Replication Machinery:
Components of Replication Machinery:
|
Mammalian enzymes- genes |
Alpha |
Delta |
Beta |
Gamma |
Epsilon |
|
Yeast enzymes (equivalents) |
Pol-1 |
Pol-III |
Pol-II |
Mit-pol |
Leading strand |
|
Yeast Genes |
Pol-I |
Pol-III |
Pol-II |
|
|
|
Mol. mass (native) |
350-356(4-subunits
|
220(3-subunits |
160-270,
|
256 105 (plastid) |
386 (4subunits) |
|
Catalytic core |
165-180 |
125 |
36-38 |
125, 35 (1:1) |
215 |
|
Other subunits |
70,(50,60 (primase) |
48 |
- - |
35, 47 |
55 |
|
Location |
Nucleus |
Nucleus |
Nucleus |
Mitochondria |
Nucleus |
|
5’>3’ pol activity |
Yes |
Yes |
Yes |
Yes |
- |
|
3’>5’ exonuclease activity |
Yes |
Yes (overt) |
- |
Yes |
Yes |
|
Primase activity |
Yes |
No |
- |
-No |
No |
|
Primer extension |
Yes, on lagging/leading strand |
DNA synthesis on lagging strands strand |
Yes in DNA-damaged |
Yes |
DNA synthesis on Leading strand |
|
Response to auxiliary factors |
Yes |
No |
No |
No |
No |
|
Response to PCNA |
No |
Yes |
No |
No |
No |
|
Template |
Yes, gapped |
Yes, poly-ntds |
Yes, gapped |
Yes to oligo’s |
|
|
Response to Mg+ |
high |
high |
high |
Respond Mn^2+ |
high |
|
Processivity |
High |
2-200 ntds |
>600 ntds |
High |
|
|
Fidelity |
High |
High |
High |
high |
High |
|
Sensitive to: Amphidicolin- ddNTPS |
High, None |
High, None |
None High |
High, High |
High |
|
N ethylmaleimide |
strong |
strong |
|
strong |
|
|
|
|
|
|
|
|
Initiation components:
Opening: ORE binding factors such as ORCs (OBFs six subunits), Cdc6, Cdt1, MCM (2-7) (minichromosome maintaining factors), Cdc7/dbf4, RF-C, RP-A, PCNA (helicase), Cdc45 and few kinases, phosphotases and transcriptional factors; all together may induce melting and stabilize the replication bubble.
ORC: Origin Recognition Complex consists of six subunits of ~400kd mass. They are transported into the nucleus and bind to ORE region and some parts of it overlaps on to B1 site of the ARS. With Cdc6 and Cdt1 it forms pre initiation complex (pic or PRC ). The factor Cdt1 activates Cdc6, which is a highly unstable protein; its half-life is 5-6 minutes, and it is acquired when nuclear membranes disassembled at the beginning of prophase. Activated Cdc6 is responsible for loading/ recruiting MCM hexamers, which sits on either side of the ORC. Cdc6 and Cdt1 phosphorylate MCMs and activate their helicase activity. This leads to the opening of the dsDNA into replication eye and replication bubble. MCM10 and Cdc7/dbf4 component are also required. Once the DNA replication initiates CDC6 are phosphorylated and released. They are degraded via ubiquitination pathway.
A Family of DNA Polymerases-General:
Prokaryotic DNA polymerases;
Bacteria have 5 known DNA polymerases:
- Pol I: Is implicated in DNA repair and has both 5'->3'(Nick translation), primer formation and 3'->5' (Proofreading) exonuclease activity.
- Pol II: Pol II is involved in replication of damaged DNA and has both 5'->3'chain extension ability and 3'->5' exonuclease activity.
- Pol III: is the main polymerase in bacteria (elongates primer in DNA replication), as such it has 3'->5' exonuclease proofreading ability.
- Pol IV: is a Y-family DNA polymerase
- Pol V: is a Y-family DNA polymerase and participates in bypassing DNA damage
Eukaryotic DNA polymerases;
Eukaryotes have at least 15 DNA Polymerases:
- Pol α: acts as a primase (synthesizing a RNA primer), and then as a DNA Pol elongating the primer with DNA nucleotides. Pol α (alpha), Pol δ (delta), and Pol ε (epsilon) are members of Family B Polymerases and are the main polymerases involved with nuclear DNA replication. Pol α consists of four subunits, two α and two-subunit primase which are encoded by the POLA1 and POLA2 genes. Once primase produces RNA primers, Pol α starts replication elongating the primer with ~20 nucleotides. Due to their high processivity, Pol ε and Pol δ take over the leading and lagging strand synthesis from Pol α respectively. After a few hundred nucleotides elongation is taken over by Pol δ and ε.
- Pol β: is implicated in repairing DNA.
- Pol γ: replicates mitochondrial DNA.
