Mechanism of Transcription:
http://technologyofbiology.blogspot.in/
DNA template + r NTPS + mg2^+ + RNAP-->
DNA template + RNA + n (pi) +RNAP
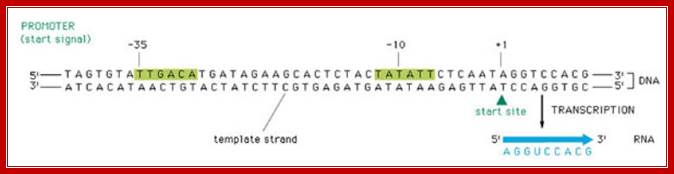
Transcription involves various stages of intricate and intrinsic mechanisms. Unlike DNA replication, transcription does not require any primer; it initiates at a particular predefined nucleotide. This is determined by the positioning of the RNAP Holozyme properly on TATAAT at –10 and also positioning at –35 sequences. The –35 sequence varies from house keeping genes to other regulated genes, with certain exceptions. The kind of the sigma factor and the –35 sequence ultimately determines which gene to be transcribed and which gene not to be transcribed. It is not just TATAAT box and TTGACA are enough to initiate transcription, it also requires, in some cases, additional proteins, may be in the form of activators or enhancers. Some times RNAP binds to the sequence in right context and activators are also bound to their upstream elements, but other proteins, called repressors, block the activation by the enzyme. Until they are removed, transcription won’t be initiated. In most of the house keeping genes the process is straightforward. It takes place in 3 basic steps, i.e. chain initiation, chain elongation and chain termination, where each of the steps require a set of factors, without which process comes to a grinding halt or it may not be executed at all.
Central dogma by F.C Crick; http://www.uic.edu/
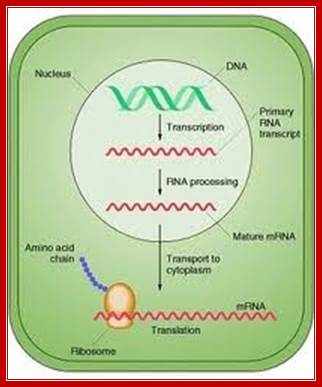
Control of Gene expression:
This diagram applies to both Prokaryotes and eukaryotes as well; http://faculty.clintoncc.suny.edu/

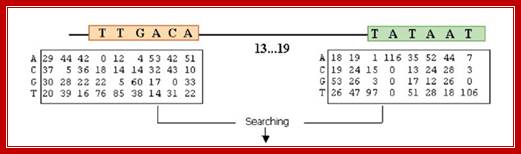
The consensus sequences of sigma 70 factor binding sites; http://www.2013.igem.org
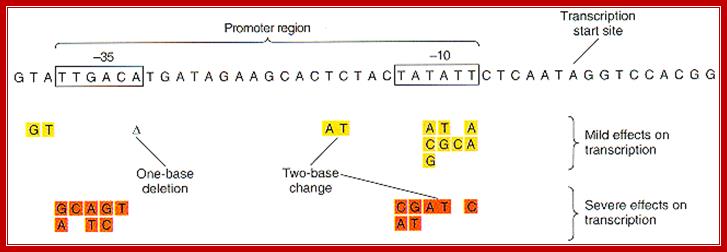
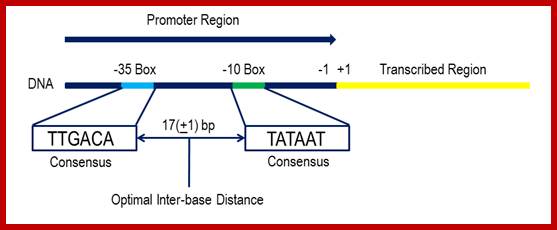
Bacterial consensus promoters include two regions of six base pairs each, at -10 and -35 bases upstream. However, no two promoters are exactly alike, and no promoter exactly matches the consensus sequence. Additional sites for environmental regulators can be found as far as -50 to -300 bases upstream.
Consensus TAAATA box; http://oregonstate.edu/ and http://biology.kenyon.edu/

http://fig.cox.miami.edu
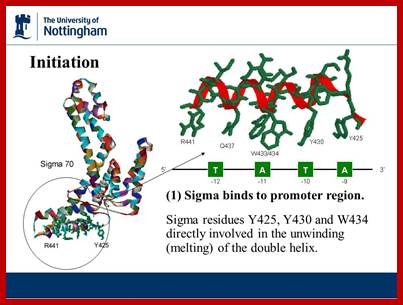
http://slideplayer.com
Protein with their specific amino acids makes contact to DNA sequences and bind. This is an illustration to show how and why certain DNA binding proteins bind to DNA only in sequence specific manner.
