Arabinose Operon:
Arabinose is another non-glucose carbohydrate source, for which bacterial cells have genetic capabilities for utilizing it as an alternative source of carbon. For utilization of Arabinose several genes operate. Few genes are required for its transport into the cell. Another set of gene products is required for the conversion of Arabinose into Xylulose. Then another set of enzymes is required for converting Xylulose into Fructose1,6-di phosphate, which becomes the part of glycolytic pathway. Multiple gene metabolic pathway is regulated by a common protein complex; so it is called ‘Arabinose Regulon’.
There are three clusters of genes that utilize Arabinose. Ara C and Ara BAD are located at 1.45 min (map unit), Ara E is at 64.2 minute and Ara FGH at 42.7 minutes. The cluster Ara G, H and Ara E are responsible for arabinose uptake from external medium into cytoplasm and utilization.
AraFGH: Uptake of Arabinose.
--pfgh—I--araF---I—araG—I—araH---I---ter-
AraE: Permease:
--pE—I-----araE-----I ter-
Regulation of these clusters is controlled by repressor C and the mechanism is similar to that of operon Ara-BAD
Ara-BAD:
pBAD—I—B--A—D—I-ter
Pathway starts with LArabinose---(A)-> L.Ribulose—(B)-->RuM’p---(D)-->Xylulose5’P, but the genes for the pathway are not organized in a sequence biochemical pathway operates. The order of cistrons in the operon is p-B-A-D. Line diagram shows the organization of AraBAD operon.
-- o2---C---//--<pC-o1—Crp/CAP—ara-i1/i2-P->----B----I---A----I---D-//-(5600-5800bp)
http://bio3400.nicerweb.com/
I-----1125bp----<I-------250------> I--B842---1--A500---I--D772---I> t/t
------42.2(32)kd----- -66.8Kd 55.8Kd 32 KD
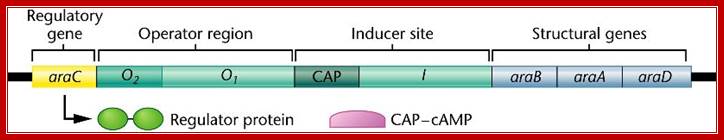
In addition to
structural genes, a catabolite-activating protein (CAP) gene, and Operator region, the ara operon contains a regulatory araC that encodes a regulatory
protein that acts as either
an inducer (in the presence ofarabinose) or a repressor (in the absence of arabinose) by binding to the Inducer site
--<-pC—o1---crp—crp- i1-i2—pBAD->
pC = GTACTG -promoter for the Gene C
pBAD=(-35)TGACG-(-10)TATACTG-promoter for araBAD operon,
o1 = CTCAN-CAC operator abutting promoter for C gene (-120---140).
o2 = operator in the middle of C gene (~ +320).
CRP = TGTGA—AGTGT CRP/cAMP activated receptor binding site-two half sites,
i1/i2 = Inducers sites to which repressor cum activators bind during repression or during activation.
<GTACTG- o2 CTCAN-CAC <o1--Pc— TGTGA—AGTGT - (P -ara(-35)TGACG-(-10)TATACTG)- TATAATà+1
Though many genes are required for the utilization of Arabinose, only 3 of them are clustered into a single unit, and others are scattered. However the AraFGH cluster consists of 3 cistrons and they are independently regulated by Ara C.
AraBAD:
-I---o2----C-----I-----<P/C-o1—cAMP/Crp--i1/i2-->P--B----I---A---I---D---I(5600-5800bp)
The genes for enzymes (all monomeric) responsible for the conversion of L-Arabinose to Xylulose-p are in the following order and the genes are clustered into polycistronic operon.