- Pol δ: is the main polymerase in eukaryotes, it is highly processive and has 3'->5' exonuclease activity; now it is confirmed it replicates Lagging strand too. Pol δ is expressed by genes POLD1, creating the catalytic subunit, POLD2, POLD3, and POLD4 creating the other subunits that interact with Proliferating Cell Nuclear Antigen (PCNA) which is a DNA clamp protein that allows Pol δ to possess processivity.
Pol ε: Instead of Pol δ, which was believed to replicate both strands, now it known it replicates leading strand. Pol ε is encoded by the POLE, the catalytic subunit, POLE2, and POLE3 genes. While Pol ε's main function is to extend the leading strand during replication, Pol ε's C-terminus region is thought to be essential to cell vitality as well. The C-terminus region is thought to provide a checkpoint before entering anaphase, provide stability to the holoenzymes, and add proteins to the holoenzymes necessary for initiation of replication.
- η, ι, κ, and Rev1 are Y-family DNA polymerases and Pol ζ is a B-family DNA polymerase. These polymerases are involved in the bypass of DNA damage.
- There are also other eukaryotic polymerases known, which are not as well characterized: θ, λ, φ, σ, and μ; there are also others, but the nomenclature has become quite jumbled. See the External Links or do a quick search of PubMed to get the most up-to-date information.
· None of the eukaryotic polymerases can remove primers (5'->3' exonuclease activity), that function is carried out by other enzymes. Only the polymerases that deal with the elongation (γ, δ and ε) have proofreading ability (3'->5' exonuclease).
Additional components of Eukaryotic DNA polymerases;
http://en.wikipedia.org/wiki
Polymerase beta, lambda, sigma, and mu
Family X polymerases contain the well-known eukaryotic polymerase pol β (beta), as well as other eukaryotic polymerases such as Pol σ (sigma), Pol λ (lambda), Pol μ (mu), and Terminal deoxy nucleotidyl transferase (TdT). Family χ polymerases are mainly found in vertebrates and a few are found in plants and fungi. These polymerases have highly conserved regions that include two helix-hairpin-helix motifs that are imperative in the DNA-polymerase interactions. One motif is located in the 8 kDa domain that interacts with downstream DNA and one motif is located in the thumb domain that interacts with the primer strand. Pol β, encoded by POLB gene, is required for short-patch base excision repair, a DNA repair pathway that is essential for repairing alkylated or oxidized bases as well as abasic sites. Pol λ and Pol μ, encoded by the POL-L and POL-M genes respectively, are involved in non-homologous end-joining, a mechanism for rejoining DNA double-strand breaks due to hydrogen peroxide and ionizing radiation, respectively. TdT is expressed only in lymphoid tissue, and adds "n nucleotides" to double-strand breaks formed during V(D)J recombination to promote immunological diversity.
Polymerase eta, iota, and kappa
Pol η (eta), Pol ι (iota), and Pol κ (kappa), are Family Y DNA polymerases involved in the DNA repair by translesion synthesis and encoded by genes POLH, POLI, and POLK respectively. Members of Family Y have 5 common motifs to aid in binding the substrate and primer terminus and they all include the typical right hand thumb, palm and finger domains with added domains like little finger (LF), polymerase-associated domain (PAD), or wrist. The active site, however, differs between family members due to the different lesions being repaired. Polymerases in Family Y are low fidelity polymerases, but have been proven to do more good than harm as mutations can cause various diseases, such as skin cancer and Xeroderma Pigmentosum Variant (XPS). The importance of these polymerases is evidenced by the fact that gene encoding DNA polymerase η is referred as XPV, because loss of this gene results in the disease Xeroderma Pigmentosum Variant. Pol η is particularly important for allowing accurate translesion synthesis of DNA damage resulting from ultraviolet radiation or UV. The functionality of Pol κ is not completely understood, but researchers have found two probable functions. Pol κ is thought to act as an extender or an insterter of a specific base at certain DNA lesions. All three translesion synthesis polymerases, along with Rev1, are recruited to damaged lesions via stalled replicative DNA polymerases. There are two pathways of damage repair leading researchers to conclude that the chosen pathway depends on which strand contains the damage, the leading or lagging strand.
Polymerase Rev1 and zeta
Pol ζ another B family polymerase, is made of two subunits Rev3, the catalytic subunit, and Rev7, which increases the catalytic function of the polymerase, and is involved in translesion synthesis. Pol ζ lacks 3' to 5' exonuclease activity, is unique in that it can extend primers with terminal mismatches. Rev1 has three regions of interest in the BRCT domain, ubiquitin-binding domain, and C-terminal domain and has dCMP transferase ability, which adds deoxy cytidine opposite lesions that would stall replicative polymerases Pol δ and Pol ε. These stalled polymerases activate ubiquitin complexes which in turn disassociate replication polymerases and recruit Pol ζ and Rev1. Together Pol ζ and Rev1 add deoxy cytidine and Pol ζ extends past the lesion. Through a yet undetermined process, Pol ζ disassociates and replication polymerases reassociate and continue replication. Pol ζ and Rev1 are not required for replication, but loss of REV3 gene in budding yeast can cause increased sensitivity to DNA-damaging agents due to collapse of replication forks where replication polymerases have stalled.
