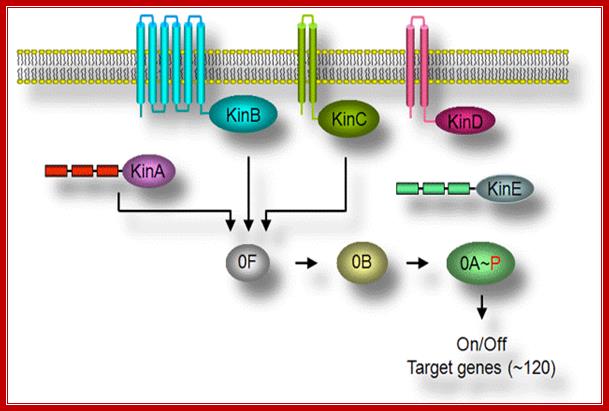
Multicomponent Phospho relay; The key to understanding the initiation of sporulation is to understanding the mechanism(s) of the signal transduction pathway. One striking feature of this signal transduction pathway is a complex phosphorelay to activate Spo0A, a transcription activator protein as a response regulator. Most response regulators in bacteria are phosphorylated directly by a cognate sensor kinase that carries out autophosphorylation on a histidine residue and then transfers the phosphoryl group to the aspartyl residue in the response regulator. Spo0A, in contrast, is indirectly phosphorylated by a multicomponent phosphorelay involving at least three kinases called KinA, KinB, and KinC. The kinases phosphorylate Spo0F, and the resulting Spo0F-P, in turn, transfers the phosphoryl group to Spo0B. Finally, Spo0B-P transfers the phosphoryl group to, and thereby activates, Spo0A; http://nsmn1.uh.edu/
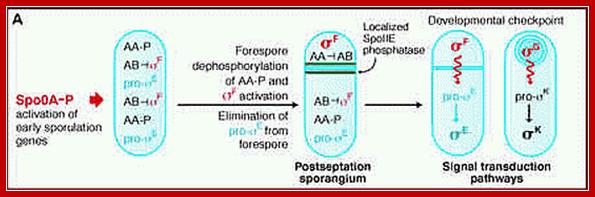
Initiation of spore development: by Josefine Jonsson,November 2009
Activation of SpoOA happens in several steps, where the first activation takes
place at the end of exponential growth and results in the so called transition
state. Transition state is associated with protease production, motility,
capability of transformation, biofilm formation and even cannibalism. Increased
phosphorylation of SpoOA is thought to be crucial in the formation of the
spore. After the sporulation induced cell division, the mother cells’ SpoOA
activity increases, whereas it declines in the prespore. Both these changes are
very important for further sporulation processes.
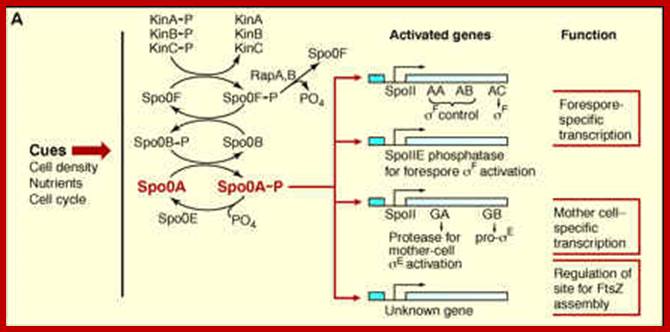
Environmental distress signals activate several protein kinases, among them, as mentioned earlier, are Kin-A (spoII J), Kin-B and KinC. Signal transduction leads to phosphorylation of SpoOF to SpoOF~p, which i.e SpoOF~p phosphorylates Spo OB. The Spo OB-p in turn phosphorylates Spo OA to SpoOA~p. These genes are transcribed early with RNA polymerase containing minor sigma-A (?). Sig-A is an housekeeping factor in B. subtilis. Spo OA is a master transcriptional regulator (Regulon) and it influences a whole set of operons, each of which are transcribed by the same RNAP but with different sig-factors. SpoOA-p directly controls ~121 genes (30single unit genes and 24 operons). SpoOA is a DNA binding protein; a Transcriptional factor. It is to be noted that B.subtilis RNAP contains an additional subunit called delta.
SpoOA~P TF binds to Promoter DNA –sequence
TTTGTCG/A AAA/TAA. (TTGTCGAA)----+1>
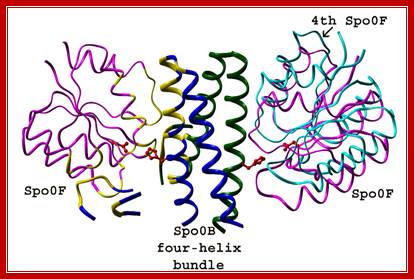
SpoOF and SpoOB protein-protein interacting domains. Josefine Jonsson
Phosphorylation of spoOA launches the activation, as earlier mentioned, of at least 10% of the genome, so it is called Regulon that sets the process going on to sporulation. In this process one can observe cascade of sigma factors and their functions.
Energy potential red-ox state activates KinA. Other kinases are also activated by similar stimuli, where Kin C, Kin D and Kin E are activated together with kinase A (KinA) phosphorylates Spo OF, which then p.lates Spo OB, that leads to the phosphorylation of SpoOA. It is a cascade of kinases where phosphotases also perform.
Kinases’ and Phosphatases’ competition in Bacterial Development:
Recognition of the diverse signals involved in the initiation of sporulation is carried out by proteins that alter the level of phosphorylation of the Spo0A, a TF. The opposing activities of kinases and phosphotases are the major players influencing the output of the system in a manner similar to eukaryotic signal transduction systems. The Spo0E family of phosphotases specifically dephosphorylates the Spo0A~P component of the phosphorelay while three members of the Rap family of phosphotases specifically dephosphorylate the Spo0F~P intermediate.
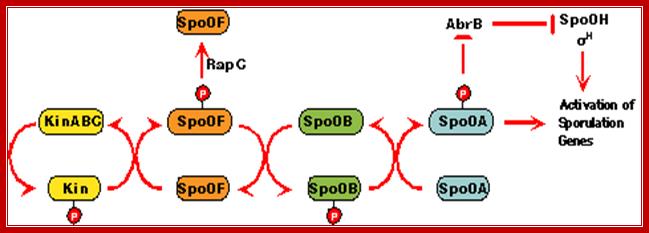
Phosphorylation and dephosphorylation-ultimately activates spoOA, which triggers the whole cascade of events.Mol.Biol www.pinsdaddy.com; https://www.ncbi.nlm.nih.gov
Initiation of spore development:
http://www.eplantscience.com/
http://www.devbio.biology.gatech.edu/
SpoOA-p triggers a cascade of gene expression, among them sig F
and sig E. Sig F primes the septal formation at the initiation of prespore
from the mother cell. With the sigF active in prespore sends signal for the
synthesis of sigE. Whatever sigF found in mother cells gets degraded similarly
whatever sigE found in prespore gets degraded. http://www.devbio.biology.gatech.edu/
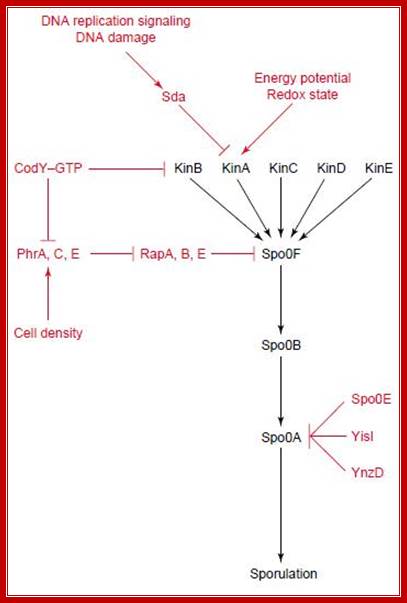
The central part of phosphorelay is in black, with various inputs in red. KinA phosphorylates SpoOF, which transfers the phosphate to SpoOB and SpoOB to SpoOA that activates transcription of sporulation genes. KinA regulates by Sda (inhibition) and one of its PAS domains (activation). Sda activates by DNA damage or incorrect DNA transcription. Whereas the PAS domain sense energy potential or redox state of the cell. Phr peptides senses cell density and inhibit Rap proteins that dephosphorylate SpoOF-PO4. CodY represses the transcription of kinB and some phr genes in the presence of GTP. SpoOE, YisI and YnzD can dephosphorylate SpoOA; Activated spoOA activates spoIIA, spoIIE and spoIIG:
http://www.studyblue.com/notes
- Once factors are triggered; replication of the bacterial chromosome takes place.