Arabinose operon- functional genes; www.spectrum.de
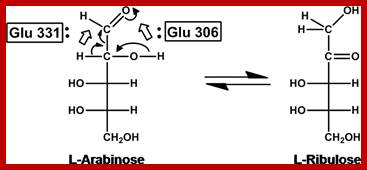
L-Arabinose isomerase isomerizes Arabinose to L-Ribulose; http://aem.asm.org
Components of Arabinose Operon:
|
Gene |
Size(BP) |
Protein |
Mol.wt (KD) |
Functions |
|
C |
1125 |
Repressor |
42.2 (32) |
Represses c and Ara-BAD genes |
|
B |
1842 |
Ribulose- kinase |
66.49kd) |
|
|
A |
1500 |
Isomerase |
55.8 (60kd) |
|
|
D |
772 |
RumP- epimerase |
32 (35)kd |
|
|
O1L |
Has two half sites |
Right to pC |
|
Located to the right of p-C |
|
O2R |
Has one half site |
250 from the pC start |
|
Located at –200 to-250 in the C gene itself |
|
CRP |
Has 2 half sites |
At -120 to 160 2 half sites |
|
Located between O1 and I-1 site |
|
i.1 |
17bp half site |
At -40 |
|
Inducer site |
|
i.2 |
17bp half site |
At -60 |
|
Inducer site |
|
Promoter-C |
|
|
|
|
|
Promoter-Ara-BAD |
|
|
|
Located at the left of Ara-BAD cluster |
Pathway:
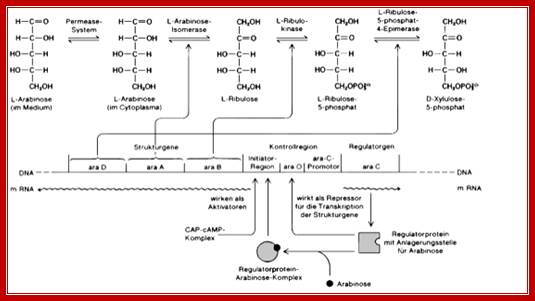
First step- Gene-A:, Arabinose>Isomerase> Ribulose.
Second step- Gene-B:, Ribulose> Ribulose kinase>Rum-5p (Ribulose monophosphate).
Third step-Gene-D:, Rump> Rump epimerase-> Xylulose-p.
There are two other established genes; one Ara-E produces an enzyme called permease for transportation of the sugar into the cell. The other one is gene-F, G and H whose products channel Arabinose into the cell. Both of them are located outside the Arabinose operon and they are regulated independently by the same repressor, so it is called Arabinose Regulon.
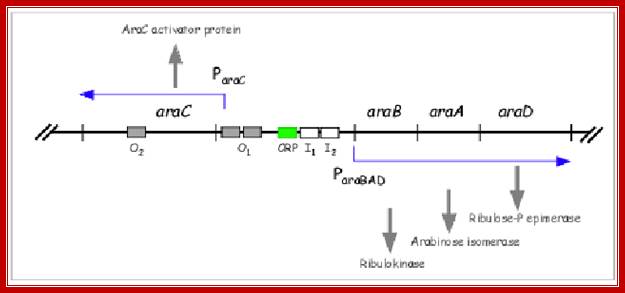
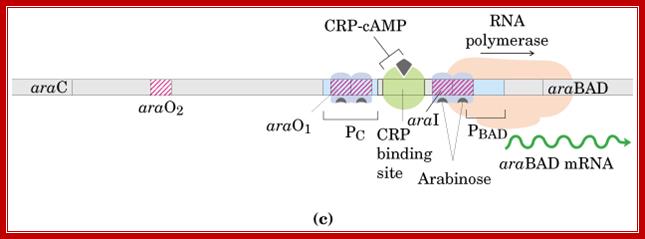
Arabinose operon organization is more elaborate. It has a regulator gene and Ara-BAD genes on opposite sides of an activator region called CRP binding site i.e -146 to -121 and -107 to -78.
On one side of the CRP binding site, there is an Ara-I (initiator), which is split into two 17 base pairs blocks called, i-1 and i-2 located in the upstream of (-78 and -40) -35 and -10> +1 of B gene. On the other side of the cap-binding site, there is a promoter pC, for Ara-C gene, which produces a repressor called Ara-C.