Family of DNA Polymerases:
Based on sequence homology, DNA polymerases can be further subdivided into seven different families A, B, C, D, X, Y, and RT.
Family A: Family-A polymerases contain both replicative and repair functions. Replicative members from this family include the extensively studied T7 DNA polymerase as well as the eukaryotic mitochondrial DNA polymerase γ. Among the repair polymerases are E. coli DNA pol I, Thermus aquaticus pol I, and Bacillus stearothermophilus pol I. These repair polymerases are involved in excision repair and processing of Okazaki fragments generated during lagging strand synthesis.
Family B: Family B mostly contains replicative polymerases and includes the major eukaryotic DNA polymerases α, δ, ε, and also DNA polymerase ζ (zeta). Family B also includes DNA polymerases encoded by some bacteria and bacteriophages, of which the best characterized, are from T4, Phi29 and RB69 bacteriophages. These enzymes are involved in both leading and lagging strand synthesis of phage DNA. A hallmark of the B family of polymerases is their remarkable accuracy during replication and many have strong 3'-5' exonuclease activity (except DNA polymerase α and ζ which have no proofreading activity).
Family C: Family C polymerases are the primary bacterial chromosomal replicative enzymes and thus have polymerase and 3'-5' exonuclease activity.
Family D: Family D polymerases are still not very well characterized. All known examples are found in the Euryarchaeota subdomain of Archaea and are thought to be replicative polymerases.
Families X: Family X contains the well known eukaryotic polymerase pol β as well as other eukaryotic polymerases such as pol σ, pol λ, pol μ, and terminal deoxynucleotidyl transferase (TdT). Pol β is required for short-patch base excision repair, a DNA repair pathway that is essential for repairing abasic sites. Pol λ and Pol μ are involved in non-homologous end joining, a mechanism for rejoining DNA double-strand breaks. TdT is only expressed in lymphoid tissue and adds "n nucleotides" to double-strand breaks formed during V(D)J recombination to promote immunological diversity.
Families Y: The Y-family polymerases differ from others in having a low fidelity on undamaged templates and in their ability to replicate through damaged DNA. Members of this family are hence called translesion synthesis (TLS) polymerases. Depending on the lesion TLS polymerases can bypass the damage in an error-free or error-prone fashion, the latter resulting in elevated mutagenesis. Xeroderma pigmentosum variant (XPV) patients for instance have mutations in the gene encoding Pol η (eta), which is error-free for UV-lesions. In XPV patient’s alternative error-prone polymerases; e.g. Polζ (zeta). Polymerase ζ is a B Family polymerase, is thought to be involved in mistakes during replication which result in the cancer predisposition of patients. Other members in humans are Pol ι (iota), Pol κ (kappa) and Rev1 (terminal deoxycytidyl transferase). In E.coli two TLS polymerases, Pol IV (DINB) and PolV (UMUC), are known.
Family RT: Finally, the reverse transcriptase family contains examples both from retroviruses and eukaryotic polymerases. The eukaryotic RT polymerases are usually restricted to telomerases. These polymerases use a RNA template to synthesize the DNA strand.
|
Family |
Types of DNA polymerases |
Species |
Examples |
|
A |
Replicative and Repair Polymerases |
Eukaryotic and Prokaryotic |
T7 DNA polymerase, Pol I, and DNA Polymerase γ |
|
B |
Replicative and Repair Polymerases |
Eukaryotic and Prokaryotic |
Pol II, Pol B, Pol ζ, Pol α, δ, and ε |
|
C |
Replicative Polymerases |
Eukaryotic |
Pol III |
|
D |
Replicative Polymerases |
Not well characterized |
|
|
X |
Replicative and Repair Polymerases |
Eukaryotic |
Pol β, Pol σ, Pol λ, Pol μ, and Terminal deoxynucleotidyl transferase |
|
Y |
Replicative and Repair Polymerases |
Eukaryotic |
Pol ι (iota), Pol κ (kappa), Pol IV, and Pol V |
|
RT |
Replicative and Repair Polymerases |
Viruses, Retroviruses, and Eukaryotic |
Telomerase, Hepatitis B virus |
Eukaryotes Polymerases (There at least 15 DNAPols): Hubscher, U.; Maga, G.; Spadari, S. (2002) Eukaryotic DNA polymerases. Annual Review of Biochemistry 71, 133-63.</ref>:
Pol
α:
acts as a primase (synthesizing a RNA primer), and then as a DNA Pol elongating
that primer with DNA nucleotides. After a few hundred nucleotides elongation is
taken over by Pol δ and ε.
