Kinds and Components of Origins:
Replicons, a Comparative Account:
|
Species |
Genome size (bp) |
Replicon numbers |
Size of a Replicon |
Rate of replication (bp)/min |
|
E.coli |
4.6 x 10^6 |
1 |
4.6 x ^6 |
50000 to 60000 |
|
Yeast (n=17) |
1.4 x 10^7 |
500 |
40,000 |
36,000 |
|
Fruit fly (n=4) |
1.4 x 10^8 |
3500 |
40,000 |
2800 |
|
House fly (n=4) |
8.6 x 10^8 |
4000 |
45,000 |
2800 |
|
X.laevis (n=) |
3.1 x 10^9 |
15000 |
200kbp |
2500 |
|
|
|
|
|
|
|
Mus musculus |
2.6 x 10^9 |
25000 |
150kbp |
2200 |
Ori-Elements, Their Size, Binding Proteins, Melting Points, Transcriptional Effects
|
Source of Ori |
Size |
Binding Proteins |
Melting Regions |
Transcriptional effects |
SV 40 |
4x5 bp-AT rich |
T-antigen, Hexamer |
Palindromes |
Stimulates |
|
HSV-1 |
2x8 bp repeats, AT rich in the center |
Gp-ul 9 |
AT region |
- |
EBV Ori-P |
4x30bp repeats, Two palindromes, 2x30bp enhancers
|
EBNA-1 |
- |
Enhancers stimulate |
|
Ori-Lyt |
2 non-contiguous regions |
- |
- |
One enhancer overlaps the other |
|
BPV |
2 noncontiguous regions |
E-1 |
- |
Similar to EBV |
|
Mitochondrial Origins: |
|
|
|
|
|
H-strand |
Promoter & RNA binding site |
D-loop |
D-loop |
Promotes |
|
L-strand |
Palindrome |
- |
- |
Promotes |
DNA replication Inhibitors:
Cancer is due to uninhibited cell DNA replication. Use of DNA replication inhibitors is useful in therapy. Isotope labeling protocol and 2-D gel Replicon mapping provide the effect of inhibitors on DNA replication events.
DNA elongation inhibitors such as Hydroxyurea, Amphidicololin, and Cytosine arabinoside act at chain elongation level, but ineffective in preventing fork movement. But Mimosine and Ciclopirox olamine (CPX) appear to inhibit initiation at origins. It is found very effective in yeast cells.DNA pol alpha is inhibited by Amphidicolin and its derivative amphidicolin glycine.,
DNA Topoisomerases are inhibited Novobiocin, Nalidixic acid, Teniposide, etoposide and 4’-9-Acridylamine metanesulfon-m-anisidide,
Sodium butyrate, 3-Aminobenzamide and nicotinamide modify chromatin structure; Coralyne and 5,6-dihydroxycoralyne inhibit Topoisomerase-1.
Dimethylzeylasterone inhibits Topoisomerase subunit II,
Some of the antibiotics have been found to act on DNA replication:
Trimethoprim (methatrexate, aminopterin) inhibit dihydrofolate reductase, Hydroxy urea- inhibit ribonucleotide reductase, 5-Ffluorouracil competes with dUMP, Dideoxynucleotide prevent nucleotide oligomerization, Mitomycin crosslinks DNA strands and prevent replication progression, Acridine dyes such as Proflavin, and Ethidiumbromide bind to major groove of DNA and prevent replication, Nalidixic acid binds to Gyrase-A, and Novobiocin acts on Gyrase B,
Rifampicin inhibits RNA pol.
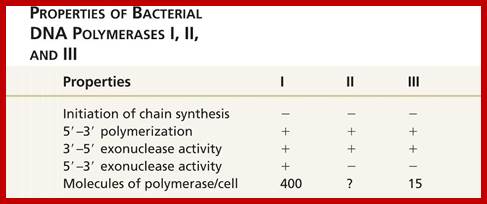
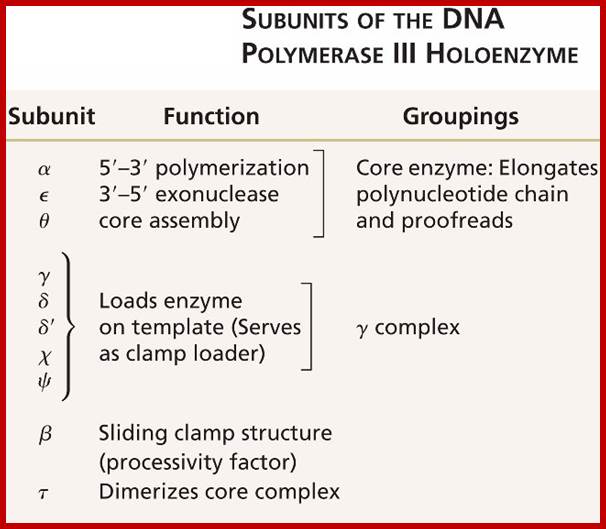
DNA Polymerases:
A list of polymerases; PolA(pol1), PolB, PolG (mitochondrial Replication), PolD1(pol3), PolE(pol2), PolZ, Pol H, Q, Pol K, PolL(polIV), Pol M Pol N and Rev1(inserts only one nucleotide in template independent manner).

