Tryptophan Operon:
The operation of Trp operon is an unusual mechanism in existence. This is not just restricted to one such operon, but found to be in operation among many operons. Examples: Phenylalanine, Histidine, Leucine, Threonine, Isoleucine Valine and others. All have attenuator mechanism in their gene regulation activities. In 1953 Jacques Monod and colleagues, discovered the trp operon in E. coli as the first repressible operon (Wiki). This is another negative regulatory operon.


https://www.studyblue.com; https://www.slideshare.net
Trp-operon; their cistrons coding for specific protein; http://www.bioinfo.org.cn/
Control Circuit for the trp Operon
P/O | L || E | D | C | B | A | Controlling Region || Structural genes|
trp Operon Gene |
Gene Function |
|
P/O |
Promoter; operator sequence is found in the promoter |
|
trp L |
Leader sequence; attenuator (A) sequence is found in the leader |
|
trp E |
Gene for anthranilate synthetase subunit1 |
|
trp D |
Gene for anthranilate synthetase subunit2 |
|
trp C |
Gene for glycerolphosphate synthetase |
|
trp B |
Gene for tryptophan synthetase subunit1 |
|
trp A |
Gene for tryptophan synthetase subunit2 |
http://www.bio.miami.edu
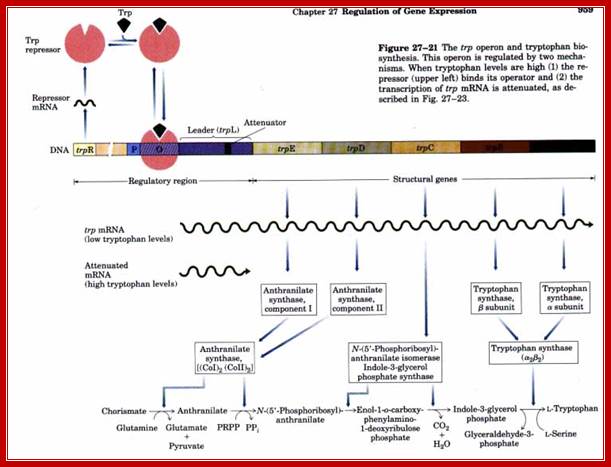
The operon consists of five genes controlled by one promoter-operator. It also has a regulator gene not in the same operon but elsewhere. In between the cistrons one finds spacers. The trpR repressor gene is located elsewhere. The gene produces 12.5kDa protein ((88)108a.a long with short helix turn helix motifs) acts as homodimeric protein binds to 22bp long operator region of the L-trp Operon. This repressor becomes active when trp amino acid binds to the Apo protein. The gene is 1041bp long. This gene is under auto regulation (C.K Singleton etal). The tac promoter contains -35 and -10 sequences as in other E.coli promoter elements.
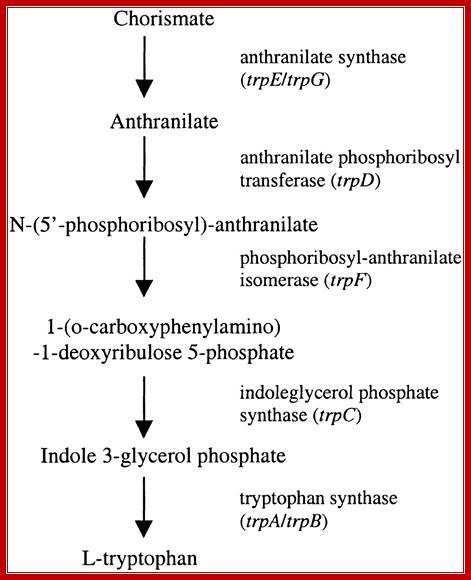
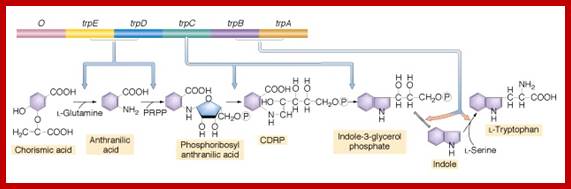
Tryptophan Biosynthetic Pathway in E.coli; http://www.biomed-search.com/
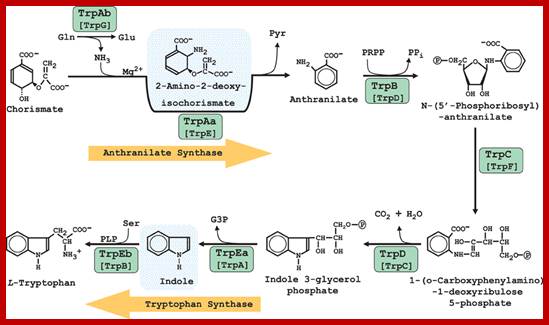
TRP operon and Enzymes in Tryptophan synthetic pathway; http://www.frontiersin.org
When cells face deficiency of Tryptophan, the trp-operon is expressed, but when Tryptophan level is adequate the operon is shut off, but with a difference. Trp-Repressor is unstable and dissociates from the operator often. This operon is leaky; often the RNAP-sig70 complex binds to the promoter operator and continues to transcribe the long upstream region of E’ gene called trL even in the presence of Tryptophan. Production of Trp transcripts when they are not needed is a wasteful expenditure of cellular resources and energy.
