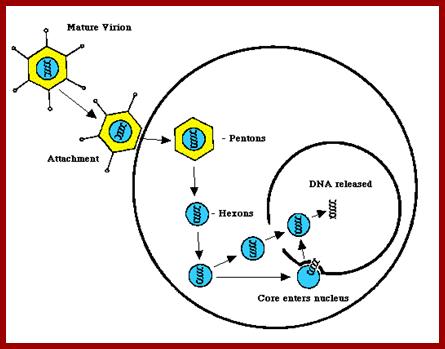
Replication of Adeno Viral DNA:
And few more:
Adenovirus:
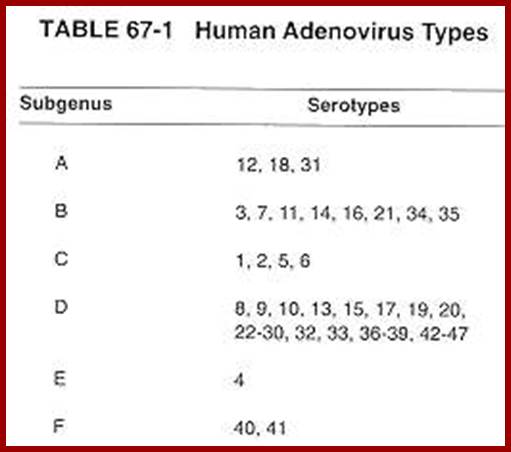
The virus, as it was isolated from adenoid tissues, called Adenovirus. There are more than ~80 different strains. In general they are classified into seven groups.
ADV. A. (12, 18, 31 strains) = produce tumors in hamsters.
Adv. B. (3, 711, 14,21,34,35, strains) = weakly oncogenic.
Adv. C. (1, 2, 5, 6) = not tumorigenic, but can transform rat cells.
Adv.D. (10, 13, 15, 17, 19, 20, 22, 32, 33) = non-tumorigenic.
Adv. E. (40) = non-tumorigenic.
Adv. F&G (40, 41) =?
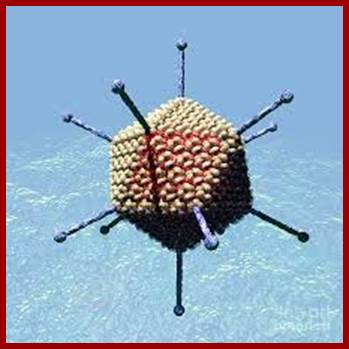
The figure is the artists diagram in all its glory. Russell Kightley; http://pixels.com/

Electron micrograph of the Adenovirus with its pentons disassembled. http://www.wadsworth.org/
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
The above diagram shows the events of infection till the delivery of the dsDNA into the nucleus. The figure depicts the newly synthesized terminal protein bound to dCMP binds to the 3’End of one strand and opens the double stranded DNA into a replication fork and provide terminal CMP as the primers for the Adenoviral DNA polymerase to extend the primers and copy the Strand. Such even takes place at the other end also. http://www.mcb.uct.ac.za/
These viruses are wide spread in nature; infect birds, mammals including human beings (Avian= avi adenoviridae, mammals = mastadenovirus), they show latency in lymphoid tissues and reactivated later. There are several strains that are oncogenic. They can cause respiratory diseases (in military personnel), pharyngitis in infants, gastroenteritis in infants, conjunctivitis in infants, pneumonia in infants, acute hemorrhagic in all age groups of people, and hepatitis in infants.
Adv. viruses are excellent examples for isometric types, 60- 80nm thick, and non-enveloped; similar to phi X 174 phages. Viruses consists of 252 capsomeres, 240 (?) of them form hexamers of each 20 triangular faces. At 12 vertices, there are 12 pentameric glycoprotein spikes, show 2-3-5 symmetry.