- Two sigma factors, residential are sigma A and sigma H, regulate gene transcription to initiate two different developmental programs in the two daughter chromosomes. Sig H is stationary phase sigma factor.
- Activated SpoOA is itself a transcription factor that modulates the expression of various genes transcribed by vegetative cell RNA polymerase and hence recognized by the regular σA and σH subunits. Sigma-H is an alternative RNA polymerase sigma factor that directs the transcription of many genes that function at the transition from exponential growth to stationary phase in Bacillus subtilis. Sigma H directs 49 promoters involving 87 or more genes leading to stationary phase.
- Compartmentalization of the cell results into the prespore then into forespore (regulated by sigma F) and the mother spore (regulated by sigma E).
Role of SpoOA in Bacillus sporulation (NCBI/NIH):
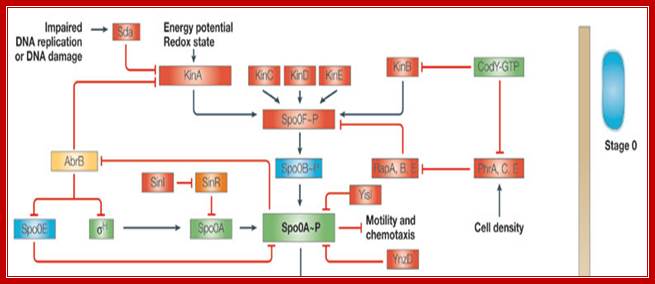
Sporulation
cascade in B.subtilis.; A simplified
representation of the sporulation cascade in Bacillus subtilis (adapted
from Refs 5,8,108,109) is shown, indicating the presence of putative
orthologues in the Clostridium
acetobutylicum, Clostridium perfringens strain
13, Clostridium
difficile, Clostridium botulinum and Clostridium tetani genomes based on existing bibliography
and BLAST110 searches.
Results regarding C.
difficile and C. botulinum have been compiled by the authors
based on the genomic data and BLASTP searches. These results will be subject to
change once a definitive annotation is released by the Wellcome Trust Sanger
Institute. Red indicates proteins that have not yet been identified in any of
the clostridia analysed. Green indicates a B. subtilis protein
or gene that has been reported to exist in all five clostridia, or that a
best–best BLASTP match can be found (>30% amino-acid identity over >60%
of both the B.
subtilis (or C. acetobutylicum)
protein sequence and the putative orthologue, as in Ref. 107). Blue indicates that it has been suggested that
such a protein or gene might be present in C. tetani, although its overall similarity to the
equivalent B.
subtilis gene is low37. Yellow indicates that an
equivalent protein or gene can be identified in all five clostridia except C. tetani,
although the presence of AbrB in C. difficile is
doubtful. Orange indicates that an equivalent protein or gene can be identified
only in C.
acetobutylicum and C. tetani.
Purple indicates that an equivalent protein or gene can only be identified in C. acetobutylicum and C. botulinum. Pink indicates that ![]() K is encoded as two fragments that require
processing to form a functional protein in B. subtilis and C. difficile,
whereas it is encoded as a single, uninterrupted gene in the rest of the
clostridia species examined. If and where the indicated signals (DNA
replication, DNA damage, energy potential, redox state and cell density) enter
the sporulation cascade is not known; motility and chemotaxis genes have been
identified in all the clostridia analysed except C. perfringens.
Each protein is roughly aligned vertically according to its putative order of
expression. Not all known regulators and regulations in B. subtilis are shown. Arrowheads indicate a
positive effect such as phosphorylation or control over the expression of the
target gene. Blunt arrows indicate a negative effect such as repression of gene
expression or dephosphorylation. When the target of the negative effect is not
a protein or gene, it means that the expression of part of that regulon is
negatively controlled..Several kinases at membrane level respond to
external signals such as Red ox, that leads to the activation of spoOA to
spoOA-p via Kinases. All sporulation genes are called spoO-genes. Carlos J. Paredes, Keith V. Alsaker &
Eleftherios T. Papoutsakis; http://www.nature.com/
K is encoded as two fragments that require
processing to form a functional protein in B. subtilis and C. difficile,
whereas it is encoded as a single, uninterrupted gene in the rest of the
clostridia species examined. If and where the indicated signals (DNA
replication, DNA damage, energy potential, redox state and cell density) enter
the sporulation cascade is not known; motility and chemotaxis genes have been
identified in all the clostridia analysed except C. perfringens.
Each protein is roughly aligned vertically according to its putative order of
expression. Not all known regulators and regulations in B. subtilis are shown. Arrowheads indicate a
positive effect such as phosphorylation or control over the expression of the
target gene. Blunt arrows indicate a negative effect such as repression of gene
expression or dephosphorylation. When the target of the negative effect is not
a protein or gene, it means that the expression of part of that regulon is
negatively controlled..Several kinases at membrane level respond to
external signals such as Red ox, that leads to the activation of spoOA to
spoOA-p via Kinases. All sporulation genes are called spoO-genes. Carlos J. Paredes, Keith V. Alsaker &
Eleftherios T. Papoutsakis; http://www.nature.com/
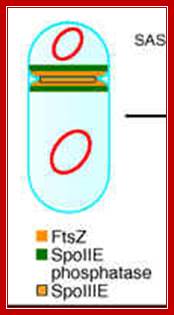
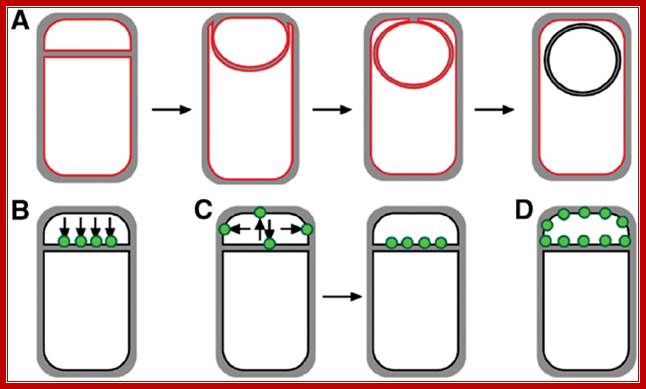
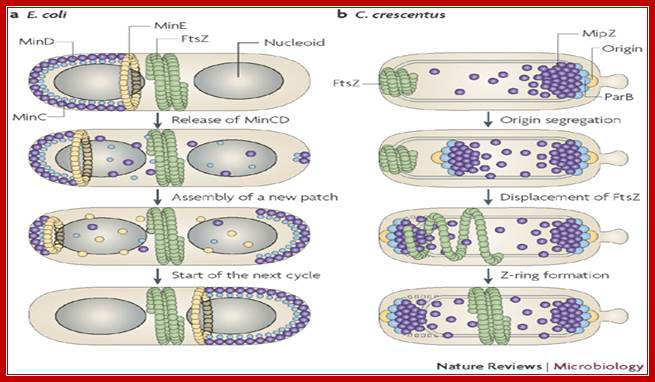
First physical change, that one can observe microscopically, is cell elongation, replication of DNA, then an unequal cell division initiates to form one smaller cell called prespore and the other large cell called pre-mother cell. During cell division cytoplasmic cleavage shifts from mid region (normal) to pole position. One of the most important proteins that accumulate in the septal region is FtsZ, which is similar to tubulins. This develops a Z-ring at the septal formation region. Some of the gene products involved are minC, minD and minE (divIVA). These are further assisted by FtsA, FsW, FtsL and DivB, DivL and PBP2D proteins.