Structure of Trp Operon; http://en.wikipedia.org/
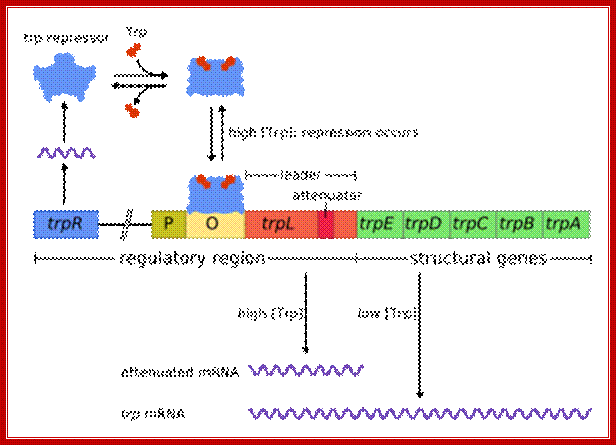
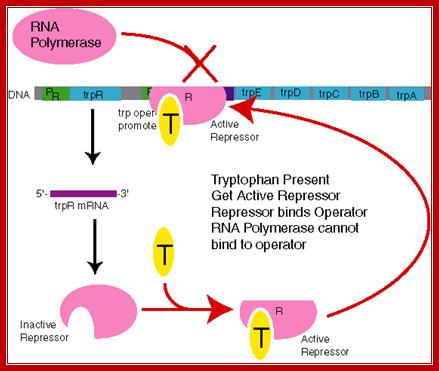
TrpR-regulator gene-repressor; it is homo-dimeric protein (helix turn helix protein), Trp- leader contains attenuator sequence. When Trp is available Trp binds to its repressor and makes it active; it as a dimer binds to binds to its operator site and blocks transcription. But this repressor is very leaky, very often if falls off of the operator and allows transcription. This repressor regulates few other operons, trpR, AroH, AroL and mtr. Consensus DNA Binding Sequence of Trp repressor is GtACTAGTTAACTAGTaC. It negatively regulates gene expression. It is a TrpR Regulon for it controls several related operons..
The conformational change enables this gene regulatory protein to bind tightly to a specific DNA sequence (the operator), thereby blocking transcription of the genes encoding the enzymes required to produce tryptophan (the trp operon). The three-dimensional structure of this bacterial helix-turn-helix protein, as determined by x-ray diffraction with and without tryptophan bound, is illustrated. Tryptophan binding increases the distance between the two recognition helices in the homodimer, allowing the repressor to fit snugly on the operator. (Adapted from R. Zhang et al., Nature 327:591–597, 1987.)
