Yeast Episomal Plasmid DNA Replication:
Yeast plasmid DNA is also known as 2u Episomal plasmid DNA. It is ds DNA, circular, 6.3kbp (about 6kbp?) long, extra chromosomal genetic element found in most of yeast in the nucleus like an Episomal plasmid. The copy numbers of them range from 50 to 100 per haploid genome. It is associated with histones and exists as nucleosomal thread and it is efficiently transmitted to the budding yeast? Its replication mode is typical of eukaryotic, but it heavily depends on host factors for its replication.
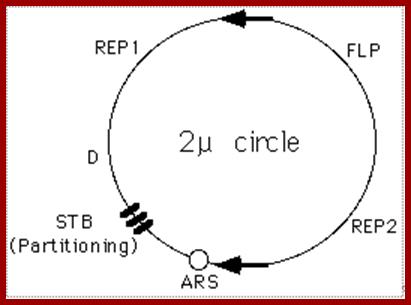
A cartoon of the yeast 2 u circle showing the ARS, the FLP gene, the three genes which encode proteins required for regulation of FLP expression (REP2, REP1, and D), and a set of small direct repeats (called "STB") required for partitioning into daughter cells during mitosis and meiosis.
http://www.sci.sdsu.edu/
The plasmid DNA contains two copies of 559 bp long sequences in inverted orientation. In them there are specific sequences for plasmid directed site specific recombination system called FLP or flip system, so it can exist in two isoforms with equal frequencies. The inversion system is required for plasmid amplification.
Four proteins are involved in replication, maintenance and copy numbers and they are encoded in the plasmid DNA. The 48kd recombinase is the product of FLP or flip gene. Rep1 and Rep2 gene products are required for the stability and stable inheritance and for regulation of the copy numbers by repressing the expression of other plasmid genes. The fourth protein coded for by the gene D, regulates the expression by antagonizing the Rep1 and Rep2 repressor activities.
In addition to flip recombination sequences, it also contains segments for autonomous replication called ARS. Another sequence for the plasmid stability, the locus is know as STB. The ARS is about 200bp long and it contains sequences for replication initiating protein binding, a region for melting and a region for enhancing replication. The binding of pre replication complex such as ORC and other accessory factors binding to ORE melt the DNA at a region called DUE (DNA Unwinding Elements).
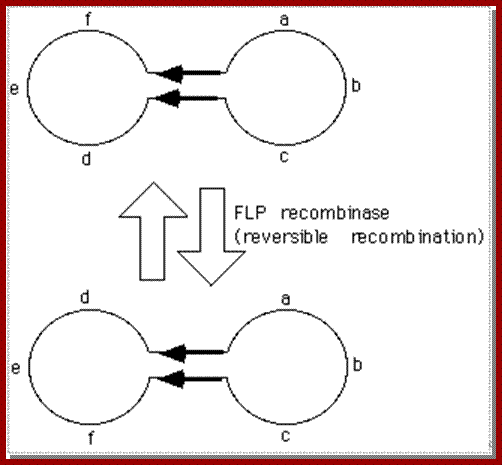
2u yeast plasmid;The 2u circle has a site-directed inversion mechanism that allows plasmid amplification. The 2u circle has two copies of a 599 bp inverted repeat sequence (called "flip" sites) and encodes a site-directed recombinase called FLP (the "flip" protein) that promotes recombination between these repeats. Recombination between the flip sequences inverts the adjacent regions of the plasmid as shown in the figure below. (The inversion is easier to visualize if you draw a crossover between the inverted repeats indicated by the arrows and follow the DNA strands with your pencil.)
http://www.sci.sdsu.edu/
The enzymes associate at replication fork and synthesize complementary strands on leading strand and lagging strand by DNA-pol delta, typical of eukaryotic replication system. The ARS like sequences as the origin of replication occur at specific sites in other eukaryotic chromosomes.
In synchronized cell division of yeast cultures, replication initiates at S phase and within 120 minutes of it, the theta structure appears and disappears in about 40 minutes. The intermediates of replication appear as intertwined, catanated circular DNAs. Between them two isoforms are equally represented in the daughter DNA population. During S phase itself decatanation occurs to yield covalently closed super coiled structures.
For efficient portioning (or portioning) of the plasmids during cell division, the plasmids should contain CEN segments. Such plasmids not only segregate equally but also remain stable. Such structures containing telomeric elements are called yeast Artificial Chromosomes (YAC). The said DNAs with ARS elements can be used to create a linear chromosome. To such structure any foreign DNA of any length (from 10^6 to 10^7bp) of DNA can be incorporated to generate artificial chromosomes, which virtually behave like any eukaryotic chromosomes.
Left to right, start from lower strand-
-----ARS—STB—D—Rep1--stem—FRT—stem---Rep2—Flp--ARS,
This is one of the isoforms.
-----ARS—STB—D—rep1—stem structure-FLP—Rep2—stem—ARS.