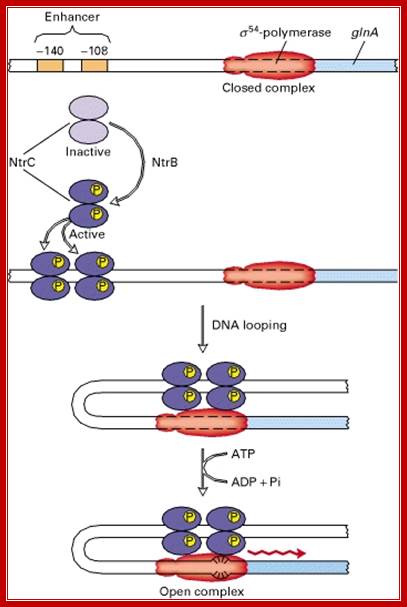
Regulation of Glutamine Synthase-A Gene:
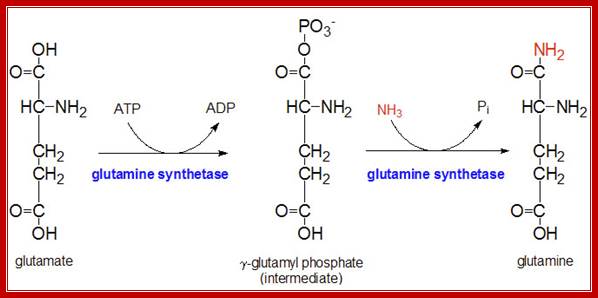
One of the well-known allosteric enzymes is glutamine synthase-A. This enzyme converts glutamate to glutamine, an amide, which acts as the reserve for excess ammonia.
Synthesis of this enzyme is regulated by a protein called Nitrogen regulator protein-C (Ntr-C). The Ntr-C is by itself inactive, but gets activated, when another protein called Ntr-B, which is a kinase, phosphorylates Ntr-C to Ntr-C-p.
The activated Ntr-Cp has an important role in enhancing transcriptional efficiency of several operons by several folds. It regulates nitrogen metabolism and its related genes, under starvation. It acts on more than 25 operons controlling more than 75 genes. The Ntr C is considered as a Regulon. The said protein becomes a dimer; it is in this state it binds to regulatory regions of the DNA of the gene.
---------I-------I----140--I--------I- -108--I -10-I+1>---------->
Enh Enh P gln A
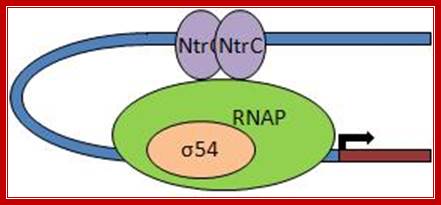
The RNAP holozyme binds to the promoter with specific sig-54 factor, but by itself it cannot initiate transcription with the required efficiency. The sig factor 54, under N2 starvation condition, gets activated and replaces sig-70. The phosphorylated Ntr-Cp recognizes the – 140 and --108 region, as dimers and bind to each site, leads to protein-protein interaction and DNA looping, thus the factors are brought in contact with RNAP Holozyme; this activates the enzyme and initiates the transcription with higher efficiency.

http://dwb4.unl.edu/ Hort Purdue - Purdue University
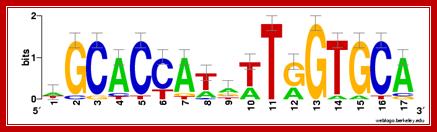
E.coli glnA promoter with palindromic sequences; http://ccmbweb.ccv.brown.edu
The polymerase binds to the glnA promoter, forming a closed complex, before being activated. In response to a low concentration of organic nitrogen, a protein kinase called NtrB phosphorylates dimeric NtrC (purple), which then binds to two enhancers (orange) centered at −108 and −140 from the transcription start site. The bound phosphorylated NtrC dimers interact with the bound σ54-polymerase, causing the intervening DNA to form a loop. The ATPase activity of NtrC then stimulates the polymerase to unwind the template strands at the start site, forming an open complex. Transcription of the glnA gene can then begin.
http://themedicalbiochemistrypage.org/
Note that NtrB and NtrC, discussed in this context, function as sensor and response regulator proteins, respectively in the two-component regulatory system that controls transcription of GlnA. Here the NtrC factors act as enhancers, a rare but an unique example for prokaryotic systems.
http://themedicalbiochemistrypage.org/
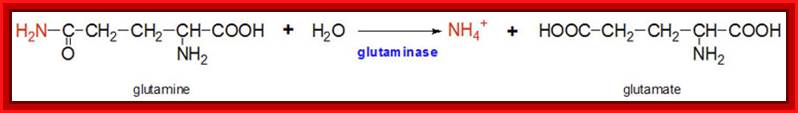
The glutamine synthetase reaction is also important in several respects. First it produces glutamine, one of the 20 major amino acids. Second, in animals, glutamine is the major amino acid found in the circulatory system. Its role there is to carry ammonia to and from various tissues but principally from peripheral tissues to the kidney, where the amide nitrogen is hydrolyzed by the enzyme glutaminase (reaction below); this process regenerates glutamate and free ammonium ion, which is excreted in the urine.