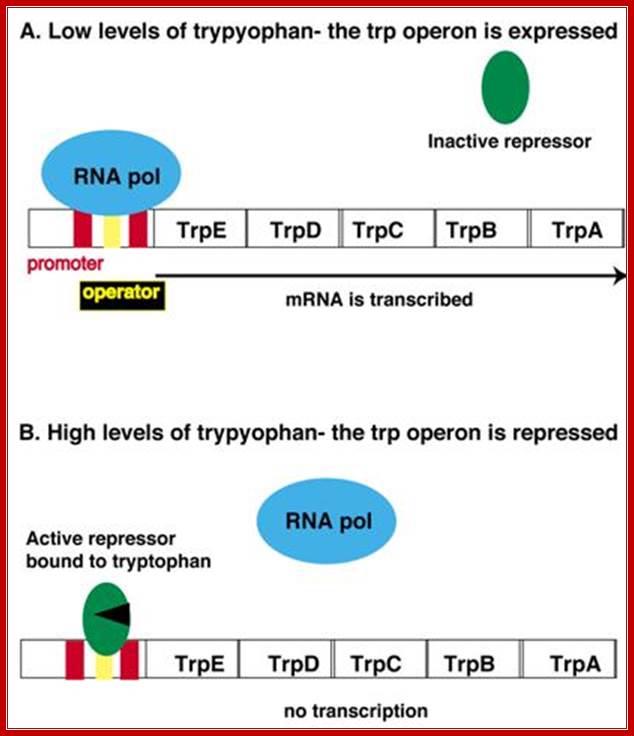
End-product repression of the trp operon.The trp operon expresses a polycistronic mRNA encoding five proteins (Trp A-E) and each catalyzes one step in the tryptophan biosynthetic pathway. These five genes are coordinately regulated by a single promoter (shown in red). Embedded in the promoter is the operator sequence (shown in yellow), which is bound by a repressor protein when there is abundant tryptophan in the cell. When tryptophan levels are low, the trp repressor (shown in green) is made, but it is in an inactive state and cannot bind to the operator. RNA polymerase (shown in blue) is free to bind to the promoter and initiate transcription (Panel A). When there are high levels of tryptophan in the cell, the trp repressor is bound to tryptophan and it is in its active state. The active trp repressor can now bind to the operator, restricting the ability of RNA polymerase to bind to the promoter and thereby repressing transcription (Panel B).Trp operator sequence shows palindromic features: https://wikispaces.psu.edu
ACTAGTT-AACTAGT
The Trp-R constitutively produces the repressor and they dimerize. This protein is in inactive form and it does not recognize the operator region. When repressor is not bound to the operator the operon is active, and Trp is synthesized.
When Trp level raises and no more Trp is required, Trp binds to dimeric repressor in one to one ratio. Binding of Trp to repressor makes it conformationally active and the protein now binds to its operator and tries to block RNAPs transcription.
So cells have evolved a mechanism called Attenuator mechanism to prevent such leaky transcription.
Mechanism of Trp- attenuation: http://en.wikipedia.org/
The attenuator mechanism:
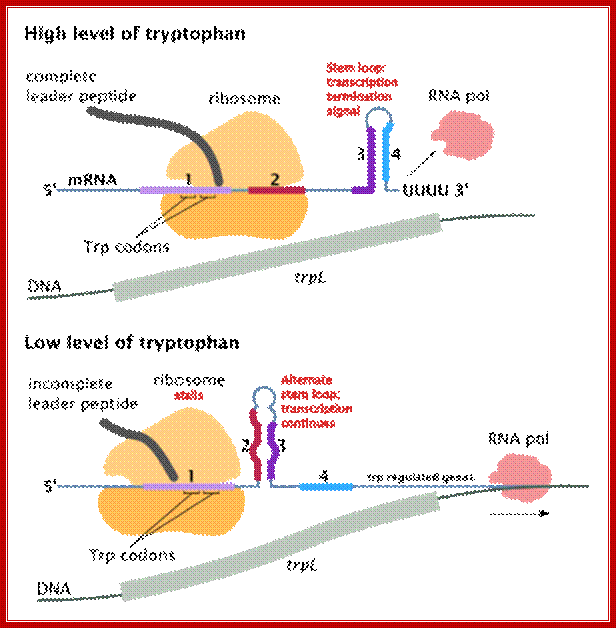
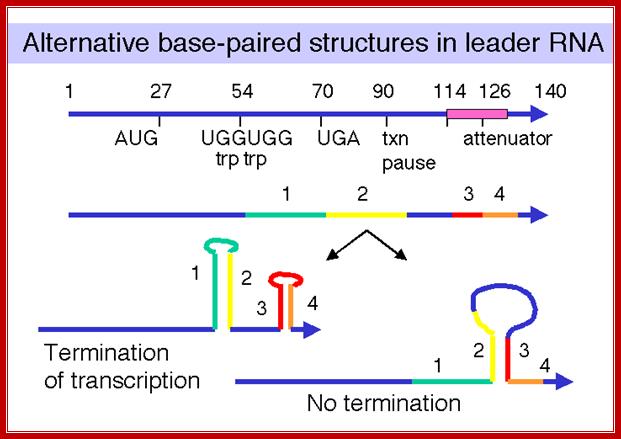
The attenuator mechanism uses the 5’ end of the leader sequence (trpL) of the transcript. The leader is about 140 ntds long; it has four segments of complementary sequences, call 1, 2, 3 and 4. These sequences facilitate partial base pairing between 1 and 2, 2 and 3 and 3 and 4 to generate two stem loop structures.