Eukaryotic DNA pols;

www.bio3400.nicerweb.ne


Pol IV; Upregulation of Pol IV by the SOS and RpoS stress responses enables it to outcompete other polymerases and have a dominant role in RDR during stress, which facilitates error-prone RDR. Pol II, however, intermittently competes with Pol IV through its exonuclease domain. D loop–dependent stimulation of Pol II exonuclease activity enables the polymerase to move in reverse and partially resect the extended D loop. This activity probably contributes to proofreading Pol IV errors and suppresses error-prone RDR. Pol IV then regains access to the D loop by displacing Pol II from the DNA.
There are more than four DNA polymerases, of which Meselson Stahl Translesion DNA pol IV in DNA repair and replication.
Notice that polymerases involved in bypass synthesis make up an appreciable fraction of all known eukaryotic DNA polymerases. Notice that Rev1 does not have a Greek name. That's because it's not considered a true DNA polymerase, since it can insert only one nucleotide at a time, usually in a template-independent fashion. Polymerases iota and kappa are related to pol eta but have somewhat different bypass specificities and are not usually error-free. Pol kappa is found in most other eukaryotic organisms, including the fission yeast, Schizosaccharomycs pombe, but is not found in S. cerevisiae. Pol iota, pol theta and pol nu appear to be found only in animals. Pol theta is especially competent at bypassing AP sites, where it inserts an A residue. Since most AP sites are consequences of purine loss, and A is a purine base, repair by pol theta is frequently error-free. Apart from replication many are involved in direct Damage repair, Mistmatch repair, Base excision repair, Nucleotide Excision repair Double strande break repair and Damage bypass repair-http://joelhuberman.net
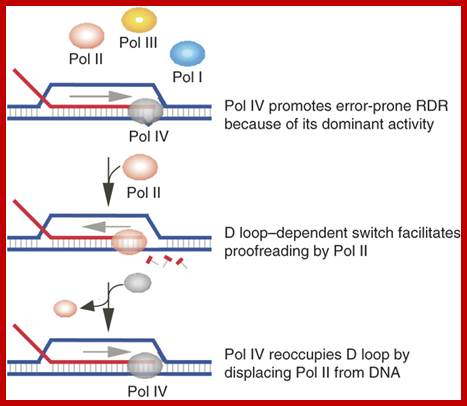
Model of translesion DNA polymerase activity at D loops during stress; Preferential D-loop extension by a translesion DNA polymerase underlies error-prone recombination
Richard T Pomerantz, Isabel Kurth, Myron F Goodman
& Mike E O'Donnell; ;http://www.nature.com