Adenoviral Proteins:
|
Gene/ Protein |
Mol.wt In kds |
Copy Numbers |
Location |
Function |
|
II |
135kd |
240 |
Hexon (H) Monomers |
Outer coat, capsid proteins Structural |
|
VI |
22 |
420 |
H.minor |
Stabilization and assembly |
|
VIII |
14 |
VIII and IX together -400 |
H.minor |
Stabilization and assembly |
|
IX |
12 |
|
H.minor |
Stability, assembly |
|
III
IIIa |
88
74 |
100 |
Penton Base (5) Penton Associated |
Viral penetration, they have Toxin like activity,
Penetration |
|
IV |
62 |
|
Fiber |
Contain terminal glycoprotein, Glycosamino-glycan, hem- Agglutinin, in penetration, Binds to receptor, |
|
V |
52 |
180 |
Core of the Virus |
Associated with Penton base And DNA, it is Arginine rich Protein, binding makes the DNA look like a chromatin |
|
VII |
19 |
1070 |
Core |
Internal histone like, Complexes with DNA |
|
X (mu) |
4.0 |
125 |
?
|
? |
|
TP (Iva) |
50-55 |
|
Bound to 5’DNA
|
It is bound 5’CMP at, Each end, involved in DNA replication |
|
|
|
|
|
|
Adeno viral Genome:
Infection is through the glycoprotein spike fiber binding to cellular receptor, including MHC class-1 glycoproteins. The viral spike protein can interact with a range of receptors and most of the cells express many such proteins which act as receptors.
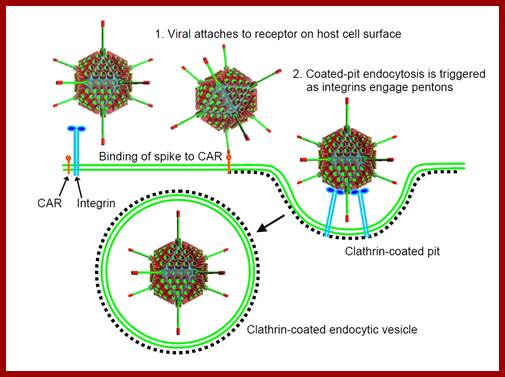
Infection; http://cronodon.com/BioTech/Adenovirus.html

http://cronodon.com/BioTech/Adenovirus.htm lnfection
· The binding facilitates pentons to interact with integrin family proteins; this allows internalization of the virus by what is called receptor-mediated endocytosis. Once inside, the fibers by their toxicity rupture the vesicle membrane and release viral particles in cytoplasm. The viral particle systematically undergoes uncoating; first pentons, then release core particles.
· The core presents to nuclear pores, where DNA is driven in, leaving the proteins outside or it may also move into the nucleus?. The replication and production of viral particles takes place inside the nucleus.
Most of the ADV viral genome size is of 36 to 38kbp long and has the potency for 30 cistrons. Do all cistrons generate protein? At least 20 or more gene products have been identified till today. The DNA inside the nucleus gets coated with histone proteins and they look like mini-chromosomes. The DNA is double stranded, blunt at ends, no terminal redundancies like T7 DNA and no cohesive ends like Lambda DNA, but it’s CMPs at their 5’ ends; they are covalently linked with 55kDa proteins. However their genomic ends contain inverted repeats.
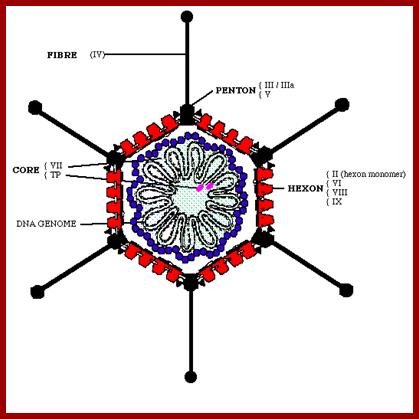
The diagram depicts capsid protein elements such as Hexons, Core proteins, Pentons, and Fiber elements and Core proteins bound to DNA and the genomic DNA; wwct.ac.zaw.mcb.u/cann
5’OC-ABCD-----DEED--------//--------DEED-------DCBA----G3’
3’-G--ABCD-----DEED--------//---------DEED-------DCBA---CO5’
This line diagram shows the end elements of the genome with inverted repeat and direct repeat elements and the central region contains all other genes. The genes are expressed early and late are depicted in next diagram.