Unlike the lac operon, the enhancer of the glnA gene is a little far from the promoter so that the bound activator does not immediately contact the polymerase. Nitrogen Regulatory Protein C (NTRC) can induce DNA looping to bring the activator in contact with the polymerase.
(a) The glnA gene is transcribed by the
Sigma-54-containing polymerase which alone cannot initiate transcription. The
unphosphorylated NTRC dimers can bind only one site at the enhancer, still
insufficient to stimulate transcription.
(b) The phosphorylated
NTRC dimers can bind both sites of the enhancer.
(c) Their binding induces
DNA looping. Contact between the activator and the polymerase stabilizes the
interaction between the polymerase and DNA, thereby initiating transcription. http://www.web-books.com/
Sigma54 regulates transcription initiation by preventing open complex formation in the absence of an activator poteinh such as NtrC. Larry Friedman et al; ttp://www.bio.brandeis.edu/
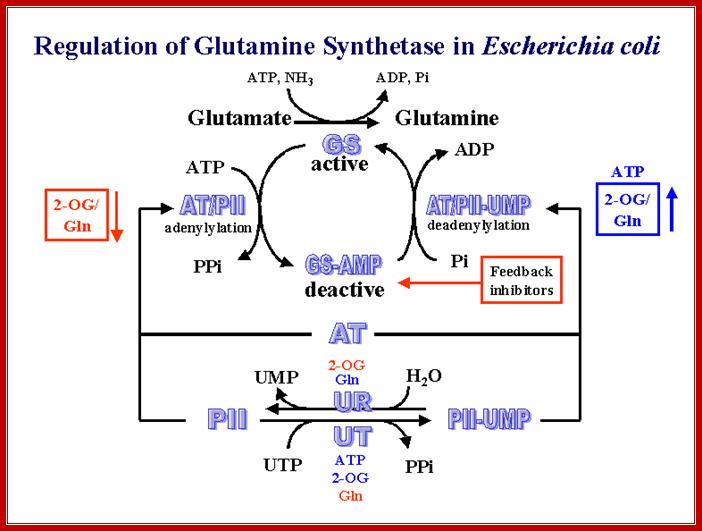
It should be known that the enzyme Gln-A synthase is inactive when it is not adenylated, but becomes active when it is adenylated. It is in this form glutamine is synthesized from glutamate; this is a major pathway in fixing ammonia generated during nitrogen and nitrate conversions.
Adenylated and non-adenylated gln-synthases (Gln-A and Gln) have very important regulatory roles to play in the regulation of Hut operon (Histidine Utilizing operon).
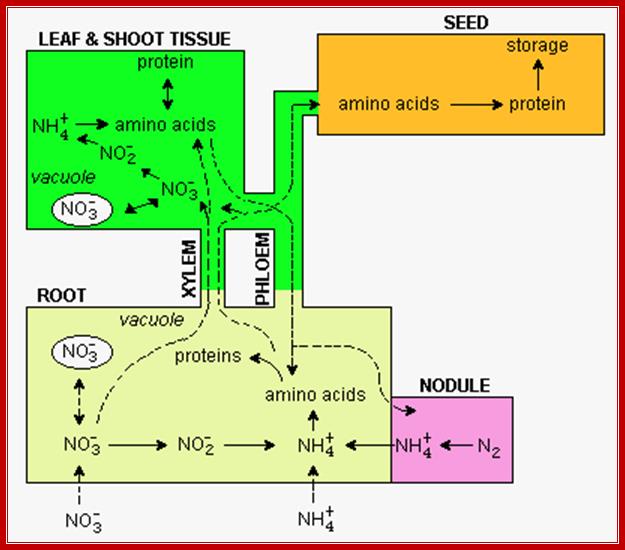
Diagram showing pathways leading to Nitrogen fixation in Plants; http://www.uky.edu
Regulation of Gln synthase-A in Vegetative and Heterocyst cells:
GlnA-synthase is also expressed in both vegetative cells and Heterocyst cells of several blue-green algae called Cyanobacteria. The expression of Gln-A enzyme depends upon the kind of sigma factor produced or made available at a particular stage.
Promoter organization is different for the gene Gln-A synthase, one for the vegetative form, which is constitutively expressed, and another promoter is used for the expression in Heterocyst cells, which develops when the algae is starved of Nitrogen source.