Pol
β:
is implicated in repairing DNA.
Pol
γ:
replicates mitochondrial DNA.
Pol
δ:
is the main polymerase in eukaryotes, it is highly processive and has 3'->5'
exonuclease activity.
Pol
ε:
may substitute for Pol δ in lagging strand synthesis, however the
exact role is uncertain.
η, ι, κ,
and Rev1 are Y-family DNA polymerases and Pol ζ is a
B-family DNA polymerase. These polymerases are involved in the bypass of DNA
damage.<ref>I. Prakash, S.; Johnson, R. E.;
Prakash, L. (2005) Eukaryotic translesion synthesis DNA polymerases:
specificity of structure and function. Annual Review of Biochemistry 74,
317-53.</ref> .
There
are also other eukaryotic polymerases known, which are not as well
characterized: θ, λ, φ, σ, and μ.
There are others too, but their nomenclature has become quite jumbled. See the
External Links or do a quick search of PubMed
to get the most up-to-date information.
None of the eukaryotic polymerases can remove primers by 5'->3' exonuclease activity; that function is carried out by other enzymes. Only the polymerases that deal with the elongation (γ, δ and ε) have proofreading ability (3'->5' exonuclease).
|
Family |
Types of DNA polymeras |
Species |
Examples |
|
A |
Replicative and Repair Polymerases |
Eukaryotic and Prokaryotic |
T7 DNA polymerase, Pol I, and DNA Polymerase γ |
|
B |
Replicative and Repair Polymerases |
Eukaryotic and Prokaryotic |
Pol II, Pol B, Pol ζ, Pol α, δ, and ε |
|
C |
Replicative Polymerases |
Prokaryotic |
Pol III |
|
D |
Replicative Polymerases |
Not well-characterized |
|
|
X |
Replicative and Repair Polymerases |
Eukaryotic |
Pol β, Pol σ, Pol λ, Pol μ, and Terminal deoxynucleotidyl transferase |
|
Y |
Replicative and Repair Polymerases |
Eukaryotic and Prokaryotic |
Pol ι (iota), Pol κ (kappa), Pol IV, and Pol V |
|
RT |
Replicative and Repair Polymerases |
Viruses,Retroviruses, and Eukaryotic |
Telomerase, Hepatitis B virus |
http://en.wikipedia.org/
Replication enzymes and factors:
DNA polymerase-α:
The enzyme is called Pol α / primase. It has four polypeptides. The total mass is 350 KD, consists of 167, 79 (70-75), 62 (55-60), and 48 (45-50) - a tetramer, one of it has a catalytic core at C-terminal and cryptic 3’—5’ exonuclease at N-terminal (?). The terminal region helps in the binding of primase and other factors into Holozyme. Concentration of this enzyme is high during cell division and very low in quiescent cells. Subunits 70-75 KD found in yeast cells may have regulatory functions. Subunits 55-60, 48-50 has primase activity. The 55kd protein has rNTP-binding site. Most of the a pol protein components are induced during cell division especially during Go to G1 stage. They have affinity to ss DNA, and rNTPs. Though they lack apparent exonuclease activity, its fidelity and the rate of synthesis is good and its error rate is one in 10^6 ntds incorporation. However it is suspected that the complex has cryptic (3’—>5) exonuclease activity, which is essential for replication.
Now it is clear about the role of DNAa-pol; its important function is to bind to both leading and lagging strands and produce a short RNAs as primers of ten nucleotides long. Once it was thought that this enzyme is responsible for laying primers on both leading and lagging strands and then to work as polymerase on lagging strand. It is now more or less accepted that pol-a / primase performs only primer synthesis. Once the primers are laid, it is displaced by RF-C and PCNA and then delta-DNA Pol are loaded. Polymerase -a is also known to be stimulated by RNase-H. It has no repair function.
Protein: N end –----Exonuclease—Pol----Zif-Zif—C end-.
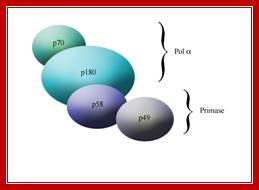
Schematic of the subunits of pol α. A depiction of the subunit organization of the two activities: polymerase and primase, that comprise DNA polymerase α.
www.etd.library.vanderbilt.edu
DNA polymerase alpha: primase is comprised of four subunits, p180, p70, p58, and p49. The two primase subunits, p58 and p49, form a tight complex. The p49 subunit contains the DNA primase activity and one role of p58 appears to be tethering p49 to p180, the DNA polymerase catalytic subunit. The fourth subunit, p70, binds p180 and may tether the DNA polymerase alpha: primase complex to Cdc45.