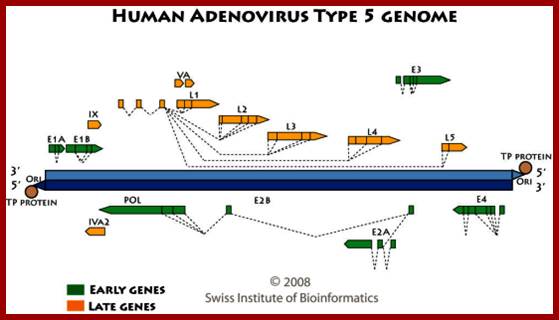
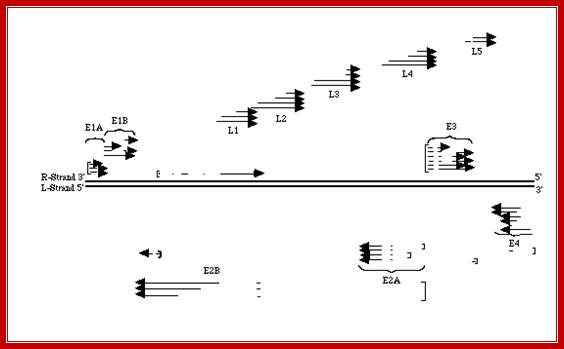
The figure shows the direction and part of the genome used for transcription at Early genes E1, E2, E3 and E4 and Late genes such as L1, L2, L3, L4 and L5; R and L represents the DNA strand used for left ward transcription and R for the DNA strand used for right ward transcription. Here both strands of DNA are used for transcription, but at different regions; http://www.mcb.uct.ac.za/cann
Replication of viral DNA initiates in about one and half hours or more after infection. Preamble for initiation of replication is that the genome has to transcribe few early genes, in temporal fashion.
· The ds Viral DNA is bound by 55kd protein covalently linked to dCMP at each of the 5’ends of the linear DNA. This protein prevents digestion from any exonucleases, thus provides protection. It also acts as the primer for the ADV DNA polymerase to use it for replication of the given strand.
· At each end of the DNA it has a 103 bp long inverted repeats called terminal repeats. Internal to it there is another 180bp long internal repeats. If the ds DNA is melted and allowed to anneal, each strands generate a panhandle shaped structures, because of the presence of inverted repeats at their ends. Though the ends have this repeats, and look similar; from their positional transcriptional activity, the DNA clearly shows polarity.
A remarkable feature of the genome is, that both strands have encoded message for different proteins and they are expressed very early, early and late. They don’t produce antisense transcripts to them.
· The expression of genes, at late stage, is the time at which DNA starts replication. The two strands are distinguished, based on the direction of transcription, as R-for rightward transcription and L-for leftward transcription.
· Early genes are expressed from different positions and use both strands at different positions. However, all the late genes are expressed using R strand transcribing in rightward direction.
The genes that expresses very early are called E1A; the host enzyme RNAP-II recognizes the E1A promoter in earnest and transcribes the single gene and doesn’t extend beyond that.
· The transcript perhaps is processed to produce a capped and poly-A containing mRNA, which on translation produces E1A, which enters into the nucleus. This factor is a transcriptional factor, but it doesn’t bind to DNA, perhaps it associates with RNAP II and recognizes specific early gene promoters and transcribes them as individual cistrons; the genes transcribed are E1B (right ward), E2A and E2B left ward using L strand and E3 and E4 right ward and leftward respectively.
· Very early expressed gene is E1A, it is a regulatory trans-activating factor, required for recognizing early gene promoters, such as, E1B, E2A, E2B, E3, and E4, also for few other virion proteins.
· Transcription of early genes takes place at about 2-3 hrs after infection. The early genes are not located in one region they are distributed all over the genome and the coding regions are found in both strands; E1B and E3 on R-strand and E2B, E2A and E4 on L- strand.
· This E1A factor also activates several host genes that may lead to transformation of host cell into an immortal cell line. When DNA replication starts, it actually triggers the whole set of late gene expression.

The diagram depicts time events and sequence of replication and transcription events from the time of infection to time of late assembly of viral particles; wwct.ac.zaw.mcb.u/cann
Early expressed genes: E1B, E2A, E2B, E3 and E4 are few of the early genes transcribed by host RNAP enzyme.
· After E1A, the next gene expressed is E1B. They are the products of the same mRNA. Combination of the products E1A and E1B has deadly effect on the host. Overall transformation frequency of the virus is very low, but the said gene products can do so with great efficiency.
· E1A- alone can immortalize primary cells in vitro.
· E1B- alone doesn’t have transformation ability, but with E1A, it is deadly.
· E1A+E1B - can transform compatible cell lines and produce tumors in a variety of animal systems.
E1A protein has been found to bind to cellular proteins such as p105-RB and p53 and such cell cycle inhibitor proteins. Binding of these E gene products make then totally disabled, so cell cycle go haywire. E1A also turns off MHC class-I genes, thus escape from immune detection.