This leads to constriction of plasma membrane and cell wall synthesis ensues and gets deposited in the space in between. Both cells contribute to forespore and its outer spore coat and exo-sporium formation. The spore walls and the stored contents are dehydrated and they can withstand extreme conditions and survive for a longer periods.

At region of septal formation it develops a ring like constriction made up of FtsZ. http://en.wikipedia.org/
It at this point of time the fore spore gets engulfed by the mother plasma membrane; thus the fore spore is a cell within cell (mother cell). It contains two membrane one of its own and the other (outer) from its mother cell. Each of them has membrane surfaces with opposite polarity to each other.
First septal wall between the prespore and mother cell is dissolved by the activity of spoIIB. It is the spoII that initiates prespore engulfment and it is facilitated by spoVG. Engulfment also requires spoIID, spoIIP and spoIIM. All of them are transmembrane proteins. This is achieved by the plasma membrane of the mother cell invaginating around the smaller prespore cell. The fusion of the membrane and release of the forespore into the cytoplasm of the mother cell requires spoIIIE, which is also involved in the transport of one of the two daughter DNAs into the prespore. Thus the smaller cell, the forespore, consists of two membranes of opposite polarity or surfaces, one of its own and the other from the mother cell. Cellular activities in both cells are different. The inner and outer membranes of the forespore have different sides. The sigmaF activated spoIIQ is an important factor in forespore formation. Mother cell nourishes the forespore cell (mother and daughter relationship) and it programs the development of endospore; in this process gene expression and the metabolic processes are different. Often they send signals to each other and facilitate the function of each other which play a very important role in the development of endospore.
Genes and gene products involved in this unequal division is not fully understood. During this event, however lot of reserve food material, in the form of poly b-hydroxy butyric acid granules are produced and the same are transported and stored in forespore.
(A) Cartoon showing B. subtilis engulfment. Following polar septation, the membrane of the larger mother cell migrates around the forespore, until it is completely enclosed. Membranes stained by FM 4-64 are shown in red; following membrane fusion, FM 4-64 no longer stains the forespore membranes, providing an assay for the completion of engulfment (B–D).
Protein localization to the septal membrane during Bacillus subtilis sporulation.
![Getting organized |[mdash]| how bacterial cells move proteins and DNA](Gene_Expression_I5G-Regulation_of_Gene_Expression_During_Sporulation_files/image047.jpg)
During sporulation (a), B. subtilis undergoes an asymmetric cell division that generates two offspring of unequal size and developmental fate — a larger mother cell (green) and a smaller forespore (blue). The mother cell subsequently engulfs the forespore in a phagocytosis-like process, thereby surrounding it with a second membrane, the so-called outer forespore membrane. During this process, several proteins specifically localize to the interface between the two compartments (b). In several cases, their positioning is mediated by diffusion and capture, as exemplified by the recruitment of the freely diffusible mother cell protein SpoIVFB (FB; purple spheres) by a pre-localized complex including the forespore protein SpoIIQ (Q). The interaction of proteins across the mother cell and forespore membranes involves a zipper-like mechanism (c). Using this principle, SpoIIQ engages the mother cell factor SpoIIIA (A), which, in turn, is responsible for the septal localization of other proteins, among them SpoIVFB. Martin Thanbichler & Lucy Shapiro; http://www.nature.com/
Models for targeting forespore-expressed membrane proteins:
Proteins (green) can either be directly inserted into the forespore septal region (B) or inserted elsewhere and migrate to the septal region (C). Then, they can either be retained during engulfment (C) or freely diffuse throughout the forespore membrane (D).
Engulfment pathway of sporulation in B. subtilis and models for targeting of proteins synthesized in the forespore. (A) Cartoon showing B. subtilisengulfment. Following polar septation, the membrane of the larger mother cell migrates around the forespore, until it is completely enclosed. Membranes stained by FM 4‐64 are shown in red; following membrane fusion, FM 4‐64 no longer stains the forespore membranes, providing an assay for the completion of engulfment. (B–D) Models for targeting forespore‐expressed membrane proteins. Proteins (green) can either be directly inserted into the forespore septal region (B) or inserted elsewhere and migrate to the septal region (C). Then, they can either be retained during engulfment (C) or freely diffuse throughout the forespore membrane (D).Septal localization of forespore membrane proteins during engulfment in B.subtilis; Aileen Rubio, Kit Pogliano ; http://emboj.embopress.org/
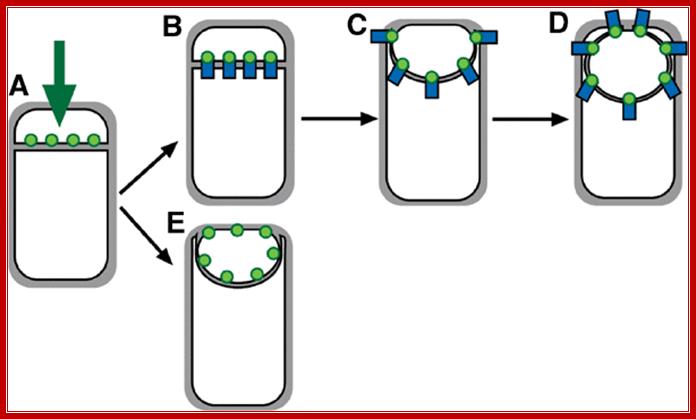
Model for localization of forespore membrane proteins. (A) Any membrane protein (green) synthesized by a σF‐dependent promoter inserts directly into the forespore septal domain. (B–D) Localized proteins (such as SpoIIQ) are retained through an interaction with a partner protein (blue) made in the mother cell or the septal peptidoglycan. (E) Nonlocalized proteins (such as MalF‐GFP) freely diffuse throughout the forespore membrane. Aileen Rubio, Kit Pogliano http://emboj.embopress.org/
Four steps in prespore engulfment, and the proteins involved in each step.

The protein names have been abbreviated by omitting the Spo prefix. SpoIIB and SpoIIQ are placed in parentheses as their functions are not completely essential for engulfment. SpoIID, SpoIIM and SpoIIP (also probably SpoIIQ) are needed throughout stages 2 and 3. SpoIID, SpoIIM and SpoIIP are needed to drive regression of the second polar septum, as well as for engulfment. SpoIIIE is involved in the final step of membrane fusion. Modified with permission from Ref. 64 © (2002) Cold Spring Harbor Laboratory Press. Jeff Errington; http://www.nature.com/

A model for coat morphogenesis: successive waves of spore encasement;
Coat morphogenesis begins with the assembly of a scaffold containing half of all coat proteins on the mother cell proximal (MCP) pole of the forespore. Assembly of the basement layer (blue), which consists of proteins that are solely under the control of the sigma factor σE for transcription, is dependent on SpoIVA. Encasement by SpoIVA is likely to be driven by multimerization and happens concomitantly with engulfment. Assembly of the inner (orange) and outer (purple) coat layers is dependent on SafA and CotE, respectively. Proteins of the inner and outer coat appear to have delayed encasement kinetics in comparison to the basement layer, and although they begin to assemble on the MCP pole during engulfment, they only form a cap on the mother cell distal (MCD) pole of the forespore after engulfment is complete. Most proteins in this class show dependency on combinations of σE and σK, as well as the transcription factor SpoIIID for expression. CotZ, a key component of the crust (red) belongs to the last encasement class and is dependent on σE, σK and the transcription factor GerE for expression. All four layers contain late-expressed σK-dependent proteins that are not part of the initial scaffold. These proteins presumably diffuse through the permeable coat matrix to reach their final location within the coat. Transcriptional regulation in the mother cell controls the kinetics of spore encasement, in particular the two sigma factors — σE (for the control of early gene expression during engulfment) and σK (for the control of late gene expression post-engulfment) — and two mother cell-specific transcription factors — SpoIIID (which is turned on by σE and modulates the σE regulon) and GerE (which is turned on by σK and modulates the σK regulon). RNAP, RNA polymerase; Peter T. McKenney, Adam Driks & Patrick Eichenberger; http://www.nature.com/
Normal cell division- role of Ftz proteins:
Normal cell division and the role of FtsZ proteins; the septa is formed in the middle of the cell. Getting organized — how bacterial cells move proteins and DNA; In recent years, the subcellular organization of prokaryotic cells has become a focal point of interest in microbiology. Bacteria have evolved several different mechanisms to target protein complexes, membrane vesicles and DNA to specific positions within the cell. This versatility allows bacteria to establish the complex temporal and spatial regulatory networks that couple morphological and physiological differentiation with cell-cycle progression. In addition to stationary localization factors, dynamic cytoskeletal structures also have a fundamental role in many of these processes. In this Review, we summarize the current knowledge on localization mechanisms in bacteria, with an emphasis on the role of polymeric protein assemblies in the directed movement and positioning of macromolecular complexes.