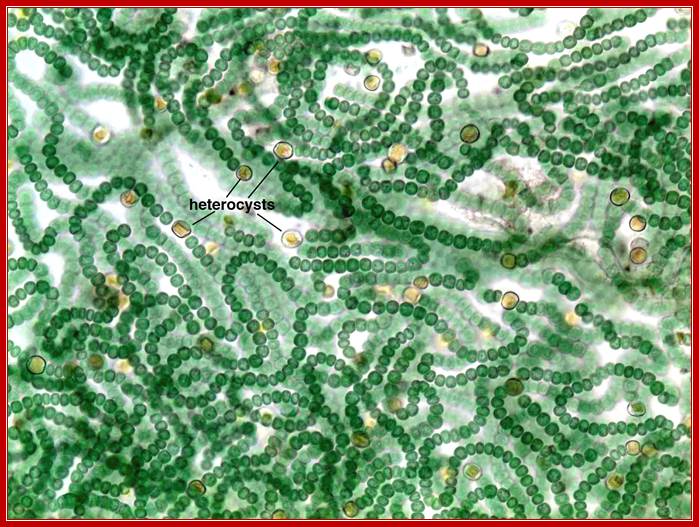
Micrograph of Nostoc filaments with specialized nitrogen-fixing cells known as heterocysts. http://academic.reed.edu/

Heterocyst metabolism and nitrogen fixation. Abbreviations: AcCoA, acetyl coenzyme A; Arg, arginine; Asp, aspartate; b/f, cytochrome b6f complex; F6P, fructose 6-phosphate; PetF, vegetative cell type ferredoxin; Glu, glutamate; Gln, glutamine; OAA, oxaloacetate; 2-OG, 2-oxoglutarate; 6PG, 6-phosphogluconate; PGA, 3-phosphoglycerate; Pi, inorganic phosphate; R5P, ribose 5-phosphate. (Böhme,1998); http://www.intechopen.com/
Gln Synthase under N2 Starvation:
In E.coli, gln-synthase gene has two distinct promoters, located at different positions in the upstream of the start site. Also it has an enhancer region at about - 400 bp from the start.
[P1]-I—EH-I---//--I-35-I-p1-I--10—I+1>- [P2]>--I---I-35-I-p2---I-10-I+1>------>-
RNAP + sig 70 RNAP + sig 54
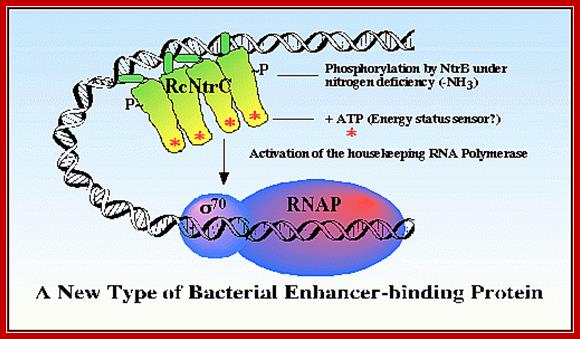
Phosphorylated NtrC binds to upstream of the promoter called Enhancer, interacts with RNAP- sig 54 instead of sig70and activates the transcription; http://image.3sir.net/
Expression of glnA in Escherichia coli is regulated at tandem promoters
Abstract
We have determined that the glnA gene of the complex glnALG operon of Escherichia coli is transcribed from tandem promoters. Expression from the upstream promoter, glnAp1, requires the catabolite activating protein, is repressed by nitrogen regulator I (NRI), the product of glnG, and produces a transcript with an untranslated leader of 187 nucleotides. Expression from the downstream promoter, glnAp2, requires NRI as well as the glnF product; full expression also requires growth in a nitrogen-limited environment. The downstream transcript has an untranslated leader of 73 nucleotides. We also provide evidence that the function of the glnL product is to mediate the interconversion of NRI between a form capable of activating glnAp2 and an inactive form in response to changes in the intracellular concentration of ammonia. The function of the two minor promoters of the glnALG operon, glnAp1 and glnLp, is to maintain the products of glnA, glutamine synthetase, an essential enzyme, and of glnG, NRI, an activator of nitrogen-controlled genes, during carbon-limited growth.
“Operons transcribed by RNA polymerases containing σ70, σ38, σ32, or σ28 are regulated by repressors and activators that bind to DNA near the region where polymerase binds. In almost all cases, repressors bind between +30 and −50 and activators between −30 and −65 in the control region, as in the lac operon.. On the other hand, operons transcribed by RNA polymerases containing σ54 are regulated solely by activators that generally bind between −80 and −160 from the start site. The σ54-activators can activate transcription even when their binding sites, called enhancers, are moved more than a kilo base upstream from the start site.