Both ABCD and DEED are symbolic representation of certain sequences. The ABC is 103 bp long inverted repeats and DEED is 180 bp long palindromic sequences. At 5’ end of each strand terminal protein (Tp), with its serine residue is covalently linked to CMP, which is paired to G in the opposite strand.
Replication Process:
Replication origin:
The origins for replication are located at 3 ends of each DNA strands and replication termination takes place at 5’ends. There are three sequence blocks, called A, B and C domains in sequence from their 3’ ends.
· Block A: Consists of 18 ntds long sequence, all sero-types share a 10ntd long sequence 5’ATAATATACC3’, the terminal protein (TP) recognizes this sequence and binds to it. Adeno viral DNA polymerase also binds to this region.
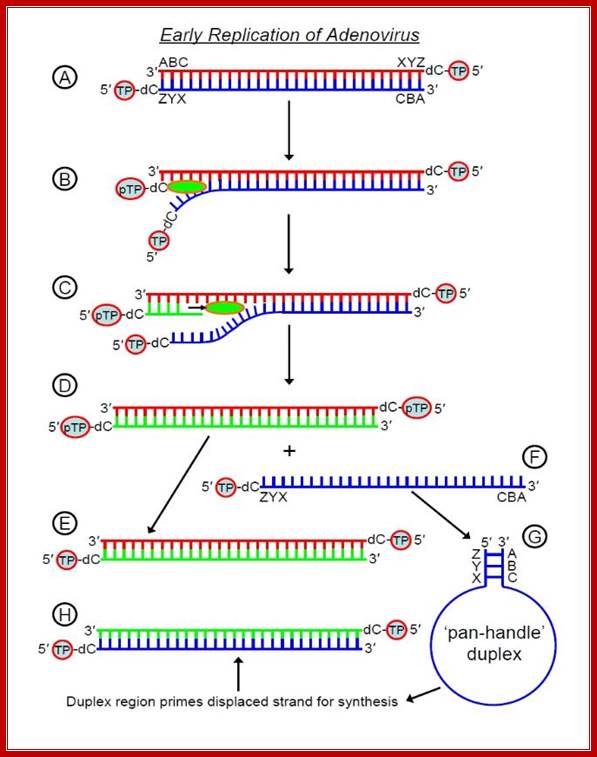
The displaced 5'-3' strand then forms a 'pan-handle' duplex -
the genome has inverted terminal
repeats that are complementary to
one-another (depicted here by ABC and ZYX) and this creates a
short dsDNA region (duplex) for initiation of synthesis - adenovirus polymerase
can bind this duplex, read
the strand from 3' to 5' and hence synthesise the complementary 5'-3' strand.
Thus we begin with one
linear dsDNA (A in the diagram above) and finish each replication cycle with
two (E and H). Recall that in
prokaryotes and eukaryotes, DNA synthesise is bidirectional, beginning at an
ori (origen of replication)
somewhere in the middle of the DNA duplex and proceeding in both directions. In
contrast, adenovirus
DNA synthesis is unidirectional and occurs at one or other end of the molecule
(there is an ori at each
end) and hence requires the TP mechanism to initiate replication by polymerase;
http://cronodon.com/BioTech/Adenovirus.html.
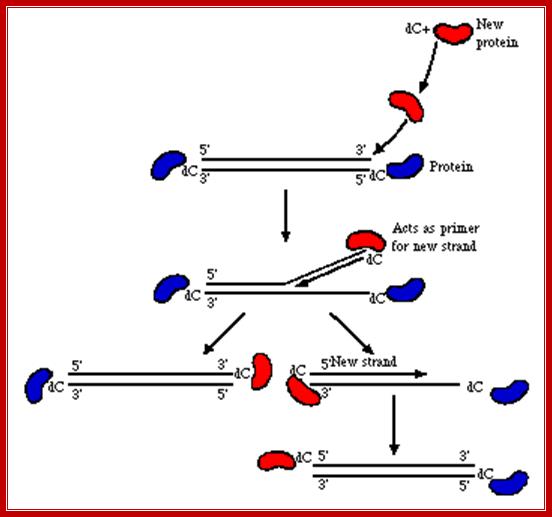
Mode of Replication, using 3’bound protein; wwct.ac.zaw.mcb.u/cann
The line diagrams represents the binding of terminal proteins covalently bound CMP interact with each of the terminal regions and open up the strands by the base pairing of TPs CMP with 3’ GMP containing strand. This opening is further facilitated by other nuclear factors and replication is facilitated by ADV’s DNA polymerase.