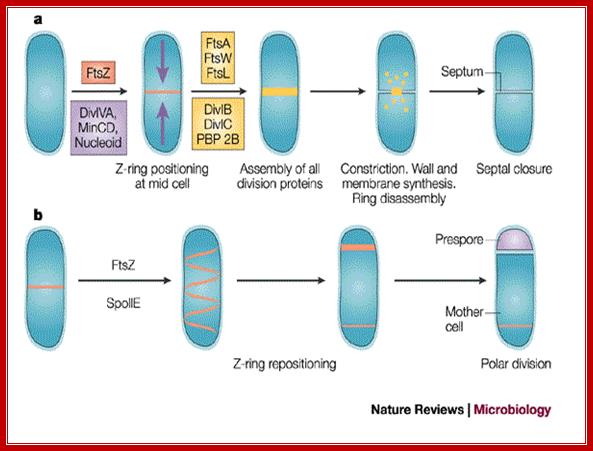
Cell division during growth (a) and sporulation (b) of B. subtilis.; The first key step in division is assembly of a ring of FtsZ protein (light red) at the future division site at the mid-cell. Positioning of the Z-ring is directed by the combined actions of the Min system and the nucleoid occlusion effect (purple arrows). Various other division proteins (yellow) are recruited to the Z-ring. At some point, the division machinery constricts the cell, coordinately organizing the synthesis of new membrane and cell-wall layers, and, in parallel, the machinery disassembles. During sporulation (b), two effects — increased FtsZ accumulation and synthesis of the sporulation-specific SpoIIE protein — lead to the repositioning of FtsZ into two separate rings, one near each of the cell poles. The switch in position occurs by a helical redistribution of the FtsZ protein from the mid-cell to subpolar positions. The Z-rings near the two poles are usually unequal and one of them is 'chosen' for the formation of a septum, which represents the defining moment in determining the fates of the prespore and the mother cell.; Jeff Errington; http://www.nature.com/
DNA segregation:
While the septal formation going on, the replicated chromosomal DNA is segregated with OriC anchored by DivIV and pulled towards their respective poles; SpoOJ is found to bind at the vicinity of OriC. Rec A is another protein interacts with DivIVA bound at OriC. Even if one of the DNA is trapped in the septal membrane, the daughter DNA is threaded linearly by spo IIIE hexamer transporter across the septum while it is still forming where Ori C of DNA is directed forward. The spoIIIE, as a hexamer, has an ATPase activity as well as a transporter function. Individual monomers contain a trans-membrane domain, and a motor protein (spoIIIEC). It threads one of the daughter DNAs into the fore spore in a directional manner. The spoIIIE’s C-terminal interacts with short DNA sequences called spoIIIE recognition sequences (SRS) and binds and propels the DNA in directional manner i.e. OriC heading forward. Transport takes about 10 minutes. In this process Div IVA is involved. Spo OJ also binds near oriC and facilitates efficient DNA segregation of replicated DNA.
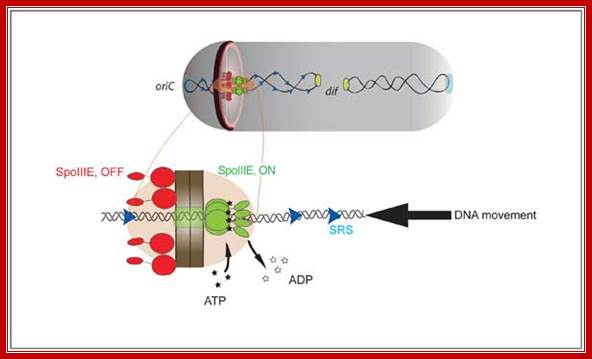
Single-molecule methods were used to show that translocation directionality of SpoIIIE is affected by the presence of short sequences (SpoIIIE recognition sequences, or SRS) that are specifically oriented in the B. subtilis chromosome pointing from the origin to the terminus of replication. Biochemical ensemble experiments allowed us to establish that interactions between SpoIIIE and SRS are orientation specific. Further genetic experiments showed that interactions between SpoIIIE and SRS are also orientation-specific in vivo. Our experiments finally showed that mutations or deletions in the γ-domain not only affected directional DNA movement in vivo but also abolished sequence recognition in vitro, suggesting that this domain is responsible for directly mediating SpoIIIE/SRS interactions. Our findings thus indicated that DNA transfer by SpoIIIE is regulated by direct and orientation-specific SpoIIIE interactions with short chromosomal sequences that tell SpoIIIE in which direction it has to translocate. These and previous data (Sharp, 2002) thus suggested that the establishment of a compartment specific complex had to be regulated by SpoIIIE/SRS interactions. We tested this prediction by using a recent fluorescence microscopy assay developed by Eric Becker to test in vivo for compartment-specific SpoIIIE assembly. Consistent with our model for sequence-directed DNA export, the γ-domain of SpoIIIE was essential for the assembly of compartment-specific complexes. This unified model reconciles previously proposed simple exporter and sequence-directed models for directional DNA transport by the SpoIIIE/FtsK/Tra family DNA translocases.; Involvement of SpoIIIE as a translocator in the transport of daughter DNA molecules. http://www.cbs.cnrs.fr/

The SpoIIIA proteins in the mother cell and SpoIIQ in the forespore form a secretion complex.
(A) Schematic diagram showing the SpoIIIA-SpoIIQ secretion complex in the two membranes that surround the forespore. SpoIIIAA (A), SpoIIIAB (B), SpoIIIAC (C), SpoIIIAD (D), SpoIIIAE (E), SpoIIIAF (F), SpoIIIAG (G), SpoIIIAH (H), SpoIIQ (Q) are shown. SpoIIIAA (A) is shown as a hexamer by analogy to other traffic ATPases [25]. SpoIIQ (Q) is shown as a multimeric pore based on the experiments of Meisner et al[22]. The actual stoichiometry of proteins in the complex is unknown. (B) The feeding-tube model. In wild-type cells (wt), the SpoIIIA-SpoIIQ complex secretes an unknown metabolite/osmolyte (red circle) into the forespore that maintains forespore integrity and σG activity (indicated by a green forespore). In the absence of the SpoIIIA (ΔIIIA) or SpoIIQ proteins, the forespore looses metabolic potential; the forespore collapses and σG activity cannot be maintained. David Z. Rudner etal; http://www.plosgenetics.org/
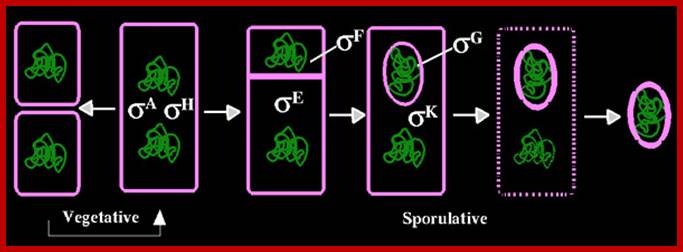
Forespore and mother cell formation also leads to the segregation of sig F in fore spore and sig E in mother cell. Before initiation of asymmetric cell division housekeeping sigma factors found in mother cells are sig A and sig H. Note B.subtilis sigA = sig70 (E.coli), http://www.biology .kanyon.edu
Cascade of gene expression:
During sporulation, a set of genes are activated in fluxes and a set of genes are inactivated in the same time frame resulting in differential gene expression. It is a par excellent example to understand molecular basis of development or say morphogenesis at gene level of a unicellular prokaryotic system. From the start of spore formation to the end, nearly 40% of cellular genes are expressed; most of them are sporulation specific.