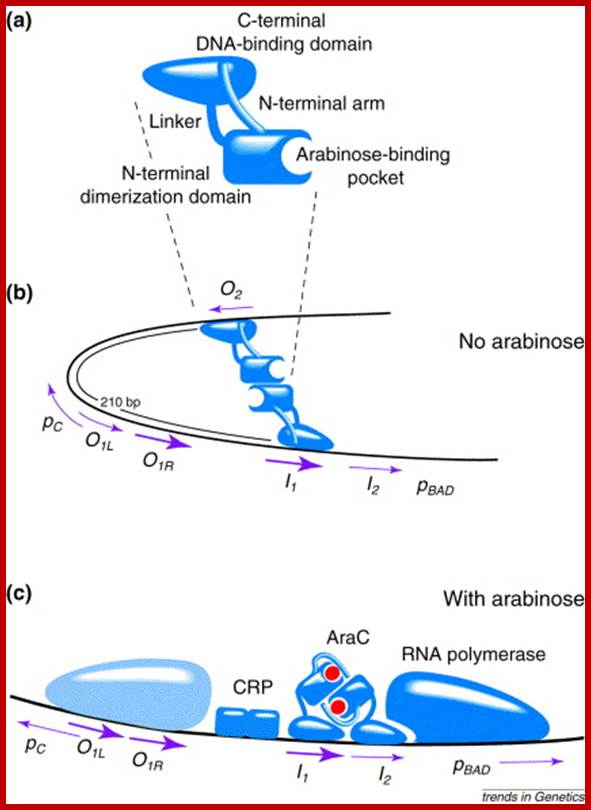
On the right side of pC there is another operator site called o1 which split into o1L and o1R. The repressor has a DNA binding domain, protein-protein interacting domain and a sugar-binding site.
There are two operators; one operator O1 is abutted to right of the pC and the other O2 is located at -250 to -270 from the start of the Ara-C gene.
Structure of Arabinose Operon:
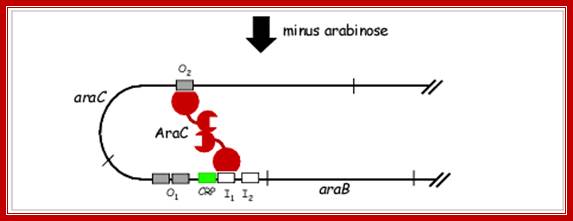
Negative regulation of the arabinose promoter PBAD. In the absence of L-arabinose, AraC functions as repressor by forming a dimer, which inhibits the binding of the RNA-polymerase; http://2013.igem.org/Team:Bielefeld-Germany.
The diagram shows the regulatory elements, pC and pBAD operon; http://faculty.clintoncc.suny.edu/.
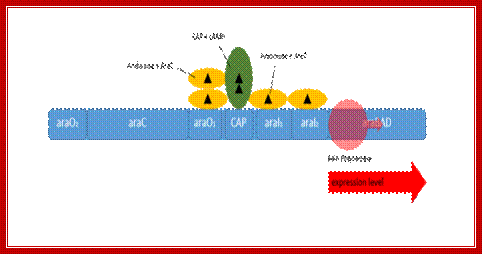
Positive regulation of the arabinose promoter PBAD. When L-arabinose is present the dimer-structure of AraC is relaxed and the AraC protein functions as activator. The activation can additionally be enhanced by the CRP-cAMP-complex;
http://2013.igem.org/Team:Bielefeld-Germany
I--o2--<<IpC—o1—crp-crp—i1-i2—p-BAD--+1>>--B---I—A—I—D-t/t
pC- promoter for C <<GTACTGT at -160 from the START, it produces a repressor as well as an activator
O2- Operator 2 is located in the middle of C-gene at +/- 300.
O1-Operator1 is located next right of promoter for C
Crp-cAMp-binding sequence is located at -100 next to o1-GTG ATTATAG A CAC
i1 and i2: located at -75 from start site, 17bp each
pBAD-promoter for BAD -35 GACGCTTT-------- -10—TCTACTG----9--+1A
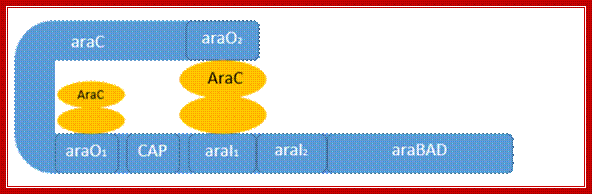
C-Repressor: 42.2 (32) KD, contain two domains. The N domain is protein-protein interacting dimerization domain and it to this L-Arabinose binds. And C domain binds to DNA. The N terminal and C-terminal domain linked by a hinge, which facilitates the bending n looping thus, facilitate interaction between the distal repressors as dimers, but prevents interaction between AraC bound to adjacent sites on DNA. In the absence of arabinose the ara C bound at O2 and ara C bound at i1 interact and the DNA reforms a loop as shown in the figure.