· Block B: This has 19-39 bp palindromic sequence, 5’TTGGC (N5) GCCAA. Nuclear factor, NF1 binds to this sequence. This factor acts not only as an initiation factor but also acts as transcriptional factor. This sequence stimulates replication by 30 fold.
Block C: It consists of 29-51 base pairs, have some repeats of the following sequence 5’TATGATAAT3’. A transcriptional initiation factor called NFIII binds to this domain. This sequence also stimulates replication at least by 3 fold.
Tp L=A=B=C=====//======C=B=A Tp R.
Block A = 18bp; 5’ATAATATACC3’,
Block B = 19-39bp; 5’TTGGC (N5) GCCAA,
Block C = 29-51 bp; 5’TATGATAAT3’,
The domains B and C are bound by nuclear factors, which are identified as transcriptional activators. The spacing of the domains and their composition is very critical for their function in initiation of replication. Among the early genes expressed, the most abundant of the products is ADV made DNA-polymerase; it is a monomer of 140kd molecular mass. Another protein that is also found abundantly is single strand binding protein SSB. The other product is 88kd protein called Terminal Protein (TP). At four hrs after infection the said products are detectable. At 6 hrs, virion capsid proteins are made.
Proteins required for replication:
Viral DNA coded proteins:
· Pre terminal protein (PTP)-80kd, it is processed to 55kd fragment, during which it get linked to CMP.
· DNA-pol-is 140 KD, monomer, responsible for replication.
· SSB protein- 59kd binds to ssDNA.
Host factors:
· Nuclear Factor NF-I (52 to 66kd) and CTF factor (CAAT binding factor) are required for replication initiation; they also act as transcriptional activator.
Nuclear factor, NF-II; 30kd protein, it is a topoisomerase, involved in relaxing super coiled DNA.
· Nuclear factor NF-III; a 92kd protein, it is involved in replication initiation as well as transcriptional activation. Besides the above some other factors such as MLTF, USF, POU, RNA polymerase and TFs are also involved.
The terminal protein, as it is synthesized it is processed and; it’s one of the serine residue is covalently linked to the 5’ end of CTP where pyrophosphates is released. It is believed that ADV-DNA Pol performs this linkage? This precursor protein recognizes the domain A at either end. Now it is very clear that the replication is unidirectional, though both strands can be replicated simultaneously. This mode of replication does not violate semi conservative mode of replication.
As the CMP linked protein contacts, it displaces the 5’CMP linked strand and places the CMP against the first 3’ base Guanine. This base pairing of 5’CMP with 3’GMP provides 3’OH group at CMP as a primer (there are other forms of primers such as short RNA or oligos). T he nuclear factors NF-I and NF-III bind to domain B and C respectively. This virtually displaces the 5’end of the DNA strand by about 40 to 60 ntds. The ADV DNA-polymerase binds to domain–I next to the 3’OH group of the CMP. Now the ADV polymerase, NF-I and NF-III are in an assembly line, to this NF-II, a topoisomerase also joins. As the DNA polymerase extends CMP in complementary strand synthesis, the upper strand is continuously and progressively displaced. The displaced strand is immediately coated with ADV-SsBs.
5’P-Cmp/-A-I-B-I-C-----------------------------------------------G-3’OH
G---------------------------------------------------C-I-B-I-A-/pmC-P3’
During progressive synthesis, if any super coiled regions impede the polymerase; it is relieved by the NF-II, which is a topoisomerase. This way replication proceeds unhindered to the other end.
· Replication can be initiated simultaneously at both ends. Or one strand is replicated first and the other displaced strand can assume panhandle structure; still this can be used for initiating replication.
As the linear DNAs are bound by TP at their respective 5’ends, by protein –protein interactions they can join and show circular permutations. Such circular structures have been observed in infected HeLa cells.