[Frick & Richardson 2002, Copeland & Wang 1994]; http://etd.library.vanderbilt.edu/
RF-A (RPA): It is a ssDNA binding protein, also called RPA, a trimer, a heteromer (70, 32 and 13 KD) similar to bacterial SSB protein. It also binds to helicase as well as polymerase. RF-A is phosphorylated in cell cycle dependent manner.
RF-C: Pentameric complex of proteins helps in extending the primer into I-DNA. Its subunits are 140, 40, 38, 37 and 36 (A, B, C, D and E). It is also involved in loading PCNA and DNA polymerase, so this is called clamp loader. It binds to 3’end of the primer. Binding and association with Pol-alpha induces iDNA (initiator) synthesis.
As soon as the dsDNA opens, they are bound by SSBs (RPA) and DNA Pol-alpha lays primers, both 10 ntds long RNA primer and 100 ntds long iDNA with the help of RF-C.
PCNA: Mol. mass 36 (28.7) KD, trimeric protein (homo-trimer), its function is to displace Pol a/primase and assist pol-d and holds enzyme on to the template, similar to the beta clamps of E.coli. It holds to the 3’end of the I-DNA. While PCNA loading, ATP hydrolysis makes the clamp open for loading on to the template. It increases pol delta processivity. It acts as sliding clamp. PCNA protein complex is called Proliferating Cell Nuclear Antigen. RF-C is the loader of the PCNA clamp in ATP dependent manner. It simulates the activity of bacterial beta clamp loader.
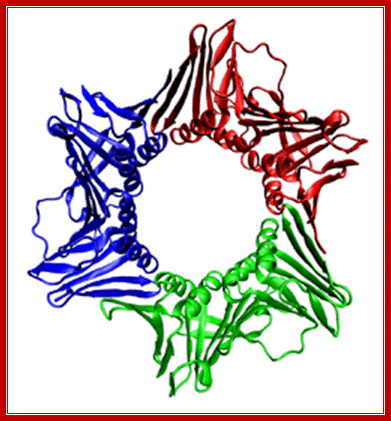
The assembled human DNA clamp, a trimer of the protein PCNA. http://en.wikipedia.org/
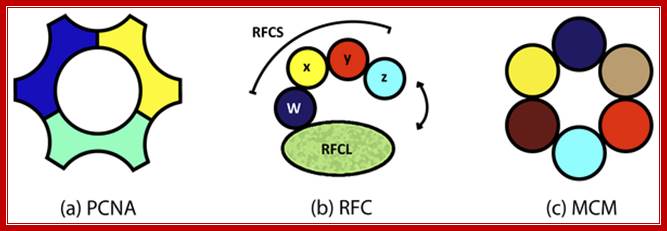
Three subunits act as PCNA-clamp, RFC as clamp loader and MCM helicase respectively; http://labrat.fieldofscience.com/
DNA Pol d:
It is located in the nucleus along with DNA pol-a. Catalytic core consists of four subunits such as 125, 515, 40 and 12. According to some the delta consists of 3 subunits (125, 55, 40). The enzyme has polymerase activity and an overt 3’->5’ exonuclease activity, which means it, performs proof reading. The d Holozyme requires RF-C and PCNA (proliferating cell nuclear antigen) to displace the a-DNA-pol and loading the delta-polymerase on to the template. P40 interacts with PCNA. It is responsible for the synthesis of leading strand, but it is now discerned that d -pol is responsible for the synthesis of complementary strand on lagging strand. Interestingly pol-delta is also responsible for filling up the gaps during primer removal and repair functions.
DNA polymerase delta sub unit; blogqpot.com
Protein: N end----Exonuclease—Pol----Zif -Zif—C end
Pol ε: The pol epsilon is a nuclear protein, tetramer (255, 79, 29,23 kd), it is now confirmed that it is involved in polymerizing leading strand of DNA. It has 5’-3’ pol activity and 3’-5’ exonuclease activity.
Protein: N end---Exo---Pol-----essential---zif Zif c end.
Now it is known in yeast epsilon performs the synthesis of leading strand and delta synthesizes lagging strand.
Pol-b: It is a nuclear enzyme; Mol.wt is 36 to 38 KD. DNA damage induces the synthesis of b enzymes. It is implicated in high fidelity DNA repair functions such as base excision.
DNAP beta; https://es.wikipedia.org
Zeta (ζ): This is complex of heteromeric involved in transletion repair i.e. Thymine dimers bypass. The enzyme is a heteromer.