Differential gene expression in bacillus is due to activation or synthesis of new set of sigma factors. The sigma factors are sporulation specific. The prevalent sigma factor in Bacillus is sig43 = sigA which transcribes most of the genes required for house keeping activity; even during sporulation the said genes operates for cell requires metabolic inputs.
In parental cell two primary sigma factors found operating are sig A and sig H perhaps transcribed by resident RNAP. It is the activated SpoOA-p is a transcription factor that binds to its specific promoters containing the mentioned DNA sequences
SpoOA binding promoter sequence:--
--------------TTTGTCG/A AAA/TAA-----à+1---
Binding leads to activation of several genes and produce several proteins, among them the two are important factor sigF and sigE. They are synthesized early i.e. before the onset of septal formation and engulfment; they are segregated how? But sigF is selectively degraded in mother cell and sig E is degraded in forespore by proteolytic activity. To begin with both are precursor proteins and they require to be activated. Thus these two sigma factors and their activity is restricted to their respective cells, however their activated gene products do have role in communication between each other across the space found in-between septal membranes.
Measuring Gene Expression on a Genome-wide Scale:
The elucidation of the complete sequence of the Bacillus subtilis genome, and the recent developments in micro-array technologies, provide the means to measure gene expression on a genome scale. Currently, membrane filters are available containing arrays of PCR fragments of all known B. subtilis open reading frames. Hybridization of total mRNA isolated from B. subtilis cells grown under different growth conditions, or from B. subtilis mutant strains, with these DNA-array filters one can understanding global expression profiles. Such complete gene expression profiles enable to study cellular processes in a detail, formerly it was impossible. This new and powerful technique is very useful to understand the regulatory processes, underlying the cellular differentiation processes occurring during starvation of B. subtilis cultures, such as competence development, peptide antibiotic production and protein secretion.

https://www.slideshare.net
The diagram shows the expression of proteins analyzed in 2-D polyacrylamide gel. The first dimension is done by using isoelectric focusing, then it is laid on polyacrylamide gel and stained with coomassie blue and radioactivity of labeled proteins is shown.
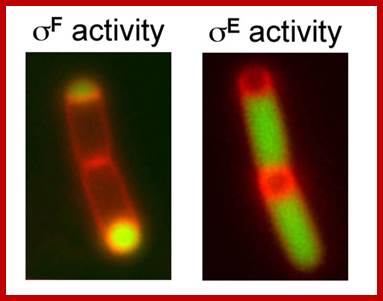
Position of sig F activity in fore spore and sig E in the mother cell; www.sfsoo.com/diagram/diagram-of-protein-secretion
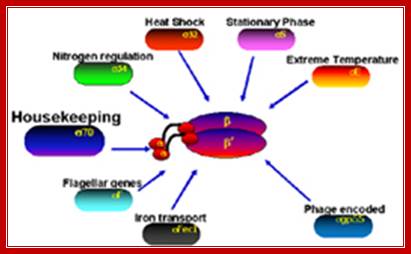

E.coli sigma factors? chinaaoju.blog.163.com
B.subtilis generates more than ten sig factors. Each of them bind to common RNAPs but bind to specific promoter elements. The RNAP of B.subtilis contains an additional subunit called delta (21kDa); this is supposed to invoke gene specificity.
Overview of Bacillus subtilis sigma factors and their functions ( from subtiwiki):
· SigA: household sigma factor
(list of SigA controlled genes)
· SigB: general stress sigma factor
· SigD: sigma factor for chemotaxis and motility genes
· SigE: early mother cell-specific sporulation sigma factor
· SigF: early forespore-specific sporulation sigma factor
· SigG: late forespore-specific sporulation sigma factor
· SigH: sigma factor that controls genes of the transition phase
· SigI:
· SigK: late mother cell-specific sporulation sigma factor
· SigL: enhancer-dependent sigma factor (Sigma-54 family)
· SigM: ECF sigma factor, controls genes required at high salt concentrations
· SigV: ECF sigma factor
· SigW: ECF sigma factor, mediates the transcriptional response to cell wall stress PubMed
· SigX: ECF sigma factor
· SigY: ECF sigma factor
· SigZ: ECF sigma factor
· Xpf
· YlaC
· YvrI-YvrHa: (composite sigma factor)
Vegetative:
Sig A-DNA binding sequences----TTGACA---17---TATAAT,
Sig B—DNA binding sequences---RGGCTTRA—14---GGGTAT,
Sporulation-
Sig E- DNA binding sequences—ZHATAXX-14—CATACAHT,
Sig F-DNA binding sequences—GCATR—15—GGHRARHTX,
Sig G- DNA binding sequences—GHATR—18—CATXHTA
Sig K-- DNA binding sequences---AC-xx-----17---CATANNTA
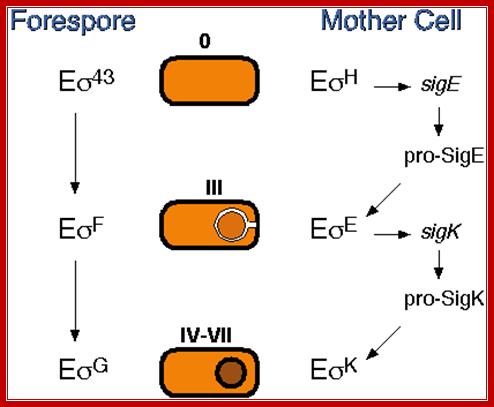
Prespore and Forespore events:
In forespore SpoO-A~p (~23. kDa) activates sig-A (sig43) -RNAP complex that transcribes hundreds of genes required for early stages of sporulation. The important products of this gene expression are sig factor F (sig-F) and sig E and they are segregated into prespore and mother cell respectively; i.e. pre-sigF in prespore and pre-sigE into pre-mother cell. The pre-sigF is produced just before the onset of fully formed cross septum.
Initially activation of sigF is important. But this sig-F is inhibited by anti sig factor spoIIAB (which is interestingly a kinase). But this inhibition is overcome by another anti-anti sig factor spoIIAA, by series of phosphorylation and dephosphorylation events.
If spoIIAA is phosphorylated it cannot interact with spoIIAB. The spoIIE is a phosphatase found in fore spore compartment is responsible for dephosphorylation of spoIIAA. The non phosphorylated spoIIAA interacts with sigF-spoIIAB (which is bound to sigF), and releases sigF from inhibition.
SigF-spoIIABà Inactive,
SpoIIAA àKinase-à SpoIIAA~p (inactive)
SpoIIAA~p (inactive) à SpoIIE Phosphatase > active SpoIIAA.
SpoIIAA (active) + sigF- SpoIIAB (inactive)à active sigF + SpoIIAB
The activated sig-F binding to RNAP activates ( as a Regulon) a group of 48 genes required for the early sporulation steps, yet other genes required for house keeping remain active by the sig43 (= sigA).
In forespore activated sig-F displaces sig-43 (sigA) and transcribes some early sporulation genes. One of the products is pro sig-G; but it is in inactive state. The pro sig-G is activated by components (spoIIIA) of mother cell through cross talk across the septal membranes. The sig-G displaces sig-F and transcribes late sporulation genes.
In mother cell spoOA activates several genes using RNAP with sig 43/sig A complex. One of the products is pro-sig-E; and it is in inactive state. Activation of this is facilitated by the forespore products. One of the early forespore gene product activated by forespore sigF is spoIIR. It is secreted across the membrane into space between two membranes found in between mother cell and forespore. The spoIIR activates a membrane bound spoII-GA in the mother cell, which cleave C-end of pro sig-E by its protease activity. The pro-sig-E remains inactive till the fore spore is formed. Remember that any sig-E produced in forespore is immediately degraded. Similarly any sig-F synthesized in mother cell is degraded.