The 4th segment when pairs with the 3rd, it forms a stem loop similar the rho independent transcriptional terminator structure with terminal Us.
In the presence of Trp, formation of this structure prevents the transcription initiated by the RNAP to proceed further and the transcription is terminated. So this is a fail-proof operation.
http://www.bx.psu.edu
When Trp is absent, the leader sequence can still generate this chain termination stem loop structure and terminate transcription, but transcription of the full-length transcript is required for Tryptophan synthesis. In order to produce full-length transcript the terminator stem-loop formation should be prevented by change in base pairing alignments among the four blocks.
http://2010.igem.org/
http://physics.scsu.edu/
By favoring the formation of base pairing between 2 and 3, the terminator stem-loop structure fails to form, so the enzyme can progress unhindered to generate polycistronic mRNA. Here is how and why hairpin structure develops or don’t develop; they are the key points in understanding attenuator mechanism of regulation.
Trp-R P.O E D C B A t
I------------//----- I-----E-----I------D------I-----C----I------B------I----A-----I
140-162. 1560 1620 1353 1191 804
60kd 60kd 45kd 50kd 29kd
Operator-Promoter-Operator ---------E-----
+-->
CGAACTAG TTAACTA GTACG C AAG
Components of Tryptophan Operon:
|
Gene |
Size |
Protein |
Mol.wt (KD) |
Functions
|
|
Trp-R |
|
|
12.5 monomer |
Repression |
|
P-O |
140-162nt |
Leader sequence |
|
|
|
E |
1560 |
Anthranilate synthase (component 1) |
60 |
|
|
D |
1620 |
Anthranilate synthase (component-2). |
60 |
|
|
C |
1353 |
Phosphoribosyl Anthranilate isomerase/ Indole 3 glycerol phosphate synthase. |
45 |
|
|
B |
1191 |
Tryptophan synthase b subunit. |
50 |
|
|
A |
804 |
Tryptophan synthase a subunit. |
29 |
|
|
|
|
|
|
|
The size of the operon is approximately 6756 bp. The promoter is slightly extended, for its operator segments flank on both sides of the TATAAT box; here in this operon the Pribnow box sequence is TTAACTA not a perfect/ consensus sequence for sigma 70.
On either side of the Pribnow box there are six base pair AACTAG- [-P-]-AACTAG sequences, which are the sites for the binding of trp-repressor.
From the start of the operon, it produces a 140-162 long leader sequence, with two initiator codons, one at 26 ntd from the 5’end of the messenger, another after 14 to 16 ntds from a poly (U) stretch. The second one is actually the correct initiator for the synthesis of first enzyme. The leader sequence has four segments, which can alternately base pair and produce stem loop structures.
Leader L- 162 ntds long: the start and end of cistrons as shown below some overlap.
26
5’AAG- - - - TATCGACAACA ATG AAA GCA ATT PAL VAL-
1 TER 2
--LEU LYS GLY UGG UGG ARG THR SER UGA A ACG GGC A
2 3
-AGUGUA UUC ACC AUG CGU AAA GCA AUC AGA UAC----->
3 4 140
>CCA GCC CGC CUA AUG AGC GGG CUU UUUU - - - - - - - ->
162 Start of E end of E and start of D
-->- ATTAGAGAATAACA /ATG CAA - - - - TTTCTGATGGCT- ->
End of D start of C
- - - - - -CACGAGGG TAA ATG CAA- - - ------------>
End of C start of B
- - - - - - - - - - - - - -TAA GGA AAG GAA ACA ATG- - --->
End of B start of A
---> -- - CGA GGG GAA ATC TGATG CAA - - - - -----
End of A 6710 -------
---> - - -TAA---TCCCAG- - - -AGG- //------TTTTA (end of operon).
Some of the cistrons have overlapping sequences, example TGATG, TGA Ter codon and ATG Initiator codon, where sequences overlap.
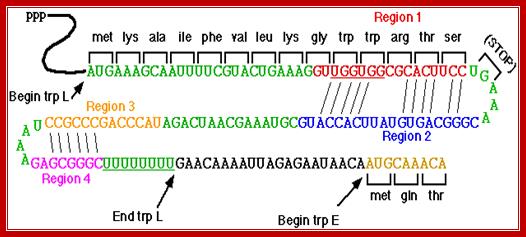
The Trp Operon; http://academic.evergreen.edu/
Formation of the transcriptional terminator stem loop structure or preventing it from formation is the key to the regulation.