Pol u, n (eta), k (kappa), ί (iota) & Rev: They are considered as low fidelity repair enzymes. All these are involved in transletion repair mechanism. Eta is a monomer, involved in base pair damage repair, iota is again a monomer, required for DNA repair at meiotic stages, kappa is a monomer, performs repair of deletion and base substitution reactions
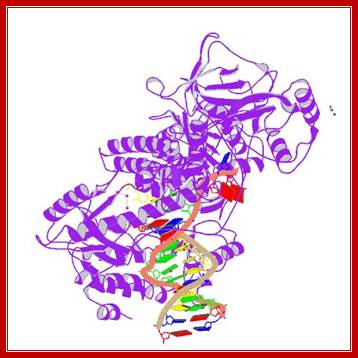
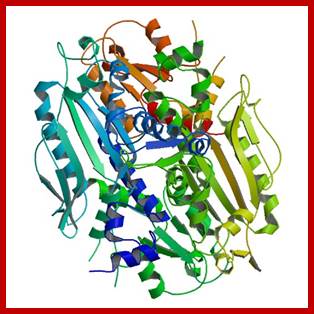
Pol ¡: It is a mitochondrial enzyme, synthesized in the nucleus and transported into mitochondria. Mol. mass is 140 and 55 KD organized into Holozyme by associating themselves in 1:1 ratio. It uses a variety of templates and primers, gapped DNA; ss primed DNA and circular DNA as well. It has polymerase as well as 3’—>5’ nuclease activity. It has high processivity and fidelity. A similar type of Polymerase is also found in plastids but its Mol.wt is 105 KD.
DNAPol-gamma subynit; http://www.rcsb.org
Positions of Polymerase subunits in the complex; http://www.biochem.umd.edu/
Mp4 protein kinase: Mol. mass is 34 KD and it has phosphorylation function?
Topoisomerases: Topoisomerase-I, 110 (125kd); and Topoisomerase-II (IIa-170, IIb 180KD) responsible for relaxing the super coiled DNA ahead of the replication fork, involved in decatenation. Among several hundreds of proteins found bound to chromatin, the most abundant chromosomal protein is topoisomerase. One can visualize the importance of it in maintaining chromosomal DNA intact.
Type 1 topoisomerases:
These enzymes remove super coils through a mechanism that involves breaking only one of the two phosphodiester backbones. As a result, these enzymes change the linking number by 1 each time.
The best-characterized member of this class in E. coli is Topoisomerase I. This enzyme is 864 amino-acids in length and is monomeric; it is encoded by the topA gene. The amino-terminal part of the enzyme (aas 2-590) has been crystallized:
The C-terminal 14 kD fragment of topoisomerase I interacts with ssDNA and with dsDNA and functions to enhance the binding of the holozyme to DNA. The protein folds into 4 domains which are proposed to contain a dsDNA molecule. The three images above show three slightly different views of the structure. You can see how the enzyme might bind DNA in the central image. From the left hand view, you can also see how it might wrap around a DNA molecule. If you look carefully at the right hand image, you will see on the upper left part of the structure that the blue colored domain and the green colored domain are quite separate; it is proposed that this part of the protein opens up to permit the entry and exit of a double stranded DNA molecule.
Topoisomerase II:
Structure Topoisomerease 2-http://reasonandscience.heavenforum.org
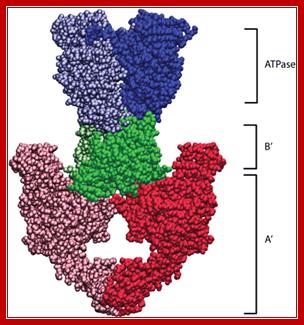
Structure of yeast topoisomerase II: Structures of Saccharomyces cerevisiae Topo II (PDB 1PVG and 1BGW) were computationally docked together to illustrate a possible representation of the enzyme. ATPase domains, shown in blue, form the N-gate. The central B′-sub fragment forms the DNA gate (green). The C-terminal A′ portion (red) encloses a hole large enough to accommodate T-segment DNA, which is thought to exit through the C-gate.
Helicase: Assemble at fork. In SV40 T-antigen (an hexamer) acts as an helicase in SV40. In most of the eukaryotes such as Xenopus and lily others MCM as a hexamer with SRF and p-loop motifs acts as an helicase; this complex is activated by kinases and it moves in 5’à3’ in direction and unwind the DNA. The MCM is recruited to ORC along with Cdc6 and Cdt1
MPF1: Mitosis promoting factor; it plays a role in progression of Mitosis.
DNA ligase: It ligates nicks with 5’phosphorylated with 3’OH groups; its Mol.wt is 125, there are four ligases in human systems and they use ATP energy. Eukaryotic ligases are different from that of prokaryotes, and they are responsible for joining the nicked ends by Phosphodiester bond formation. They use ATP for activation.
FEN 1 and RNase H: They have a role in nibbling the RNA primers (5’-3’exonuclease) in removing the primers. FEN1 has endo nuclease activity too. RNase H removes 5’triphosphorylated ribonucleotides. PCNA stimulates its activity.