Promoter recognition by amino acids of the α subunit and σ70. Orange and blue arrows indicate recognition of promoter regions as double-stranded DNA elements by the α and σ70 subunits, respectively. The two red arrows delineate a region of the nontemplate strand DNA recognized by σ70 subsequent to strand separation. In the linear representations (not drawn to scale) of both σ70 and α, the N-termini are on the right. Only the sequence of the nontemplate strand is shown (5′ end on the left). A typical E. coli promoter does not have all elements shown and exhibits deviations from the consensus sequences shown here for the –10, –35, and UP21 elements, as well as the consensus spacer length (17 bp) , NIH Public Access.
The sigma factor 70 in association with RNA polymerase makes sure the RNAP binds to specific promoter sequence of DNA, otherwise RNAP perse cannot bind to specific promoter site. Namrata Chhabra; http://www.namrata.co/
The consensus sequence shown above has not been found upstream of any genes in bacteria, and was compiled by looking at the expression levels of a range of strong E.coli promoters to determine the optimal base to use at each position would be. The current hypothesis is that the use of this sequence in E.coli is detrimental to bacterial growth because transcription from this loci would be so efficient that the cell would suffer a growth burden if it were to use it. However, some promoters come very close. For example, the RecA promoter is a very strong promoter that under normal circumstances is suppressed by a protein called LexA. However, during times of stress and DNA damage the LexA repressor can be cleaved to allow RecA production. The similarity between the RecA promoter and the consensus means that the promoter mediates very high expression levels. The RecA promoter -35 box is TTGATA and the -10 box is TATAAT, with an inter-box gap of 16bp. This means that the promoter is only one base different from the consensus. ;https://www.oxfordgenetics.com
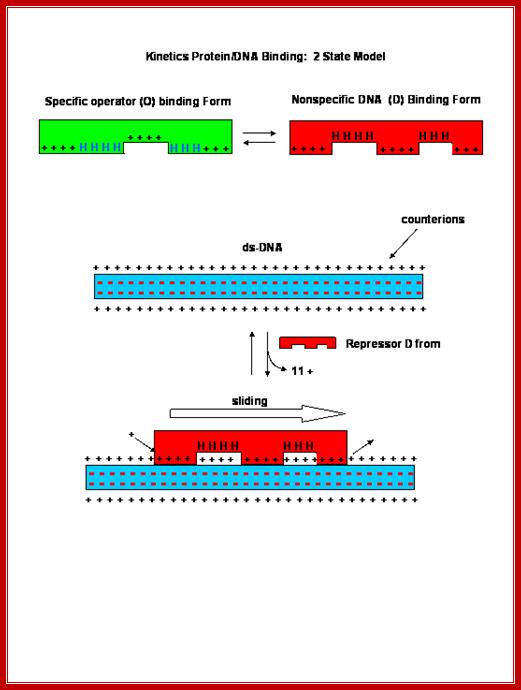

Thermodynamics of DNA Hybridization; The formation of DNA/DNA duplexes is what makes the hybridization reaction work! ;;This graphic diagram shows interaction between negatively charges DNA surface interacting with positively charges amino acid residues of the protein and bind.
![]()

https://blog-biosyn.com
RNA pol is transcribing the DNA, shows the entry of DNA and the exit of transcript; http://www.conservapedia.com/
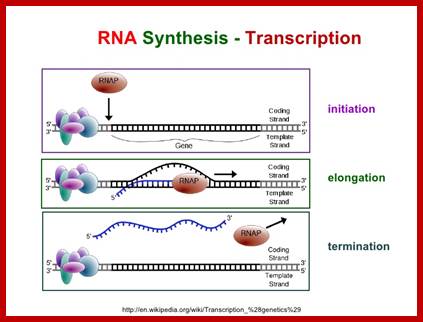
Chain initiation:
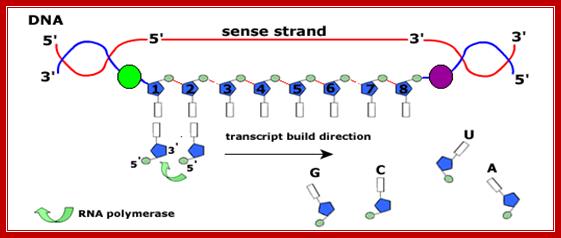
RNA-polymerase though holds both DNA strands, it uses only one of the strands as the template. Which template to be used is determined by the nucleotide sequence found in one of the strands found in the promoter region? For any given gene, only one of the strands is used and both the strands cannot be used for the opposite strand generates anti-sense to the transcript. But the opposite strand can become a template for transcription for some other gene to produce a different transcript. The entire length of DNA running to millions of base pairs don’t use only one strand for transcription. Transcription of both strands are used for generating different products such as mRNA, tRNA, rRNA an small molecular wt. RNAs. A good example from mitochondria, where the H and L strand produces different transcripts.
One strand is used for some products. The other strand is used for another product, but not the same dsDNA segment. If bot strands are used they generate anti strands. I some cases such anti-strands, called antisense strands are produced for gene regulation.