Linear, non-segmented, d/s DNA, 30-38kbp (size varies from group to group) which has the theoretical capacity to encode 30-40 genes. Genome structure (cross-hybridization, restriction map) is one of the characters used to assign viruses to groups (70-95% homology within groups, 5-20% homology between groups).,The terminal sequences of each strand are inverted repeats, hence the denatured single strands can form "panhandle" structures (100-140bp).There is a 55kD protein covalently attached to the 5' end of each strand. wwct.ac.zaw.mcb.u/cann
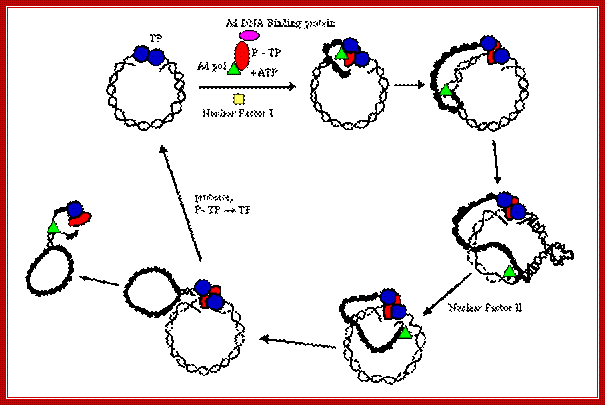
Interaction between Terminal binding proteins makes the replicating Adenoviral DNA look like circular. All the replicating components are labeled. http://www.mcb.uct.ac.za/cann
As the replication starts, late genes are expressed. The transcription generates a 28,000ntds long transcript by R strand. Alternate splicing generates at least 13 individual mRNAs, each having a common 3 partite leader sequence with a cap at 5’ end and a poly-A tail at 3’ end.
Transcripts are transported out of the nucleus and translated. Some of the early gene products such as E2B and E4 facilitate the transportation of viral mRNA and the movement of host mRNAs is blocked by certain design, which is not known. So the viral mRNAs are the only transcripts that are transported and translated.
· The translated viral capsid and other associated proteins move into the nucleus. There the DNA is packed into the prohead and the viruses mature. But they are not released but accumulate as visible esonophilic inclusion bodies. Though the infected cell has stopped its most of the activities, cellular machinery is still used to viral advantage and produces all the proteins for the virus, yet the cell is intact.
· The capsid proteins start pre-assembling as protomers, hexons and pentons and then they move into the nucleus, where they finally assemble, but not released. The viral particles are released by accidental damage to cell or death to the cell.
Adeno viral DNA as vector:
Removal of E1A region from the ADV genome facilitates for cloning the required gene into its place. The cloned gene in ADV DNA can be encapsidatd by using helper cells and thus one can generate large number of live recombinant viruses. Such viruses can directly used for infection of tissues, where the desired gene is targeted too a tissue.
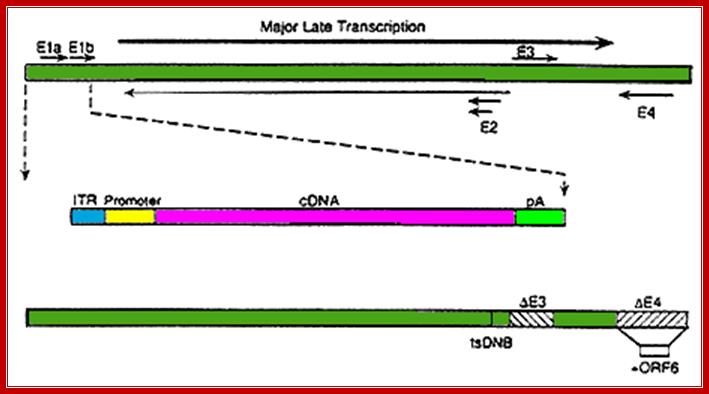
Adenoviral genomic sequences are chosen to develop a vector that can be used to transfer or express a specific cloned gene into human or any eukaryotic mammalian systems; coronodon; www.microbiologybytes.com.
The wild type of viral DNA is about35kb long and nearly 30kb can be replaced with the desired recombinant DNA. The E1,E2,E3 and E4 have regulatory functions. The designed vector contains either E1 or E3 segments inactivated. One can use Tm sensitive E2a mutant or E4 can be deleted. The vector is designed have inverted terminal repeats and packaging sequences. For it to work, use helper viruses. The required gene can be inserted in ITR sequences with packaging sequences around. Such designed vector can be loaded into viral proteins and can be used to infect cells and transfer the desired gene and make it to express, for the adenoviral recombinant DNA has the ability to get inserted in the host genome and express.