When cells are grown in the presence of glucose, the RNAP holozyme with sigma factor 70 binds to Ara-C promoter and initiate transcription and produces Ara-C protein; it is a repressor. This protein goes on accumulating in the presence of glucose.
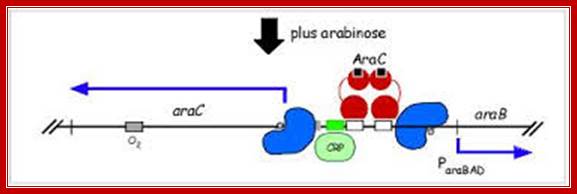
Derepression by ara C. Arabinose bound araC changes its conformation and it frees from O2 and binbds to I1 and interacts with another araC bound to I2. The adjacent ara bound repressors become activator, but require another activator i.e.CRP-CAP;
Regulation of the l-arabinose operon of Escherichia coli; Over forty years of research on the l-arabinose operon of Escherichia coli have provided insights into the mechanism of positive regulation of gene activity. This research also discovered DNA looping and the mechanism by which the regulatory protein changes its DNA-binding properties in response to the presence of arabinose. As is frequently seen in focused research on biological subjects, the initial studies were primarily genetic. Subsequently, the genetic approaches were augmented by physiological and then biochemical studies. Now biophysical studies are being conducted at the atomic level, but genetics still has a crucial role in the study of this system. Robert Schleif; http://www.cell.com
When the concentration of repressor is sufficient it binds to O.2 and i.1 as monomers. Binding to O.1 represses its own transcription (autogenous). Moreover, binding of repressors to i.1 and O.2, leads to protein-protein interaction and the DNA loops results in tight repression. This totally abolishes transcriptional initiation at the promoter region of the Ara-BAD and also at Ara-C promoter. This is when the inducer is absent.
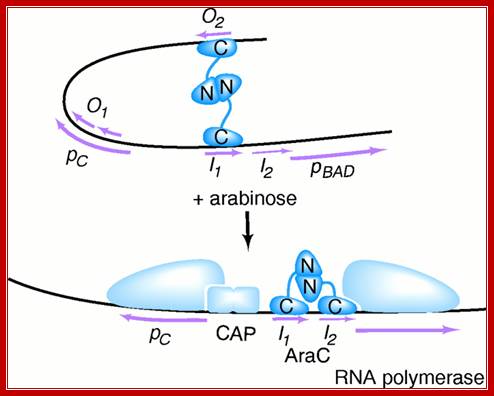
In the absence of arabinose, the Ara C protein binds to both ara I and ara O regions, forming a DNA loop. This binding prevents transcription of the ara operon.;http://biosiva.50webs.org/
In the absence of glucose and in the presence of Arabinose, several inductive events happens instaneously.
A repressor becomes an activator; http://www.sciencemag.org/
The binding of transported Arabinose to the repressor, induces conformational changes in ara Repressor; now it frees from o2 and binds to ara i2 as dimers side by side. At the same time cAMP bound receptor protein, as dimer binds to the activator site and induces a change in the repressor; now the repressor become activators and activates RNA polymerase sigma for induction of transcription.
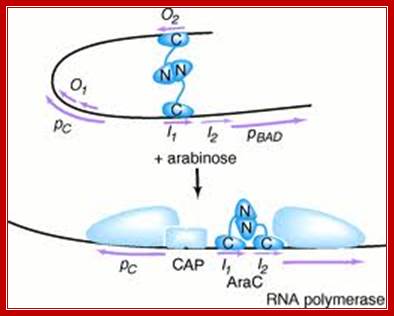
The red circles in the AraC pocket denote the inducer,L.Arabinsoe (Schleif, 2003); When arabinose is present, it binds to the AraC protein and this destabilises the AraC protein binding to the araI1-O2 half-site looped complex, but stabilizes binding to the adjacent half-sites araI1 and araI2, which are upstream of the pBAD promoter. This then ‘straightens’ the DNA loop, (Carra and Schleif, 1993) allowing the activation of the transcription of pBAD (Schleif, 2011). http://www.sciencemag.org/
How does one protein, araC, act to both promote or repress operon activity?. A schematic of the arabinose operon and how the presence of arabinose alters activity of the operon. http://2011.igem.org/; www.Nature.com
In the absence of Glucose cAMP concentration builds up. cAMP binds to CAP binding protein or cAMP receptor (CRP).