Replication Components found to Participate During SV 40 DNA and other Replication Systems (generalized).
|
Factor |
Mol.wt (KD) |
Function |
|
T-antigen |
90, hexamer, MCM in other metazoans; viral product, |
3’>5’ Helicase ploughs thru leading strand DNA and opens |
|
RPA- single strand binding protein |
70, 34, 11- host cell product, |
Binds to ssDNA, promotes ds opening, stimulates helicase, RFC, PCNA and pol-delta |
|
DNA-pol alpha |
167(170),79, 62, 48 (60, 50) [167,83,58,48] [165,67,58,48] 58&48 –primase; host cell product, |
Lays primers on both leading and lagging strands, has high fidelity, but no 3’>5’exonuclease activity |
|
DNA pol delta |
125, 55, 40, [12], host cell derived, host cell |
125 subunit has pol and 3>5’ exonuclease activity, 40 interacts with PCNA |
|
DNA pol epsilon,
|
215,79,23,22- host cell
|
Both have DNA repair activities, function not known |
|
RFC |
140,40,38,37,36-host cell |
Complex acts a loader of clamp and pol on to the template at fork |
|
PCNA |
36, homo-trimmer, host cell |
Acts like a clamp holds pol-delta at primer-template joint |
|
Topoisomerase-I |
110, host cell |
Relieves torsional strain ahead of the fork movement, breaks and reseals single strands |
|
Topoisomerase II |
II-A-170 dimer II-B-180, dimer Host cell |
Breaks double strands and ligates after removing super coils. |
|
FEN-1 |
43 |
Acts as 5-3’ exonuclease removes primers from 5’ end |
|
DNA ligase |
125 |
ATP dependent, ligates the nicks |
|
RNase-H1 |
89 |
Removes RNA in cells that express retro-transpons |
|
DNA ligase |
125 |
Four types, require ATP |
|
ORC |
Hexamer |
Binds to origin |
|
Cdc6/cdt1 |
Licensing factor-2 |
Loads Mcm |
|
Mcm 2-7 and MCM10 |
97.2,108.7,104.4,86.9,120.5,94 Kds respectively |
Helicase |
|
Cdc7, Dbf4-p |
Kinase |
Cdc7activates dbf7, which p.lates Mcm |
|
Cdc45 |
|
Require for pol assembly |
|
GINs,(GCN- independent), Geminin |
Geminin- sequester cdt1/cdc6 |
Sequester cdt1 |
A list of Yeast Genes and Gene Products Involved in DNA Replication:
Yeast is a unicellular system; it can be grown very easily to high density. It exhibits Haplo, Diplo and Haplo-Diplobiontic type of cell cycle. One can generate a large number of conditional, nutritional mutants, which can be sustained and maintained. It is easy to genetically manipulate the cells. Identification, isolation and purification of the products are relatively better, but not easy. Regarding regulation of cell cycle events, yeast has provided mutants to each of the steps. At least not less than hundred genes have been implicated and identified in cell cycle events. Following are the few.
Yeast Genes and Gene Products Involved in Replication:
|
Gene |
Product |
Function |
|
Pol-I |
DNA-pol-I |
=Pol-alpha/primase |
|
Pol-II |
DNA-pol-II |
=Pol-beta |
|
Pol-III |
DNA pol-III |
= Pol-delta |
|
Pol-30 |
PCNA |
clamp |
|
DPB2 |
DNA-p-lI subunit-B |
|
|
DPB3 |
DNA-p-lI subunit C |
|
|
PRI-1 |
DNA primase |
|
|
PRI-2 |
DNA-primase 2 |
|
|
CDC 8 |
Thymidine kinase |
|
|
CDC-9 |
DNA ligase |
|
|
CDC-21 |
Thymidine synthase |
|
|
CDC-25 or 28 |
Phosphotase 1 |
|
|
RNAR-1 |
Large subunit of ribonucleotide reductase |
|
|
RFA-1=RPA |
Replication factor-A, an SSB protein |
|
|
RFA-2 |
Replication factor |
|
|
RFA-3 |
Replication factor |
|
|
CI B5 |
Cyclin B5 |
|
|
CI B6 |
Cyclin B6 |
|
|
CI N1 |
Cyclin N1 |
|
|
CI N2 |
Cyclin N2 |
|
|
CDK |
Cyclin dependent kinase |
|
|
CCBF-swi-6 |
|
|
|
CCBF.swi-4 |
|
|
|
|
|
|
|
|
|
|
DNA Polymerase α and its Associated Proteins.
|
Interacting protein |
Pol α subunit(s) |
Function |
|
T-antigen |
All subunits |
Binds to replication origin; Initiates of SV40 DNA Replication |
|
RPA |
SSDNA binding, Catalytic and primase subunits |
Stimulation of Pol α activity and processivity |
|
Cdc45 |
B-subunit |
Recruitment of Pol α onto origins? |
|
p12DOC-1 |
All subunits |
Regulation of Pol α activity and phosphorylation |
|
PARP |
Catalytic |
Coordination of DNA replication and DNA damage response? |
The role of Pol α in DNA repair is still unclear. In yeast, the repair of UV- or X-ray induced DNA damage does not require Pol α (Budd et al. 1989, Budd & Campbell 1995) and in Xenopus oocytes Pol α is not needed for base excision repair (Matsumoto et al. 1994). However, a recent report shows that Pol α may be involved in double strand break repair in yeast by homologous recombination (Holmes & Haber 1999).