Transcription map of human adenovirus serotype 5. The ~36 kb genome is divided into four early region transcription units, E1–E4, and five families of late mRNA, L1–L5, which are alternative splice products of a common late transcript expressed from the major late promoter (MLP) located at 16 map units. Four smaller transcripts, pIX, IVa, and VA RNA’s I and II, are also produced. The 103 bp inverted terminal repeats (ITRs) are located at the termini of the genome and are involved in viral DNA replication, and the packaging signal (ψ) located from nucleotides 190 to 380 at the left end is involved in packaging of the genome into virion capsids.;ADV’s genome map; https://www.omicsonline.org
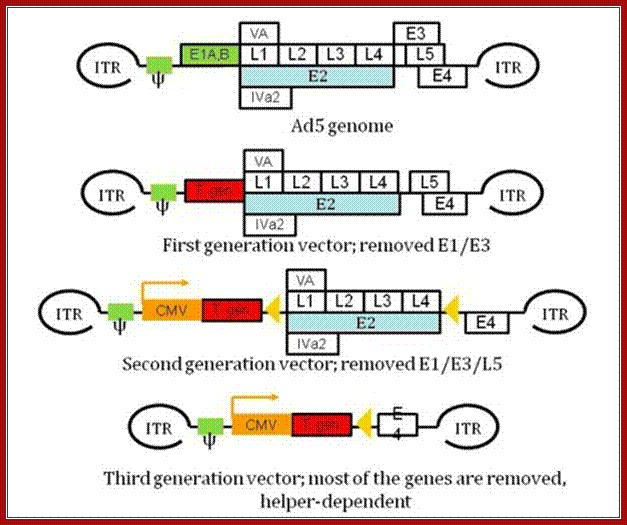
Molecular therapies: Dr. Zoltán Balajthy, et al; http://www.tankonyvtar.hu/
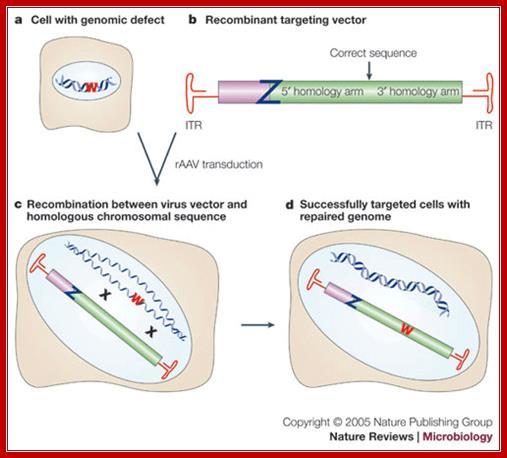
Gene targeting with recombinant adenoviral vectors:
A nonfunctional mutant of a reporter gene (W in the figure) is introduced or is present in a cell (a). In the recombinant targeting vector (b), the viral genes are replaced by a sequence that is homologous to the chromosomal locus targeted for modification. Usually, the modification is introduced between stretches of homology called 5' and 3' homology arms. Z designates an inactivating mutation in the viral repair template. This recombinant adeno-associated virus (rAAV) is used to transduce the cells. Recombination with the chromosomal target (c) can result in repair of the defect and recovery of a healthy cell (d). The frequency of gene targeting is determined as the fraction of infected cells that expresses a functional reporter. Stable integration is confirmed by antibiotic selection and Southern analysis. (In the figure, crosses [X] mark regions of homology between the chromosomal and viral DNA, and ITR indicates an inverted terminal repeat.)