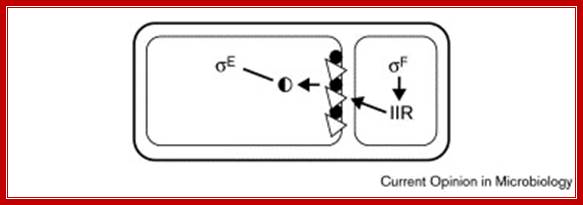

Activation of pro sig E; Mother-cell-specific processing of pro-σE in response to a signal from the forespore. σF RNAP (σF) transcribes the spoIIR gene in the forespore. SpoIIR (IIR) is secreted from the forespore and activates SpoIIGA (triangles) to cleave pro-σE (filled circles) to σE (half-filled circle) in the mother cell. http://chinaaoju.blog.163.com
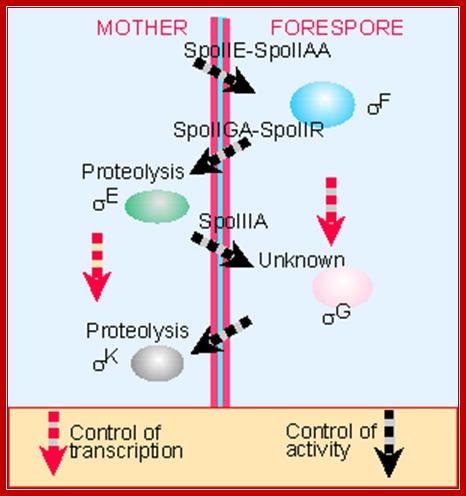
Cross talk between forespore and mother cell during spore formation; http://flylib.com/books
http://www.microbiologia.altervista.org
This diagram shows spore formation goes though several stages at least four, so also four fluxes of gene expression, activation and inactivation, takes place
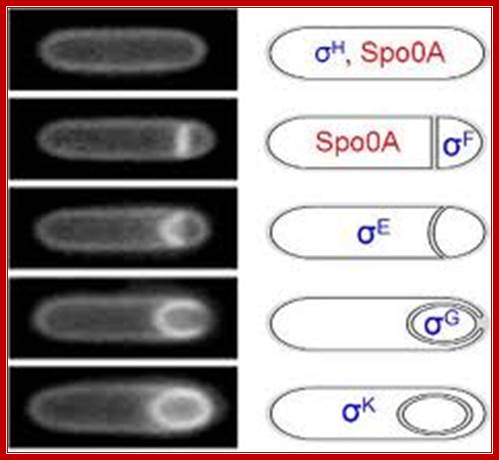
Coupling of gene transcription and morphogenesis; http://nsmn1.uh.edu/
http://www.nature.com/
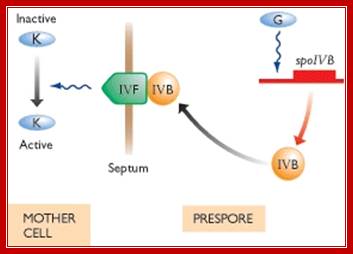
In mother cell sigE activates hundreds of genes; one of the genes that is transcribed among many is sig K. The sig K remains as precursor for in the middle of its gene a 4kbp long phage DNA segment is found. This segment is removed by a recombinase (spoIVCA) and the active sigK is generated. Activity of sig-G is required to generate signals that are transmitted across the septal membrane to activate pro sig-K in mother cell by spoIVFB, which is a protease.
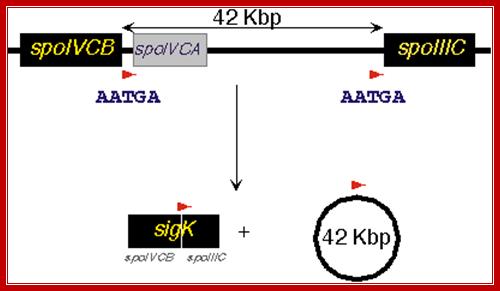
The diagram shows recombination to generate active sig-K
Sig-K is a recombinant product. The active sig-K displaces sig-E from the sigE-RNAP complex and sets activation of many other genes for the late stage spore formation. The sig-G activated by sig-K, replaces sig-F and induces many more genes, at least 81 genes, required for the late spore development stages.
In all these activations, sig-43 is displaced by several newly formed sig factors. The sig-43 though displaced by other sigma factors, is yet active for the expression of genes required for normal cellular activities. Thus one can observe a fascinating display of cascade of sigma factors and their power in the development of spores. In fact mother cell contributes to spore wall formation, the required components are synthesized and secreted across the membranes.
Summary of the role of cell and sigma factors in spore formation;
Environmental signals received at the membrane level are transduced to activate the process of sporulation in cascade manner of gene expression. The kinases involved are KinA, KinB, KinC, KinD and KinE. One of the early events is the activation of Histidine kinase called Kin-A and Kin-B, which p-lates amino acid aspartate of Spo OF, which in turn p-lates spoOB in successive manner; ultimately to produce activated form of Spo OA-p.
KinB and Kin A-p>>spo OF-p .>> spo OB-p >> spo OA-p >>
http;//www.mmbr.asm.org

An outline sigma factor cascade; http://biology.kenyon.edu

http://biology.kenyon.edu/

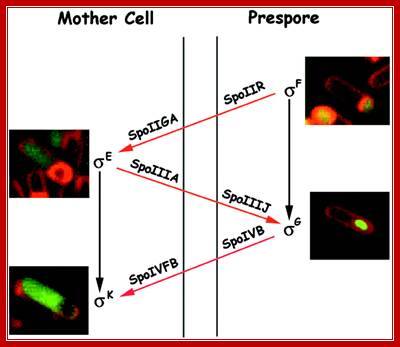
Note here the interaction between the products of forespore and mother cells. Sig-F activated factors interact with mother cell factors and vice-versa. Sig E activates sig-G and sig-G activates sig-K; http://www.flandershealth.us/
|
Sporulation Cascade: |
|
|
Stress Signals such as pH change, cold, heat, starvation are impinged upon the cell surface receptors, which activate kinases (Two components Histidine kinase?); kinases go through a cascade of events ending in activation of spoOA. |
|
|
KinB and Kin A-p>>spoOF-p .>> spoOB-p >> spoOA-p* >>
|
|
|
Activated spoOA~p interacts with RNAP containing sigma A (43) and sigma H (minor factor), activate the transcription of 10% or more of cellular genes; among them are sigF and sigE; they are expressed even before prespore engulfment. Septal formation segregates sig F to forespore and sig E in mother cell; a copy of DNA is transported into prespore. SpoIIIE is responsible for threading the DNA into the fore spore. SigF is the first to be activated. Plasma membrane induced septal formation divides the cell into polar smaller prespore and larger mother cell. Both sig factors exist in precursor forms. |
|
|
Events in Mother cell- (sigE)
|
Events in Forespore cell- (sigF) Prespore matures into Forespore |
|
RNAP with sig-A transcribes to produce pro sig-E (inactive form) |
Host sig H expresses Sig F, but it is inhibited by anti sigma factor SpoIIAB, so sig F is inactive state; later SpoIIE by its phosphotase activity dephosphorylates spoIIAA ; the activated spoOIIAA (anti-anti sig factor) removes anti sig spoIIAB from the complex ; |
|
|
Thus Sig-F becomes active; |
|
Any sigF produced in mother cell is inactivated or degraded |
Any sigE produced in forespore is degraded |
|
|
|
|
|
Now sigF activates transcription of several genes (at least 48 genes), some of them are sporulation specific. |
|
Components of mother cells communicate with forespore cell components and vice versa |
Components of forespore cells communicate with mother cell components and vice versa |
|
|
One of the products of sig-F is spoOII-R, which is secreted into space between forespore and mother cell; this activates spoIIGA located on the side of mother cell membrane |
|
Inactive Sig E requires proteolysis at its C-end which is localized in the membrane. Activated spoIIGA cleaves the C-end of sigE end and sig E becomes active |
|
|
sigE is a Regulon activates a large number of genes; GerR and spoIIID are important for they inturn activate many and inactivate many, |
|
|
|
SigF activates several genes, one of them is sig-G, precursor, it is inactive , |
|
Active sigE in mother cell is required for the activation of sigG in the fore spore cell. |
Sig-G is activated by mother cell components, perhaps the process is similar to that of sigF (spoIIIA, spoIIJ) |
|
One of the genes expressed is sigK. The sigK is released by recombinase (spoIVCA) and activated by the action of specific proteases (spoIVFB). |
Active sigG in forespore cell is required for the activation of sigK in the mother cell.