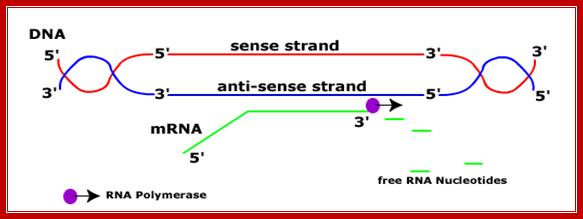
Sense and Antisense DNA;http://dnamismatch.com/
Antisense-DNA; https://lookfordiagnosis.com
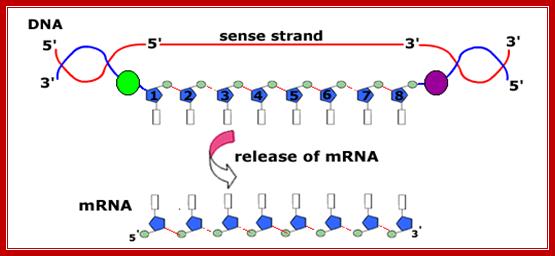
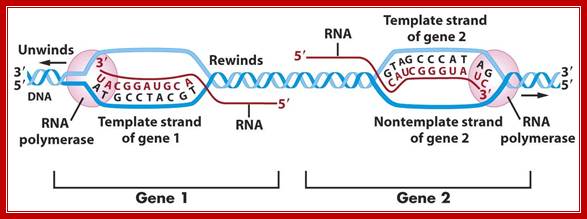
The figure shows transcriptional chain initiation at specific site and progresses (elongation) and terminates at specific site; Top.fig:http://biosiva.50webs.or/;http://click4biology.info/c4b
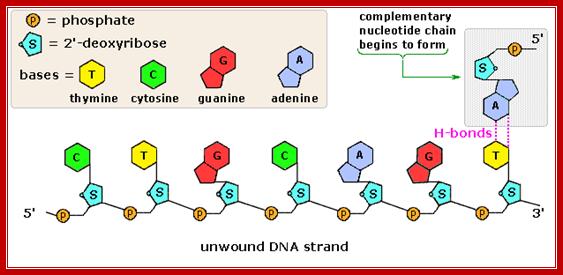
The polymerization mechanism described here is constant. It always extends the developing DNA segment toward the 3'-end (i.e. when a nucleotide triphosphate attaches to the free 3'-hydroxyl group of the strand, a new 3'-hydroxyl is generated). There is sometimes confusion on this point, because the original DNA strand that serves as a template is read from the 3'-end toward the 5'-end, and authors may not be completely clear as to which terminology is used./www2.chemistry.msu.edu.
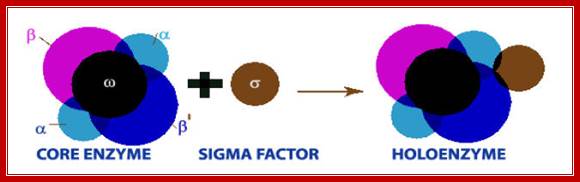
· The core enzyme by itself can bind to the DNA and can diffuse linearly but fails to recognize the promoter regions so as to make a closed complex.
When the polymerase is bound with a specific sigma factor, the complex binds to DNA and linearly diffuses along the sequences of no relevance, but once the sigma factor associates, when the enzyme diffuses along the DNA and when it reaches promoter sequences, it halts and contacts a specific sequences, then activates the core enzyme to conformationally change from loose complex to tight complex with DNA.
Thus the enzyme exhibits two conformational forms, one for linear diffusion and another for tight holding on to the DNA.
· The enzyme does not use any primers for transcriptional initiation unlike DNA-polymerase. The unique aspect of it is that it initiates at a predefined nucleotide without a primer.
The important feature of this recognition is, that the enzyme complex binds to DNA but when it encounters specific sequences, it strongly binds with great affinity (thousand to ten thousand times more than to the other regions).
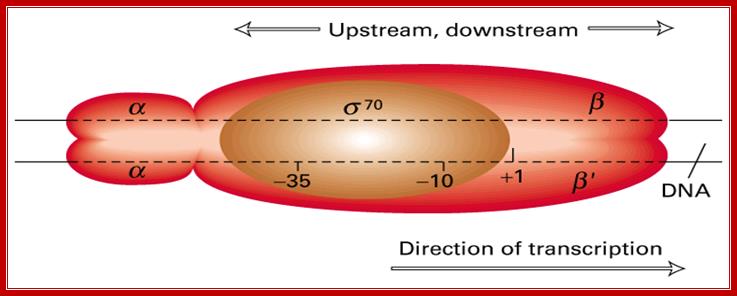
· E.coli genome is 4.6 million base pairs long but the RNA pol binds only to the -10 and –35 sequences of a gene recognized by the sigma factor and binds with the great affinity, and it does not have that type of affinity to other regions; it means the binding is sequence specific. Sigma factor by its self cannot bind to DNA in any form; it requires RNAP for its act. Both RNAP and Sigma are mutually inclusive not exclusive in their action.