Logic of regulation:
In the presence of Trp, transcription operon should not operate, but occasionally it operates; so it is leaky. In the absence of Trp, full length mRNA is produced and the synthesis is 700 times faster than the normal. Hence Trp promoter is considered as efficient for high level of expression.
In the presence of Trp, it binds to inactive repressors; and dimers bind to operator, there by blocking RNAP to bind to its promoter. But the repressor that is bound is not stable, often falls off and RNA-Pol initiates transcription and produces many full-length transcripts.
Repressor is coded separately by trp-R gene. The protein is ~12.5kd and 108 amino acids long. The protein has a helix turn helix motif for the binding to operator region of the DNA in sequence specific manner. It also contains another site for the binding of co-repressor i.e. Tryptophan. The protein also contains domain for protein-protein interaction to produce dimers. In this state they can bind to direct repeat regions in the DNA called operator sites.
In the absence of Trp, repressor does not bind to operator and operator is free which allows RNAP to bind and initiate transcription.
In both the cases the mRNA leader sequence of Trp E gene is 162 ntd long. The leader sequence has two translator initiation sites. The two are separated by a long noncoding sequence.
The first translational region codes for 14 amino acids. At 10 and 11th position two UGG codons for Trp are located. At the end of 14th codon a terminator codon UGA is present, which is actually located in 1st and 2nd segments (which are used for generating stem loop structures) region.
The first segment of coding region is futile in the sense it does not produce any meaningful polypeptide. The second initiator codon is located at 140-ntd position and it is the start of (ORF) real reading frame for the production of trp-E product.
Leader sequence has four segments, which have complementary bases, so segment 1 and 2 can pair, and 3 and four can pair, under one circumstance.
Formation of secondary structure between 3 and 4 favors the formation of transcription terminator stem-loop structure. If a segment 1 and 2 pairing is prevented, and favor 2 and 3 pairing, terminator hairpin structure does not form and transcription will proceed to full length.
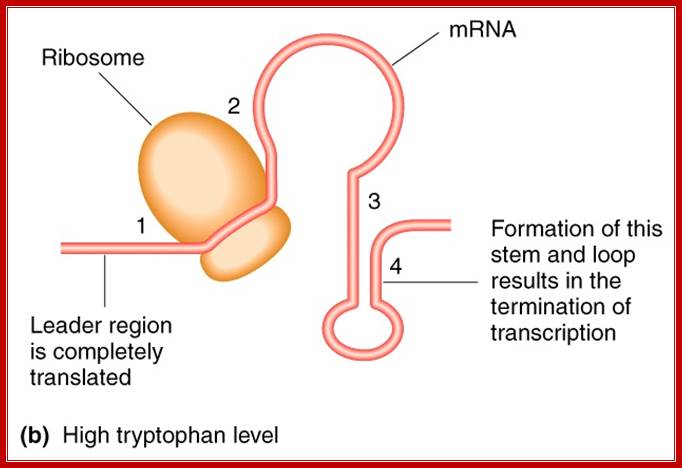
In the absence of Trp, repressor doesn’t bind to operator, and RNAP with sig-70 binds to the promoter i.e. to both –10 and –35 regions and the enzymes initiate transcription. As it proceeds, ribosomes bind to the 5’ end of the transcript and initiate translation.
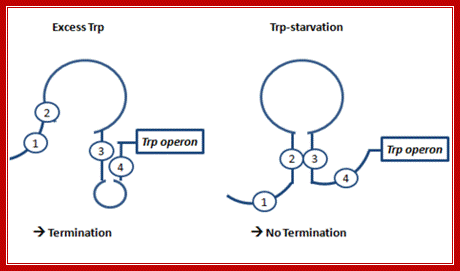
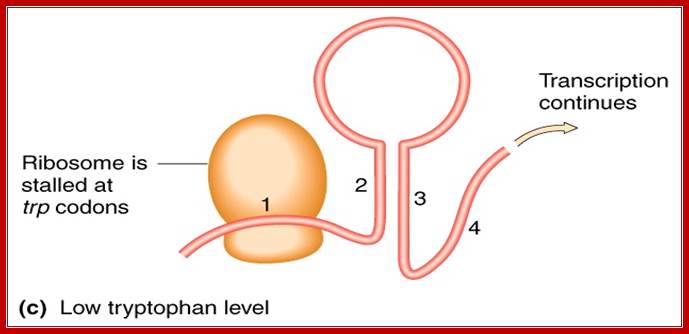
While the transcription is going on, ribosomes continue translation, but when the ribosome reaches two Trp codons it halts for there no trp bound-tRNAs. As ribosomes are large and can occupy about 50-60 nucleotide long spaces, the pairing between 1 with 2 is prevented. This favors pairing between 2 and 3, because of strong affinity between these segments. This pairing prevents pairing between 3 and 4, and it also obviates the formation of terminator stem loop structure; so the transcription can proceed to full length. This is attenuator strategy employed in Trp operon when Trp is absent.