|
|
Sig-K displaces sig-E and transcribes late sporulation genes. |
Sig-G displaces sig-F and sponsors activation of late sporulation genes |
|
Sig E expresses 263 genes(88 operons) |
|
|
SigK activates >75 genes |
sigG activates at least 81 genes. |
Cascade of sigma factors:
|
Sigma factor |
Gene |
Sig binding sequence |
function |
|
Sig43(sigA) |
Sig-a 43KD |
TTGACA-TATAAT |
Principle factor |
|
SigB
|
Sig-b |
TTGAA-TATAAT |
unknown |
|
Sig C |
Sig-c, 32KD |
|
Sporulation specific |
|
SigD |
Sig-d |
|
Flagellar gene transcription |
|
SigE |
Sig-e(spoTTGB) 24KD |
AATC-TAATGCTTT |
Sporulation specific in mother cell |
|
SigF |
Sig-f (spoIIAC), 28kDa |
|
Gene expression in fore spore |
|
cSigG |
Sig-g(spoIIIG) |
pYGNATpu-cANT/A |
Gene expression in fore spore |
|
SigH |
Sig-h(spoH),32KD |
|
Transcription of early sporulation genes |
|
SigK |
Sig-k(spoIVCB& spoIIIC |
|
Late gene expression in the mother cell |
Table: Functional Roles of Subsystem- Spreadsheet.
|
|
Specific Sigma factors for cellular functions and sporulation |
|
RpoS |
RNAP Sigma A |
|
FliA |
Sigma- flagellar operon |
|
SigE |
RNA polymerase sporulation specific sigma factor SigE |
|
SigF |
RNA polymerase sporulation specific sigma factor SigF |
|
SigG |
RNAP sigG |
|
Sig K |
sporulation |
|
HrdA |
principal RNA polymerase |
|
HrdC |
principal RNA polymerase |
|
HrdD |
principal RNA polymerase |
|
SigW |
principal RNA polymerase |
|
SigV |
principal RNA polymerase |
|
SigZ |
principal RNA polymerase |
In addition there are External Cytoplasmic Factors (ECF) as sigma which perceive external stimuli; ECF sigma factors constitute a diverse family of proteins. Most members of the family studied to date are known to regulate gene expression in response to stress conditions. The Bacillus subtilis genome encodes at least 17 distinct sigma factors, seven of which are members of the ECF subfamily. Among these, five sigma factors, namely SigV, SigW, SigX, SigY and SigM, are encoded by the first genes of the cognate sigma operons.
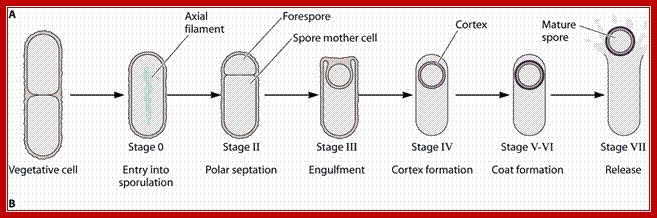
Sporulation cycle; http://www.studyblue.com/
Carefully observe the invaginating and membrane and after enclosure the topology of each membranes. https://micro.cornell.edu
Endospores:
The mother cell in which endospore is produced is called sporangium. Position of the endospore can be any where, ultimately sporangium dissolve leaving endospore free. Endospores consists of three layers, in some fourth layer called Exosporium is also present. Spore coat consists of outer spore wall and the inner spore wall and the middle cortex. It acquires resistance because it has a very thick coat with calcium dipicolinic acid; it has specialized proteins (small mol.wt) which bind to DNA and protect it from damage; dehydration of cortex material protect the cell from various environmental stress like heat, cold, UV and chemicals. A kind of metabolic turmoil sets in the cell. Some low molecular weight proteins produced bind to DNA and protect it from any damage to it. Besides cells produce Dipyridine carboxylic acid (DPA) with Ca2 and excess Ca^2 is accumulated. They also generate dihydropicolinic acid; diaminopimilinic acid and generate lot of peptidoglycans. They are deposited in their three cell wall layers. The mother cell also contributes to forespore formation of outer spore coat and exo-sporium formation. As spores mature, the spore wall and the stored contents get dehydrated and now they can withstand extreme conditions and survive for a longer periods.
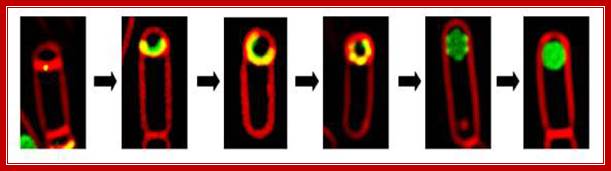

Sporulation in Bacillus subtilis involves an asymmetric cell division followed by differentiation into two cell types, the endospore and the mother cell. The endospore coat is a multilayered shell that protects the bacterial genome during stress conditions and is composed of dozens of proteins. Recently, fluorescence microscopy coupled with high-resolution image analysis has been applied to the dynamic process of coat assembly and has shown that the coat is organized into at least four distinct layers. In this Review, we provide a brief summary of B. subtilis sporulation, describe the function of the spore surface layers and discuss the recent progress that has improved our understanding of the structure of the endospore coat and the mechanisms of coat assembly. http://www.nature.com/
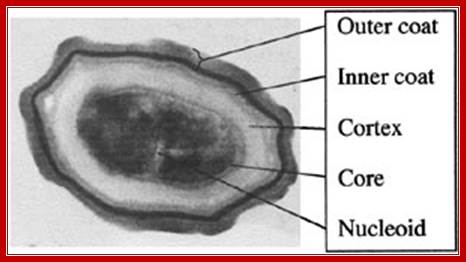
Electron microscope cross-section of a spore of Bacillus subtilis, showing the cortex and coat layers surrounding the core (dark central area). The spore is 1.2 microns across, about 100 times smaller than the width of a human hair. (Credit: S. Pankratz); Robert Sanders; http://www.berkeley.edu/
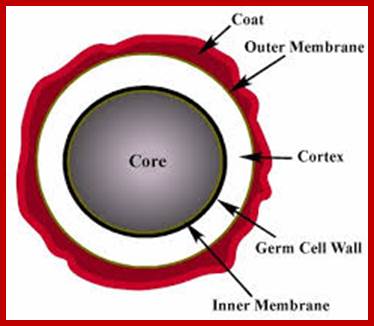
http://www.microscopesblog.com

B.anthracium; http://textbookofbacteriology.net
The mother cell in which the spore is formed is called sporangium. The position of the spore is mostly at one of the ends in the cell; ultimately the mother cell is lysed/dies and the endospore is released. This endospore can remain for long-long time weathering all hostile conditions up to 7500 yrs (record!).
Spore coat will be impregnated with calcium, DPA and far excess amount of cysteine. Nearly 80% of the material accumulated looks like keratin substance. It is very hard, tough and resistant to various factors. Even the DNA is bound by special proteins, leads to dehydration and solidification, thus resistant to desiccation. Cytoplasm also stores DNA repair enzymes and reserve food materials and eventually protoplasm gets dehydrated. Some coat proteins identified on SDS gel are cot A65Kd, cotB 59kd, cotG37Kd and cotC 12Kd.
labs.mcb.harvard.edu/burton/research.html
In the case of B. anthracis nearly 750 proteins are produced; this is analyzed by DNA micro array and multidimensional chromatographic analysis of proteins (proteome). The spore coat itself consists of 25-30 proteins. During spore formation nearly 33% of the genome is expressed and used.