The best-characterized σ54-activator — the NtrC protein (nitrogen regulatory protein C) — stimulates transcription from the promoter of the glnA gene. This gene encodes the enzyme glutamine synthetase, which synthesizes the amino acid glutamine from glutamic acid and ammonia. The σ54-containing RNA polymerase binds to the glnA promoter, forming a closed complex, but cannot form an open complex and initiate transcription until it is activated by NtrC. NtrC, in turn, is regulated by a protein kinase called NtrB. In response to low levels of glutamine, NtrB phosphorylates dimeric NtrC, which then binds to two enhancer sites upstream of the glnA promoter.. Phosphorylated NtrC bound at the enhancers stimulates the σ54-polymerase bound at the promoter to separate the DNA strands and initiate transcription. This process requires ATP hydrolysis by phosphorylated NtrC, as shown by the finding that mutants defective in ATP hydrolysis are invariably defective in stimulating open-complex formation by σ54-polymerase. It is postulated that ATP hydrolysis supplies the energy required for melting the DNA strands. In contrast, as we saw earlier, σ70-polymerase does not require ATP hydrolysis to separate the strands at a start site, indicating that the molecular mechanisms of open-complex formation by the two forms of E. coli RNA polymerase differ significantly”.
The promoter P1 is recognized by RNAP and sig-70, this is when conditions are normal, but under N2 starvation sig-54 gets activated and replaces sig-70 in the RNAP and the complex recognizes P2 and binds; systems are different..
It is the phosphorylated form of Ntr-Cp binds to the enhancer region at four sites. The enhancer bound factors; interact with one another then by DNA looping they are brought in contact with RNAP. The looping of the DNA is further facilitated by IHF (Integration Host Factor) binding to DNA in between enhancer and promoter region.
This interaction with the RNAP activates the enzyme to form tight open complex to initiate transcription. This, process of activation perhaps requires ATP. The rate of transcription is very high.

http://www.web-books.com/
The enhancer of the glnA gene is located about 120 bp from the start site, containing two binding sites for the transcription factor nitrogen regulatory protein C (NTRC).Unlike the lac operon, the enhancer of the glnA gene is a little far from the promoter so that the bound activator does not immediately contact the polymerase. Nitrogen Regulatory Protein C (NTRC) can induce DNA looping to bring the activator in contact with the polymerase.
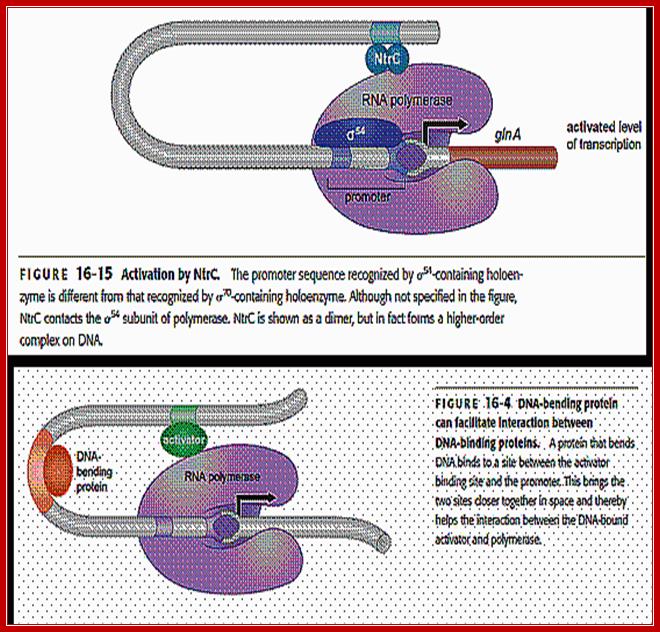


Activastion of GlnA transcription by NtrC; http://www.web-books.com
For NtrC to bind, it requires another protein like IHF which is responsible for bending the DNA so as to facilitate the NtrC to bind to RNAP-sig54 and activate the gene;Enhancer is the DNA element that, upon binding with transcription factors (activators), can enhance the rate of transcription. It may be located either upstream or downstream of the transcriptional initiation site. However, most of them are located upstream. In prokaryotes, enhancers are quite close to the promoter, but eukaryotic enhancers could be far from the promoter.
Unlike the lac operon, the enhancer of the glnA gene is a little far from the promoter
so that the bound activator does not immediately contact the polymerase.
Nitrogen Regulatory Protein C (NTRC) can induce DNA loopingto bring the
activator in contact with the polymerase. The
mechanism of transcriptional activation by NTRC.
(a) The glnA gene is transcribed by the
Sigma-54-containing polymerase which alone cannot initiate transcription. The
unphosphorylated NTRC dimers can bind only one site at the enhancer, still
insufficient to stimulate transcription.
(b) The phosphorylated NTRC dimers can bind both
sites of the enhancer.
(c) Their binding induces DNA looping. Contact
between the activator and the polymerase stabilizes the interaction between the
polymerase and DNA, thereby initiating transcription. https://www.slideshare.net.