At the same time residual exoplasmic Arabinose permease transports Arabinose into the cell. The small inducer molecule i.e. L-Arabinose binds to repressor proteins, which are already bound to O.2 and i.1.
The repressor has DNA binding domain and protein-protein interacting domain. It also contain Arabinose binding site
It is important to remember that O.2 has one half binding site, but and Ara-i have two half binding sites each.
Binding of Arabinose to repressors, induces conformational changes in such a way, the repressor by conformational change becomes an activator. Now the repressor is free from O2 site and binds to I2 and another next to I1; the DNA becomes linear. At the same time, the activators associate with CRP dimeric protein, which is also bound to CAP site. The CRP alone is unable to activate RNAP Holozyme with sig70, which is already bound to its promoter but inactive for the reason that the pribnow box sequence and -35 and -10 sequences are not consensus.
The repressors allosterically activated interact with CRP, then with bound RNAP at pBAD to activate the enzyme. It is speculated that the CTD tail of alpha subunit interacts with Arabinose bound repressor and the other CTD tail interacts with CRP, this leads to the activation RNAP.
The stimulated form of RNAP enzyme initiates transcription with very high efficiency. Here is an excellent example of one kind of protein acting as both negative and positive regulator, remarkable indeed,
Salmonella typhimurium: in salmonella typhimurium, the Arabinose operon is extensive and consisting of seven cistrons such as A, B, O, L, M, N, P, Q and abfa. They too have elaborate regulatory elements.
The Bacillus subtilis L-Arabinose (Ara) operon : nucleotide sequence, genetic organization and expression
Isa be1 Sd-Nlogueira,l Teresa V. Nogueira,'t S6nia Soares'If:
and Herminia de Lencastre1t2
The Bacillus subtilis L-arabinose metabolic genes araA, araB and araD, encoding L-arabinose isomerase, L- r i bulokinase and ~-ribulose-5-phosphate4 -epimerase, respectively, have been cloned previously and the products of araB and araD were shown to be functionally homologous to their Escherichia coli counterparts by complementation experiments. Here we report that araA, araB
and araD, whose inactivation leads to an Ara' phenotype, are the first three ORFs of a nine cistron transcriptional unit with a total length of 11 kb. This operon, called ara, is located at about 256" (mpu) on the B. subtilis genetic map and contains six new genes named araL, araM, araN, araP, araQ and abfA. Expression of the ara operon is directed by a strong &like promoter identified within a 150 bp DNA fragment upstream from the translation start site of araA. Analysis of the sequence of the ara operon showed that the putative products of araN, araP and araQ are homologous to bacterial components of binding-protein-dependent transport systems and abfA most probably encodes
an a-L-arabinofuranosidase. The functions of araL and araM are unknown. An in vitro-constructed insertion-deletion mutation in the region downstream from araD allowed us to demonstrate that araf, araM, araN, araP, araQ and abfA are not essential for L-arabinose utilization. Studies with strains bearing transcriptional fusions of the operon to the E. coli lacZ gene revealed that expression from the ara promoter is induced by L-arabinose and repressed by glucose.
B.subtilis: L-Arabinose Regulon/operon.
----P-/O--+1>-araA>araB> >araD >araL> araM> araN> araP> araQ >abfA.
This operon is 11kbp long located 256’ mpu on the B.subtilis genome map; araB, araB and araD are similar to E.coli araBAD cistrons, but araN, araP, araQ are involved in the transport of Arabinose; in E.coli they are located in separate operon with a common promoter. Ara E located as another cistron with its own promoter. Cistrons abfA codes for arabino-furanosidase, but ara L and araM’s function is yet to be determined which are located as separate operon in E.coli. The araR codes for a regulatory protein of 42.5Kd. Its binding sites are located to the left of A cistron at oRA1-oRA2. The ara repressor also binds to operator region found in area and also its own operators such as oRR3 and to area operator oRE1 and oRE2.