Non-specific interactions or binding is mainly due to electrostatic attractions, but in specific binding both electrostatic and specific hydrogen bonding takes place, because the DNA structure provides a specific geometry, for the side chains of amino acids interact and bind to bases arranged in a particular sequence; here the binding force is hydrogen bonds. Sequences in the DNA act as landmarks for identity.
Once the binding is proper, the sigma factor trigger conformational change in the enzyme; now it tightly binds and forms tight closed complex. Again it undergoes one more change to make it tight open complex, where the enzyme is bound to both the strands of DNA and pulls apart to form single stranded open complex called transcriptional bubble.
· Few isomerization steps of open and close, results in very tight closed complex; then finally it opens as a tight –open complex.
For all these conformational changes, sigma factor is absolutely essential. Sigma factor provides an input of signal for making the enzyme to change from low affinity to high affinity status.
· It is at this event the DNA helix gets unwinded and pulled apart by b andb’ subunits and create transcriptional bubble with two single strands.
· When the Holozyme, when initially placed on the promoter it covers from -65 to +15 that is about 80 base pairs. When the enzyme pulls the strands apart, it opens about 12 to 17 bp long; this is called the transcription bubble.
The enzyme, now, is in open complex form. Opening of the DNA in a specific region from -11 to +3, perhaps is facilitated by negative super coiled DNA, yet because of the opening, DNA helix, on either side of the bubble, is strained and assumes positive super coiled state, which is actually relieved by topoisomerase II (cut-relax and join) and the same enzyme can introduce negative super coil which greatly facilitate the opening of the DNA.
· The holozyme with its palm like region has a channel of 25 Å wide and 16 bp long.
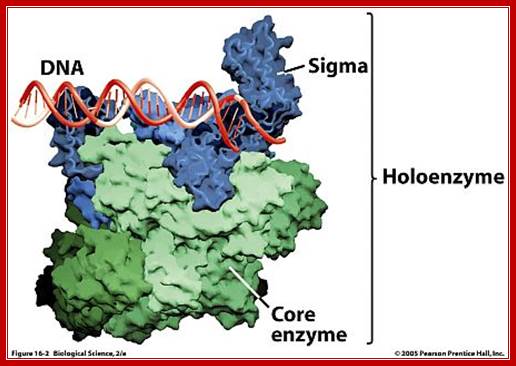
PK holozyme; http://www.uic.edu/
· The subunit b has two sites, one for the first nucleotide against the defined nucleotide what is called START (InR) site, next to it is second site can be called elongation site. These sites in the enzyme contain pocket shaped cavities, determined by the opposite nucleotide on the template. Depending upon what nucleotide present in the template strand the active site develops in the enzyme for accommodating the correct nucleotide for proper base pairing. At the other end the sites have cavity to accommodate triphosphate group of the nucleotides and the nucleotides bind to if the nucleotides are complementary, otherwise they are repelled from the surface of the enzyme.
The first nucleotide diffuses on to the first site, by conformational changes, enzyme determines, whether the nucleotide is complementary or not. When the monitoring is perfectly matched, the second nucleotide is allowed to bind. Again here, the matching by geometric complementarities determines whether to take this nucleotide or to reject.
· When the second nucleotide binds, the enzyme now gets conformationally activated to perform covalent bond formation between the 3’OH of the deoxyribose group of the first nucleotide and the 5’ end Phosphate group of the second nucleotide, where a phosphodiester bond is formed between the 3’OH of the ribose and the alpha phosphate group of the second nucleotide.
The released pyrophosphate is immediately cleaved by pyro-Phosphotases and release bond energy. And the energy is used for the movement of the enzyme along the coiled DNA.
In this monumental endeavor, the enzyme continues to add nucleotide after nucleotide, form few more bonds, but only to abort. Again, it reinitiates the process all over again. Aborting attempts by the RNAP Holozyme is one of the remarkable features of its functional attributes. RNA polymerases does not require any primers, perhaps that is the reason why they produce 9-10 nucleotide long segment early in transcription and use it as the primer.
· When it succeeds putting together 9-10 ntds, it becomes a stabilized complex and the enzyme is set to move along, and physically moves out of the promoter, a process called promoter clearance.
As the enzyme clears the promoter the sigma factor dissociates from the core enzyme. With the dissociation of sigma factor, nus-A associates with the core enzyme; these two events are mutually exclusive.
· During initiation it adds 9-10 ntds, with out any displacement of the enzyme from its hold. While it performs progressive assembly of nucleotide after nucleotide, the enzyme contracts, when it assembles 9-10 nucleotides in one grip, it leaves its hold from the front end and extends forward for another 9-10 ntds and then grips the template and the enzyme contracts from behind again to hold the DNA with a grip. This is like an inchworm movement.