Endospore Structure:
Exosporium: thin delicate covering surrounding the spore.
Ø Spore Coat: - lies beneath the exosporium
- composed of several protein layers
- may be fairly thick
- impermeable and responsible for the spore’s resistance to
chemicals.
Ø Ø Cortex: - may occupy half of the spore volume, rests beneath the spore
- Coat.
- made of peptiodgycan that is less cross-linked than the
vegetative cells.
Ø Ø Spore Cell Wall (Core Wall)
- inside the cortex and surrounds the protoplast or core
- the core has the normal cell structures such as the nucleiod, but
is metabolically inactive.
Exosporium: The outermost layer, it is thin and delicate layer, dried membrane like.
Spore coat: It is fairly thick and consists of several layers of proteins; one of them can be cross linked keratin. It is impermeable to a variety of chemicals and resistant to such toxic compounds. The coat also contains enzymes required for germination.
Cortex: It may occupy more than half the spore volume. It is filled with peptidoglycans but loosely cross linked.
Spore cell wall: the innermost layer of the spore. It covers the cell membrane.
The core: the cellular material containing, DNA, ribosomes and other cellular components; all in freezed state and metabolically inactive.
One chemical produced by endospores that is thought to lend to their high resistance is dipicolinic acid. This chemical has been found in the spore cell of all endospores examined. Dipicolinic acid interacts with calcium ions to form calcium dipicolinate, which is the main substance believed to lend endospores their resistance and represents about 10% of the dry weight of an endospore. The spore cell also contains small acid-soluble spore proteins (SASPs). These function to protect DNA from UV radiation, desiccation and dry heat, and they also serve as a carbon and energy source during the germination process (conversion back to a vegetative cell). Another component of endospores that contributes to their resistant to chemical agents is the strong spore coat, which is composed of highly cross-linked keratin. Identification of particular organisms can be aided by the presence, location and size of endospores. Endospores can be located centrally, terminally or sub terminally within a cell. Sometimes the endospore is much larger in diameter than the cell, which causes the cell to appear swollen at the location of the endospore."

Bacterial endosporeas; https://micro.cornell.edu
The resilience of an endospore can be explained in part by its unique cellular structure. The outer proteinaceous coat surrounding the spore provides much of the chemical and enzymatic resistance. Beneath the coat resides a very thick layer of specialized peptidoglycan called the cortex. Proper cortex formation is needed for dehydration of the spore core, which aids in resistance to high temperature. A germ cell wall resides under the cortex. This layer of peptidoglycan will become the cell wall of the bacterium after the endospore germinates. The inner membrane, under the germ cell wall, is a major permeability barrier against several potentially damaging chemicals. The center of the endospore, the core, exists in a very dehydrated state and houses the cell·s DNA, ribosomes and large amounts of dipicolinic acid. This endospore-specific chemical can comprise up to 10% of the spores dry weight and appears to play a role in maintaining spore dormancy. Small acid-soluble proteins (SASPs) are also only found in endospores. These proteins tightly bind and condense the DNA, and are in part responsible for resistance to UV light and DNA-damaging chemicals. Other species-specific structures and chemicals associated with endospores include stalks, toxin crystals, or an additional outer glycoprotein layer called the exosporium
Bacterial Endospores:
In B. subtilis, spores are approximately 1.2 μm in length and are ellipsoidal. Released spores have a clear protective shell known as the spore coat and is comprised of as many as 25 different protomeric components assembled into discrete layers. Panel B shows a typical SDS-PAGE (12.5%) fractionation of solubilized spore coat proteins revealing predominant species
Why the endospore is resistant to heat, freeze, and a variety of toxic materials? It has been estimated that 15% of the spore is all dipicolinic acid complexed with calcium; all localized in the core of the spore. It is believed that heat resistance is due to dipicolic acid; it is now thought that the calcium as calcium-dipicolinate provides that resistance. The calcium dipicolinate with acid soluble DNA binding proteins provide resistance to radiation, desiccation and toxic chemicals. The cell is stored with poly-beta hydroxyalkanate, of which polyhydroxy butyrate is found in abundance.
Stages in Endospore Formation:
1. Signal transduction in cascade manner and activation of sporulation genes.
2. Plasma membrane creates a septum to which one of the daughters DNA is attached in linear state, perhaps to mesosome.
3. The PM continues to grow and engulfs the smaller fore spore cell. Now the spore contains two unit membranes. The outer surface of PM now faces the fore spore. Cortex materials are laid between two membranes. Calcium dipicolinate is the prominent component. Even PHB is stored.
4. Protein coat is added around the cortex.
5. Maturation of the coat protein takes place.
6. The fore spore core is slightly acidic and dehydrated. Small acid soluble proteins (SASps) bind to DNA and protect from UV, heat and other stress factors.
7. Then lytic enzymes release the spore from the mother cell.
The whole process takes about 8-10 hrs.
Spore germination:
The spore can be dormant for a long time. Germination starts with activation, may be a kind of heat treatment with proper nutrients. Peptidoglycans get hydrolyzed and dipicolinate are released. Process of germination is very fast. Metabolic activities are due to the activation pre-existing inactive enzymes. It is then followed by the break down of the wall; this happens with enlargement of the spore, rupture of the wall with release of the young cell enveloped with bare membrane develops its own cell wall. Soon it develops its own cell wall and all the vegetative developmental genes become active.
Factors Influencing Germination of Bacillus subtilis Spores via Activation of Nutrient Receptors by High Pressure:
Elaine P. Black,1,2 Kasia Koziol-Dube,3 Dongsheng Guan,1 Jie Wei,1 Barbara Setlow,3 Donnamaria E. Cortezzo,3 Dallas G. Hoover,1 and Peter Setlow3*
Different nutrient receptors varied in triggering germination of Bacillus subtilis spores with a pressure of 150 MPa, the GerA receptor being more responsive than the GerB receptor and even more responsive than the GerK receptor. This hierarchy in receptor responsiveness to pressure was the same as receptor responsiveness to a mixture of nutrients. The levels of nutrient receptors influenced rates of pressure germination, since the GerA receptor is more abundant than the GerB receptor and elevated levels of individual receptors increased spore germination by 150 MPa of pressure. However, GerB receptor variants with relaxed specificity for nutrient germinants responded as well as the GerA receptor to this pressure. Spores lacking dipicolinic acid did not germinate with this pressure, and pressure activation of the GerA receptor required covalent addition of diacylglycerol. However, pressure activation of the GerB and GerK receptors displayed only a partial (GerB) or no (GerK) diacylglycerylation requirement. These effects of receptor diacylglycerylation on pressure germination are similar to those on nutrient germination. Wild-type spores prepared at higher temperatures germinated more rapidly with a pressure of 150 MPa than spores prepared at lower temperatures; this was also true for spores with only one receptor, but receptor levels did not increase in spores made at higher temperatures. Changes in inner membrane unsaturated fatty acid levels, lethal treatment with oxidizing agents, or exposure to chemicals that inhibit nutrient germination had no major effect on spore germination by 150 MPa of pressure, except for strong inhibition by HgCl2.
Spores of Bacillus species are germinated by a variety of agents, including chemicals such as specific nutrients, a 1:1 chelate of Ca2+ and pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]), and surfactants such as dodecylamine. Nutrients trigger germination by binding to receptors located in the spore's inner membrane and encoded by tricistronic operons. There are three such receptors in B. subtilis spores, the GerA receptor responding to L-alanine and the GerB and GerK receptors that together respond to a mixture of L-asparagine, glucose, fructose, and K+. Stimulation of these receptors triggers the release of the spore's large depot of DPA, and this triggers activation of the enzymes CwlJ and SleB, either of which can initiate hydrolysis of the spore's peptidoglycan cortex leading to completion of spore germination. Spore germination with exogenous Ca2+-DPA is by activation of CwlJ, whereas dodecylamine causes release of endogenous Ca2+-DPA from the spore core, leading to subsequent events in germination.