In the presence of trp, ribosomes initiate translation and go through two trp codons and incorporate Tryptophan, and as they move on they come to halt at a TER codon UGA, which is located at loop region of the 1 and 2 stem. Ribosomes, as they occupy substantial length in the leader sequence, the 2 nd segment pairing with 3 is prevented and 3 and 4 pairing is favored, which is what is required for transcriptional termination. Thus this attenuator mechanism is a second defense against leaky transcription, though activated repressors are present.
Other Attenuators:
Attenuator mechanism is not restricted to Trp operon alone, it is also found in many operon systems, especially the operons involved in the synthesis of certain amino acids, such as Phe, His, Leu, Thr, Ile and Val. Apart from the said examples computational analysis shown that there are 80 different orthologous groups regulated by transcriptional attenuation (www.cell.com).
In the methionine synthetic pathway, Met-repressor, which is again a dimer, complexes with s- adenosyl methionine gene and represses its own gene and also those encoding enzymes involved in methionine synthesis. The protein has 5 helical segment and two antiparallel motifs, which interact with half site in the major groove of the promoter element.
The discovery of this type of mechanism to control the expression of genes in a biosynthetic operon lead to its rediscovery in a wide variety of such operons for which repressors had never been discovered. For example:
Histidine: MTRVQFHHHHHHHPD-Histidine operator leader
Threonine: MKRISTTITTITITTGNGAG, Threonine operator leader,
Ilv GEDA: MTALLRVISLLPTAPSAAVVVVRVVVVVGNAP stop.
IlvB: MSHIVRFTGLLLLNAFIVRGRPVGGIQH stop.
Leucine: MSHIVRFTGLLLNAFIVRGRPVGGIQH; stop-leader motif
Phenylalanine: MKHIPFFFAFFFFTFP; stop-leader RNA motif.
Attenuation mechanism is also found in some eukaryotes such as Drosha 5’>3’ exonuclease XRN2 may terminate further transcription by torpedo mechanism.
Regulation of tryptophan operon expression in the Archaeon Methanobacterium thermautotrophicus: Xie Y, Reeve JN.
Conserved trp genes encode enzymes that catalyze tryptophan biosynthesis in all three biological domains, and studies of their expression in Bacteria and eukaryotes have revealed a variety of different regulatory mechanisms. The results reported here provide the first detailed description of an archaeal trp gene regulatory system. We have established that the trpEGCFBAD operon in Methanothermobacter thermautotrophicus is transcribed divergently from a gene (designated trpY) that encodes a tryptophan-sensitive transcription regulator. TrpY binds to TRP box sequences (consensus, TGTACA) located in the overlapping promoter regions between trpY and trpE, inhibiting trpY transcription in the absence of tryptophan and both trpY and trpEGCFBAD transcription in the presence of tryptophan. TrpY apparently inhibits trpY transcription by blocking RNA polymerase access to the site of trpY transcription initiation and represses trpEGCFBAD transcription by preventing TATA box binding protein (TBP) binding to the TATA box sequence. Given that residue 2 (W2) is the only tryptophan in TrpY and in TrpY homologues in other Euryarchaea and that there is only one tryptophan codon in the entire trpEGCFBAD operon (trpB encodes W175), expression of the trp operon may also be regulated in vivo by the supply of charged tRNA(Trp) available to translate the second codon of the trpY mRNA.
Bacillus subtilis- TRAP:
This bug also has attenuator mechanism and it is employed in the synthesis of Tryptophan. In this case, a repressor called Mtr-B binds to the leader sequence of the mRNA and promotes the formation of a terminator secondary structure. This is possible only when Trp is present. When Trp is present, it binds to Mtr-B protein now called TRAP (Tryptophan Activator Protein) and activates it, which in turn binds to the leader of mRNA and induces the formation of terminator stem loop structure and terminates transcription. The protein is called TRAP (Trp Activated (Attenuation) Protein).
In B.subtilis TRAP binds to four sites. The repressor regulates at least four operons such as folate operon, mtr operon (TRAP), trp P( transport) operon, at operon (anti TRAP) and aro supra operon; aro operon consists of six cistrons, cistron F is extra when compared to E.coli Trp operon. So this can be called a Regulon.