Thus it initiates transcription and clears the promoter.
· The RNAP performs two types of proof reading activities. One is called pyrophosphorlytic reaction, where it removes the nucleotide from the 3’ end if the ntd is mismatched. The second form of proof reading is called hydrolytic editing, the enzyme back tracks several nucleotides if there is any error not corrected removes the whole length and resynthesizes. The proof reading activity is stimulated by Gre factors, which also act as stimulating factors for chain elongation.
Elongation:
Elongation is an extension of initiation. Addition of successive nucleotides, and formation of phosphodiester bonds in succession, is template dependent process, like a trade milling action.

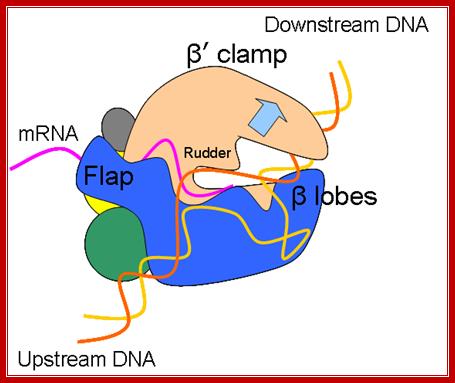
Schematic model of the elongation complex; Core RNA polymerase (RNAP) (in bacteria, a complex composed of an α-dimer, a β-subunit, a β′-subunit and an ω-subunit) is bound to the DNA duplex composed of the template strand (black) and the non-template strand (blue), and the nascent RNA (red). The α-amino-terminal domains (α-NTDs) serve as a scaffold for complex assembly; the α-carboxy-terminal domains (α-CTDs) and ω-subunit play regulatory roles during initiation. The β- and β′-subunits jointly form the active site and make all the contacts to the nucleic acids. The substrate nucleoside 5′-triphosphate (NTP) (bound to a second Mg2+ ion) is thought to enter through the secondary channel. 12–14 bp of the DNA are melted in the transcription bubble. The non-template DNA strand is exposed on the surface, where it may interact with regulatory proteins. The nascent RNA is annealed to the template strand to form 8–9 bp of the RNA–DNA hybrid, which is the key determinant of elongation complex stability7, 98, 99. The upstream RNA is extruded through the RNA exit channel formed between the β-flap and β′-clamp. Thomas J. Santangelo & Irina Artsimovitch http://www.nature.com/
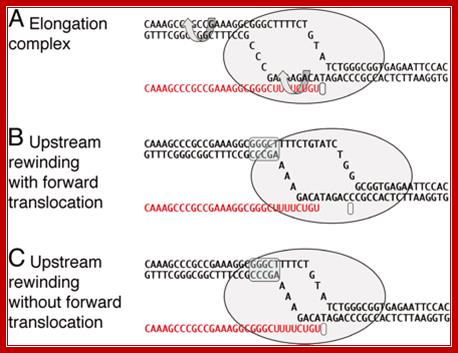
The elongation complex and models of RNA release in termination. (A) A model of an elongation complex showing helix rotation that would accompany branch migration in mechanical models of termination. (B) Termination by forward translocation. (C) Termination by bubble collapse. A shows a particular intrinsic transcription terminator poised at the site of release, but B and C are general to both intrinsic and enzymatic termination. The upstream rewound segment is indicated by the blue overscreen. http://www.pnas.org/
During full speed of elongation the enzyme contracts from initial occupation of 80 base pairs to hardly 30 to 40 bp.
· As the transcription uses only one of the strands, it moves along the template strand, like a cock screw, one round for every ~10 ntds transcribed.
Important feature of elongation is that it takes care of mismatch base pairing. This mismatch correction is done by b’ subunit. Though there is no explicit or overt 3’>5’ exonuclease activity, it possesses, nucleolysis or phosphorlysis functions and it acts from 3’ ends.
During this correction of errors the whole process of transcription comes to halt or say pause, till the mismatched base is removed and corrected.
· Some times the progression of the enzyme and polymerization can be halted or paused, by secondary structural constraints formed in the nascently synthesized RNA, preceding to the assembly site; it can also pause when the concentration of NTPs falls below the optimal concentration required or if the moving enzyme encounters a G: C rich region or other nucleotide sequences such as thymidine dimers; but the pausing is over-come by a variety of factors like anti terminator factors, which interact with Nus-A.
In the case of bulky adducts and inter-strand cross linkings, the polymerase halts and often skips or jumps hundreds of nucleotides ahead of the obstruction.
· Though the rate of transcription is 40 to 50 ntds per second in vitro, under in vivo conditions its rate is about 50000 to 60000 ntds per minute. Perhaps no other polymerizing enzyme has this ability and agility; it is a unique and par excellent enzyme.