© 2005 Nature Publishing Group Vasileva, A. & Jessberger, R. Precise hit: adeno-associated virus in gene targeting. Nature Reviews Microbiology 3, 841 (2005)
Gene targeting with recombinant ADV vectors;http://www.nature.com/
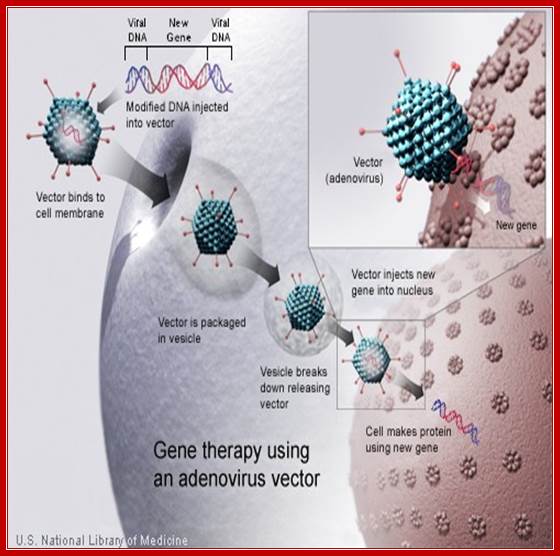 Recombinant
Virus used for human transfection; http://cronodon.com/BioTech/Adenovirus.html
Recombinant
Virus used for human transfection; http://cronodon.com/BioTech/Adenovirus.html
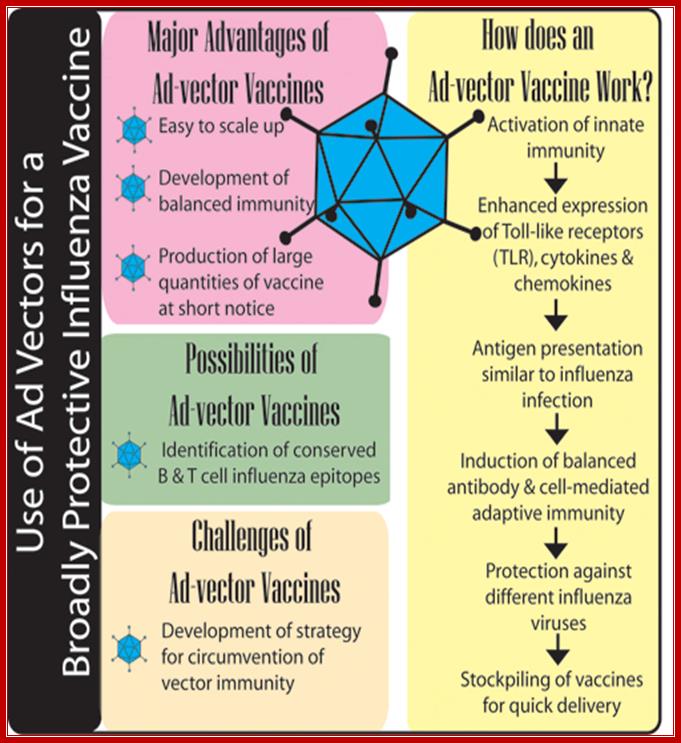
Nonhuman adenoviral vectors for gene therapy and recombinant vaccines; https://www.vet.purdue.edu
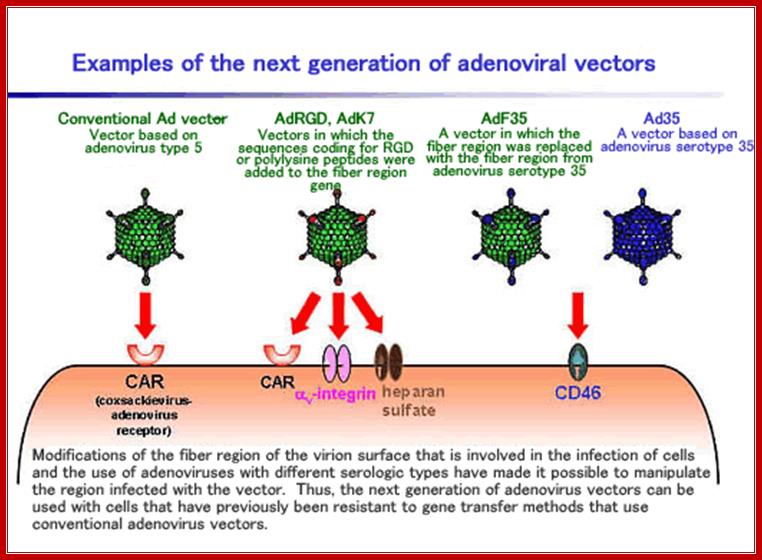
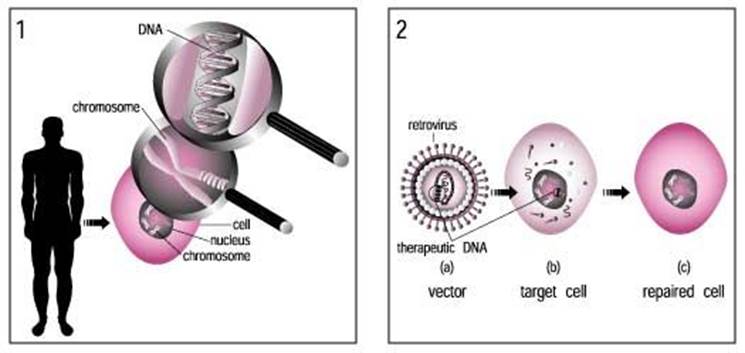
ADV vector used for gene transfer; www.ck12.org/life-science