TRP operon in B.subtilis consists of six cistrons with spacers- trpE, trpD, trpC, trpF, trpB and trpA in the same order and the TRAP binding site is located at leader region. The folate operon consists of seven cistrons - pabB-trpG-pabC-sul-folA-folK-yazB-yacF and the TRAP binding site is located between G and C.
The TRAP protein is multimer of 11 subunits and each of them bind to one tryptophan, thus the TRAP becomes active and the circularly oriented. Monomer TRAP protein is 75aa long and 8.25Kd. This binds to mRNA especially to leader sequence UAG , CAG, AAG or G/UAG repeats and the mRNA enraps the protein complex and leads to transcription termination by producing terminator loop.

Paul Gollnick; http://wings.buffalo.edu
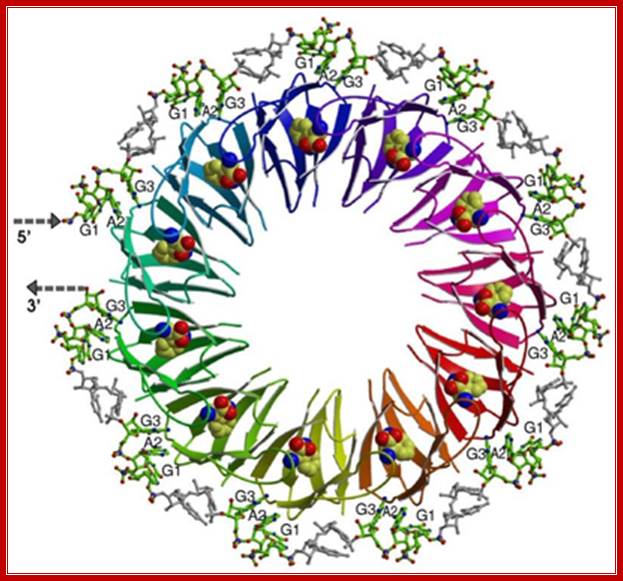
TRAP; Paul Gollnick; http://wings.buffalo.edu/
In the absence of Tryptophan anti TRAP protein coding gene transcribes, while it transcribes, uncharged trp tRNAs bind to elongating mRNA to specific sequences and prevent from chain termination, thus the transcription progresses till the end. Recently, a protein of previously unknown function, now termed anti-TRAP, was shown to bind to TRAP and prevent it from interacting with RNA. We are now investigating the nature and antagonism of TRAP/anti-TRAP versus TRAP/RNA interactions.
Model of the trpEDCFBA operon transcription attenuation mechanism:
A. Under tryptophan-limiting conditions TRAP is not activated. During transcription the 5′-stem loop and antiterminator structures form. Antiterminator formation prevents formation of the terminator, resulting in transcription read-through into the trpstructural genes.
B. under excess tryptophan conditions TRAP is activated. During transcription TRAP interacts with the 5′-stem loop and with the (G/U)AG repeats as soon as they are synthesized, thereby wrapping the RNA around the periphery of the TRAP complex. TRAP binding prevents formation of the antiterminator, which allows formation of the terminator, and hence termination of transcription before RNA polymerase can reach the trp operon structural genes. The (G/U)AG repeats are indicated in boldface type. The four nucleotides that the antiterminator and terminator structures have in common (ACCC) are indicated by the outlining type.
Molecular Genetics: RNA-Protein Interactions and Prokaryotic Gene Expression; Paul Gollnick:
Two alternative RNA secondary structures regulate transcription termination in an untranslated leader region upstream of the structural genes. A trans-acting regulatory protein called TRAP (trp RNA-binding Attenuation Protein) controls which RNA structure forms. In the presence of tryptophan, TRAP is activated to bind to a specific target in the leader RNA. This binding results in formation of the transcription terminator, thereby halting expression of the operon. In the absence of tryptophan, TRAP does not bind RNA and the alternative antiterminator structure forms allowing expression of the operon. We have shown that TRAP protein has a unique structure consisting of 11 identical 75 amino acid subunits arranged in a ring . This is the first example of an 11 subunit protein, and one of few RNA-binding proteins whose three dimensional structure is known. TRAP is activated by binding 11 tryptophan molecules in clefts between each subunit. The TRAP binding site in the trp leader consists of 11 repeated GAG and UAG trinucleotide repeats. The discoveries that TRAP contains 11 subunits and that its RNA target contains 11 triplet repeats suggested a model for RNA binding to TRAP with each subunit of the protein interacts with on repeat in the RNA (Fig. 1). Authors of this paper used alanine scanning mutagenesis to identify three amino acid residues in each TRAP subunit (Lys37, Lys56 and Arg58) that are directly involved in RNA binding. The location of these residues on the structure of TRAP suggested that RNA binds by wrapping around the protein ring.
Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader
1. Alexander V. Yakhnin, Helen Yakhnin, and Paul Babitzke*
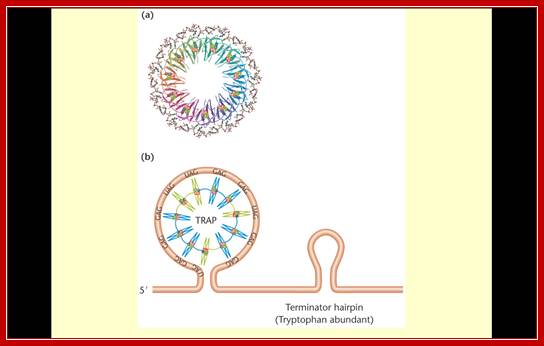
The symmetrical molecule consists of 11 subunits, each capable of binding to one molecule of tryptophan that is embedded within the subunit. The interaction of a tryptophan-bound TRAP molecule with the leader sequence of the trp operon of B. subtilis. This interaction induces the formation of the terminator hairpin, attenuating expression of the trp operon. http://www.studyblue.com/

http://www.sciencemag.org/
Models of B. subtilis trp operon regulation: Upper fig- Transcription attenuation model. During transcription RNAP pauses at U107. Under limiting tryptophan conditions, TRAP does not bind to the RNA. Once RNAP resumes transcription, the AT forms, resulting in transcription read-through. Under excess tryptophan conditions, TRAP binds to the 5′SL and the (G/U)AG repeats. Bound TRAP releases paused RNAP and prevents AT formation. Thus, formation of the T causes transcription to terminate at G140 or U141. Because termination is never 100% efficient, a fraction of RNAP molecules will not terminate despite the presence of bound TRAP.
Lower fig; trpE translational control model. During transcription of read-through transcripts, RNAP pauses at U144. Under limiting tryptophan conditions, TRAP does not bind to the RNA. Once RNAP resumes transcription, the RNA adopts a structure such that the trpE SD sequence is available for ribosome binding. Under excess tryptophan conditions, TRAP binds to the paused transcript. Once RNAP resumes transcription, the trpE SD-sequestering hairpin forms and inhibits translation of trpE. The same structure functions as the terminator and U144 pause hairpins. The 5′SL is shown only in the upper left drawing.
tRNA-mediated transcription antitermination in vitro: Codon–anticodon pairing independent of the ribosome
1. Frank J. Grundy, Wade C. Winkler *, and Tina M. Henkin †
Proposed T box antitermination mechanism: The arrow indicates the transcription initiation site. The black rectangle represents the coding region of the regulated gene. Uncharged tRNA is postulated to interact with the nascent transcript at both the specifier sequence and the antiterminator bulge, stabilizing the antiterminator and preventing formation of the competing terminator. RNA polymerase (RNAP) then continues past the terminator region, and the full-length transcript is synthesized. “Factor?” indicates putative factor(s) that could modulate the leader RNA-tRNA interaction in vivo
Protein-RNA interactions during transcription regulation:
Here we use Bacillus subtilis system involving TRAP/anti-TRAP proteins and RNA as a model to study regulatory processes involving protein interactions with RNA sequences containing multiple repeated segments. The structure of TRAP/RNA complex (shown on Figure) explained the dependence of RNA binding on tryptophan.
Trp Regulon:
The trp repressor also regulates related genes, hence it is called Trp regulon. It regulates aroF, aroA, aroP and aroG down regulated as shown below.
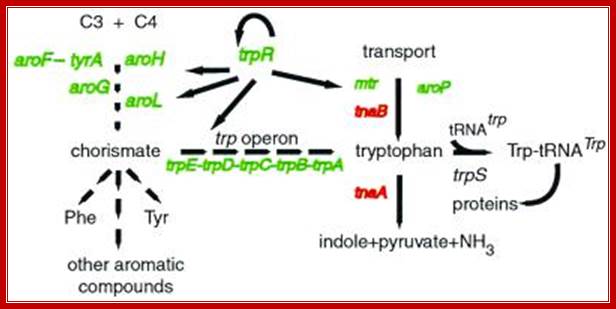
TrpR can act as multiregulator protein; so it can be considered as Regulon. http://www.pnas.org