Magnesium divalent ion is an essential component in RNA synthesis for the Mg^2+ bind to gamma and beta phosphates of dNTPs. Such magnesium bound nucleotide triphosphates act as very good substrates, for magnesium bound triphosphates provides geometrically stable and straight structural feature for the enzyme to recognize as the substrate, to bind and act.

Determination of RNA polymerase binding surfaces of transcription factors by NMR spectroscopy; In bacteria, RNA polymerase (RNAP), the central enzyme of transcription, is regulated by N-utilization substance (Nus) transcription factors. Several of these factors interact directly, and only transiently, with RNAP to modulate its function. As details of these interactions are largely unknown, we probed the RNAP binding surfaces of Escherichia coli (E. coli) Nus factors by nuclear magnetic resonance (NMR) spectroscopy. Perdeuterated factors with [1H,13C]-labeled methyl groups of Val, Leu, and Ile residues were titrated with protonated RNAP. After verification of this approach with the N-terminal domain (NTD) of NusG and RNAP we determined the RNAP binding site of NusE. It overlaps with the NusE interaction surface for the NusG C-terminal domain, indicating that RNAP and NusG compete for NusE and suggesting possible roles for the NusE:RNAP interaction, e.g. in antitermination and direct transcription:translation coupling. We solved the solution structure of NusA-NTD by NMR spectroscopy, identified its RNAP binding site with the same approach we used for NusG-NTD, and here present a detailed model of the NusA-NTD:RNAP:RNA complex; Johanna Drögemüller et al, https://www.nature.com
One of the important components of RNAP, while in elongation mode is Nus-A and its associating components.
How is it done?
During elongation, the transcription fork moves like a floating bubble and moves in only one direction. The size of the bubble is maintained at 16-18 base pair size. The forward fork movement and the unwinding is performed by the RNAP itself but DNA relaxation is aided by Topoisomerases. However the rewinding of DNA closes from behind the fork. Thermodynamically these two events compensate with each other and dynamically accepted as complaint forces operating at two different ends.
The growing transcript remains hybridized to the DNA template only up to a 12-ntd length in the transcription bubble, and the free 5’ end of the transcript is threaded through the groove in the enzyme meant for exit of the RNA.
· Movement of the enzyme, like a giant helicase (a motor protein) requires energy and it is provided by the hydrolysis of dNTPs and pyrophosphorolysis of pyrophosphate.
Most of the polymerases exhibit fair rate of movement while polymerization. However, polymerization is rNTPs dependent, similar to helicases which are ATP dependent. Most of the motor proteins are energy dependent.
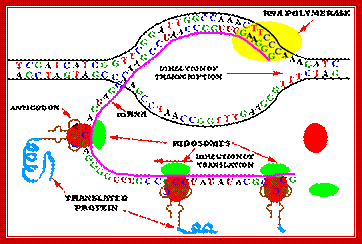
· It should be a point to keep in the mind that in bacterial systems transcription and translation are coupled. As and when the 5’ end of the transcript (of 50-60 ntds long) is made available, ribosomes binds to the 5’ end and initiate translation.
The 5’ ends should not be kept free for the ends are susceptible for exonuclease digestion.
https://www.slideshare.net
Chain termination:
· Most of the genes have ends with noncoding sequences, whose length varies from 50 or more nucleotides to several hundreds of nucleotides. More than 90% of the genes have distinct and characteristic end sequences, which, actually, are responsible for transcription termination. There are few genes whose termination depends on a Trans-acting factor, like Rho. The involvement of this factor is very apparent in lambda phage infected cells, yet this type of termination takes place only in few genes, especially of lambda ones. To distinguish between the two, they are described as rho- independent and rho-dependent termination.
Rho dependent termination: http://microbiochem.weebly.com

http://biosiva.50webs.org
In bacteria transcription and translation events are coupled. As the 5’ end of the mRNA is made available activated ribosomes bind and initiate translation. As the enzyme reaches transcription termination sites, transcription stops and the mRNAs released this is done in two ways.
Rho-independent termination:
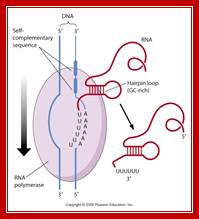
Beyond the coding region in the gene, there is a length of DNA that has palindromic sequences and ends in four to six ‘A’s.
· Such sequence in DNA generates a stem loop structure with six consecutive Us in the transcript.
When m-RNA is transcribed from the template strand (3’à5’), the resulting sequence is
5’ GGCUCCUUUUGGAGCC 3’
Because of palindromic sequences containing G: C rich region, produces a stem loop with a terminal six Us of which two are in the stem, and the other four are hybridized to DNA.
· As the stem loop structure is produced, which is very close to the enzyme, it interacts with the enzyme and stalls the progress of the enzyme.
The hydrogen bond between ‘A’s and Us is very very weak when compared to that of A s and T s.
· Because of this reason, the chain automatically falls off from the template.
When the transcript falls off, the rewinding of the DNA from behind the bubble forces the enzyme to dissociate from the template, for it cannot initiate RNA synthesis at that position.
· The stem loop formation is very important, for example if Inosine nucleotides are incorporated into the loop region, Inosine fails to from any loop and transcription fails to be terminated.
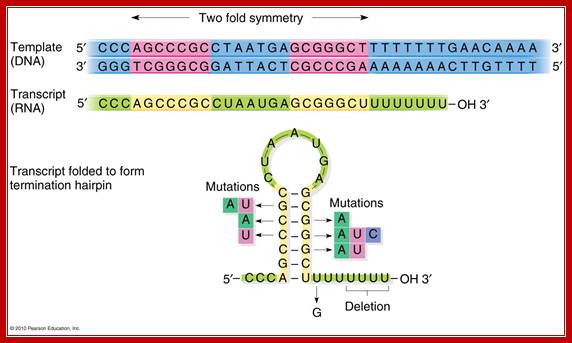
Transcript at the end is self-complementary, with two fold symmetry; so stem loop structure develops;The 2x6bp has a standard motif in the dsDNA produces a self-complementary C-G rich region in the transcript, which halts thethe progression of RNA pol https://www.mun.ca
DNA: --------GGG TCGGGCG GATTACT CGCCCGA AAAAAA-CT
RNA: 5’------CCC AGCCCGC CUAAUGU GCGGGCU- -UUUUUU—3’-
Principles of Cell Biology-http://www.mun.ca/biology; www.study blue.com
Conserved economics of transcription termination in eubacteria; Shyam Unniraman, Ranjana Prakash and Valakunja Nagaraja IISc. A secondary structure in the nascent RNA followed by trail of U residues is believed to be necessary and sufficient to terminate transcription. Such structures represent an extremely economical mechanism of transcription termination since they function in the absence of any additional protein factors.
The algorithm classifies the structures into five classes. We find that potential secondary structure sequences are concentrated downstream of coding regions in most bacterial genomes. Interestingly, many of these structures are not followed by a discernible U-trail. However, interestingly, many of these structures are not followed by a discernible U-trail. However, irrespective of the nature of the trail sequence, the structures show a similar distribution, indicating that they serve the same purpose.
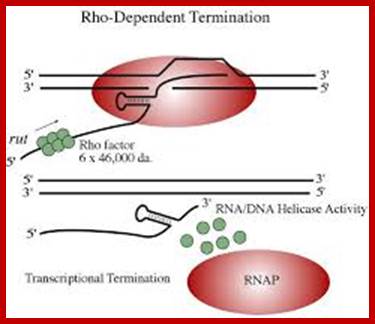
Rho-dependent transcription termination:
This mechanism requires the presence of rho protein, a RNA-binding protein that forms a hexameric complex at a particular signal site, and races up the RNA strand. Once it reaches the polymerase, the protein complex displaces the polymerase from the DNA, terminating transcription. In this particular diagram, the system is shown as requiring a hairpin to slow polymerase; for the purposes of this class, that is an extraneous detail; Transcription termination; http://utminers.utep.edu/
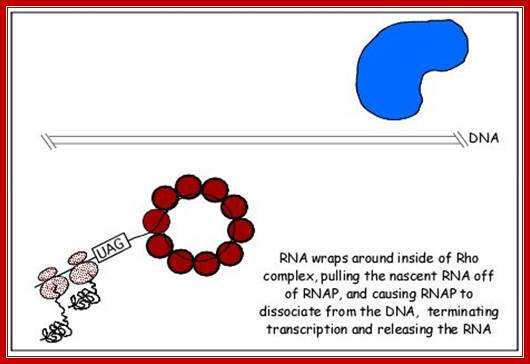
http://www.sci.sdsu.edu/
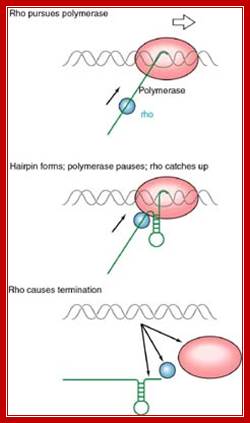
http://utminers.utep.edu/
Rho dependent chain termination is observed in lambda phage infected cells; nearly 50% of the transcripts are terminated by Rho factor.
· Rho is a 46 KD protein; its N-terminal end has greater affinity to ss RNA and its C-terminal has ATPase activity. It is an hexamer ATP dependent helicase.
· As soon as the 5’ end of the transcript is made available ,
if it is free from ribosomal assembly, rho assembles on to it as ring of subunits; and moves in 5’>3’ direction like an helicase in ATP dependent manner.
· It can unwind RNA-DNA hybrid region and also RNA-RNA hybrids.
In rho dependent genes, the terminal region of the gene does contain a stem loop sequences, but no Us at the end, instead it has a stretch of nucleotides, 100 nucleotides down stream of the hairpin structure, mostly rich in Cs and with less number of Gs; 41 % of the bases are Cs and 14% are Gs. Rho recognizes them as cryptic signals for its termination process.
5’AUCGCUACCUCAUAUCCGCACCUCCUCAAACGCUACCUC-3’
When the RNAP reaches this segment, it stalls and Rho traverse along and when it reaches the region where RNA is still hybridized to DNA (in an 12 bp segment), rho by its helicase movement unwinds RNA from the DNA and interacts with RNAP and induces the RNAP to release the transcript and also induces the RNAP to dissociate from the template.
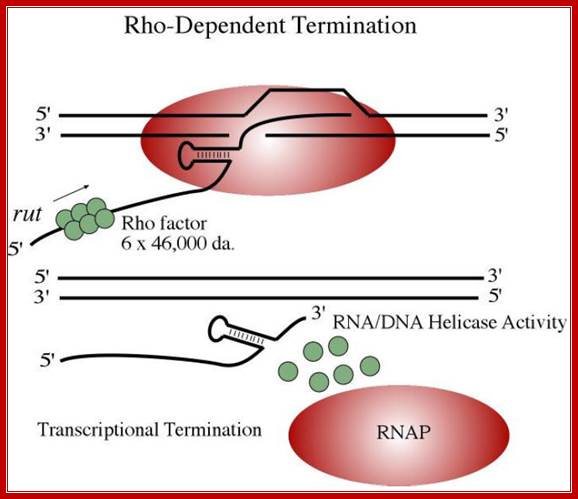
It also appears that termination is not absolutely rho dependent or rho-independent. Rather, rho-independent termination can utilize rho and rho dependent termination can proceed in the absence of the protein (biosiva); http://www.bologsu.us/

Three mechanisms of transcription termination: (A) Intrinsic termination is driven by formation of an RNA hairpin in the emerging transcript, the base of which occurs 8–9 nt from the site of release. Release also requires a Uridine-rich segment downstream of the hairpin, particularly in the region immediately adjacent to the G/C-rich end of the stem. Although not illustrated, we suggest (as described in the text) that the DNA bubble is partly rewound and that the RNA/DNA hybrid is partly unwound when the hairpin is fully formed. (B) The termination factor Rho is a hexameric RNA translocase that binds ≈60 nt of emerging transcript, moving along it in a 5′–3′ direction in an ATP-dependent reaction. This movement is believed to extract the transcript. (C) Mfd is a DNA translocase that binds duplex DNA upstream of the transcription bubble and RNAP in a region near the site of DNA rewinding. The activity of the translocase causes dissociation of the complex in conditions that do not allow the RNA chain to advance through NTP
termination (Joo-Seop Park* and Jeffrey W. Roberts).
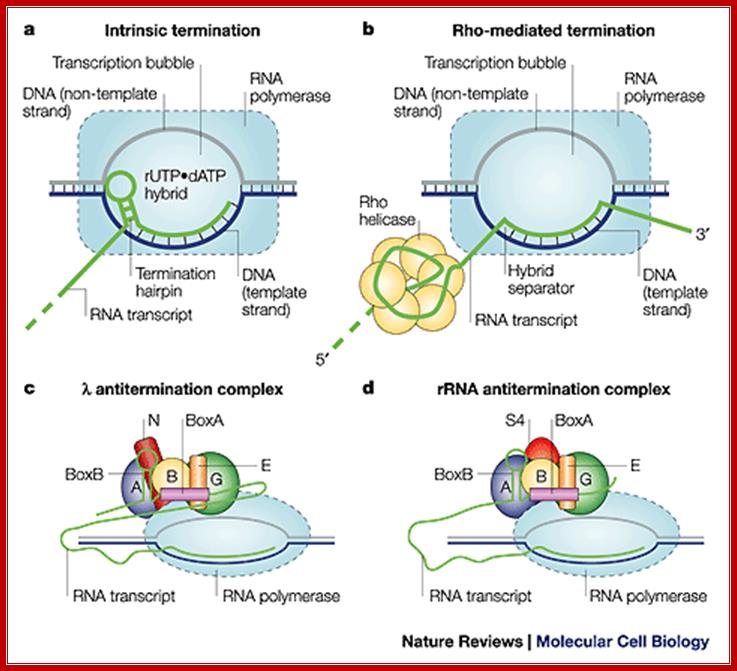
Comparison between the two systems; Robert Maxwell
The Rho dependent termination is greatly facilitated by b subunit of the RNAP.
Rho requires a specific sequence for effecting termination, but the sequence and the size in the terminal region is not always constant, it can vary from 50 to 80 ntds, some times it can be 100 ntds.
In the absence of Rho, termination takes place on its own but several hundred nucleotides down stream of the TER codon and no secondary structures are found in such regions.