In addition to DNA replication, DNA damage response and repair, Pol α has been implicated in telomere maintenance (Diede & Gottschling 1999, Martin et al. 2000), and thus may also play a role in the maintenance of chromosome stability. It has two subunits, one small produces RNA primer (10ntds) and the large extends as DNA 20-200 ntds.
Subunit structure of DNA polymerase δ.
|
Pol δ subunit |
S. cerevisiae |
S. pombe |
H. sapiens |
Function |
|
Catalytic |
Pol3 |
Pol3 |
p125 |
DNA polymerase activity |
|
B-subunit |
Hys2/Pol31 |
Cdc1 |
p50 |
DNA replication |
|
Third subunit |
Pol32 |
Cdc27 |
p68-p66 |
PCNA binding |
|
Fourth subunit |
- |
Cdm1 |
p12 |
Stabilization of the holoenzyme? |
Pol δ is stimulated by an accessory protein, proliferating cell nuclear antigen, PCNA (Tan et al. 1986). PCNA is a homotrimer of 29 kDa found only in actively proliferating cells (Bauer & Burgers 1990). The crystal structure of yeast PCNA reveals that the trimer forms a closed ring to encircle double-stranded DNA (Krishna et al. 1994). PCNA stabilizes the interaction of Pol δ with template/primer and thus increases the processivity of this enzyme (Ng et al. 1991), allowing rapid replication of chromosomes without dissociation from the template DNA. The activity of the S. cerevisiae Pol δ catalytic subunit expressed in bacteria is stimulated and the processivity is increased by yeast PCNA while deletion of 220 amino acids from the NH2-terminus results in loss of PCNA activation
Nomenclature and classification of mammalian DNA polymerases (adapted from Friedberg et al. 2000).
|
Name |
Alias |
DNA polymerase family |
Function |
|
Pol α (alpha) |
|
B |
Replication |
|
Pol β (beta) |
|
X |
Repair |
|
Pol γ (gamma) |
|
A |
Mitochondrial replication and repair |
|
Pol δ (delta) |
|
B |
Replicationof lagging strand |
|
Pol ε (epsilon) |
|
B |
Replication of leading strand |
|
Pol ζ (zeta) |
Rev7 |
B |
Translesion synthesis |
|
Pol η (eta) |
Rad30A, XPV |
UmuC/DinB |
Translesion synthesis |
|
Pol θ (theta) |
Pol η |
A |
Cross-link repair? |
|
Pol ι (iota) |
Rad30B |
UmuC/DinB |
Translesion synthesis |
|
Pol κ (kappa) |
DinB1, Pol θ |
UmuC/DinB |
? |
|
Pol λ (lambda) |
|
X |
Meiotic repair? |
|
Pol µ (mu) |
|
X |
Somatic hypermutation? |
|
Rev1 |
|
UmuC/DinB |
Translesion synthesis |
Cell Cycle Dependent Components-Cyclins and CdKs:
|
Cell cycle stage |
Components produced |
Functions Of the components |
|
Go to G1 |
Increase in Cdk4+cdclD, Cdk4+cyclD, Cdk4+cyclD |
|
|
G1 to S |
Increase in: Cdk2 +cyclE, |
|
|
S-start to S-end |
Constant levels of; Cdk2+cyclA, start of cyclinB |
|
|
S and M |
Increase, Cdc2+cyclA, Cdc2+cyclin B (MPF), |
|
Some Cdks and Cyclins Involved in Different Stages of Cell Cycle:
|
Organism |
CDK |
Cyclins |
Stage dependent activity |
|
S.pombe |
Cdc2 |
Cdc13 |
G2àM transition, early M |
|
S. cerevisiae |
Cdc28 |
Cyc1, 2, 3. Clb5 and 6 Clb 1,2,3,4 |
G1àS transition Active in S stage G2àM transition/early M
|
|
Mammalian |
Cdk2
Cdk4, Cdk5, Cdk1 Or p34 Or cdk1 |
Cln-D1, D2, D3. ClnE Cln A. Cln D1, 2,3 Cln D1, 2,3 Cln A, B |
Active in G1 G1, Active in S stage. G1 G1 G2àearly M |
|
|
|
|
|
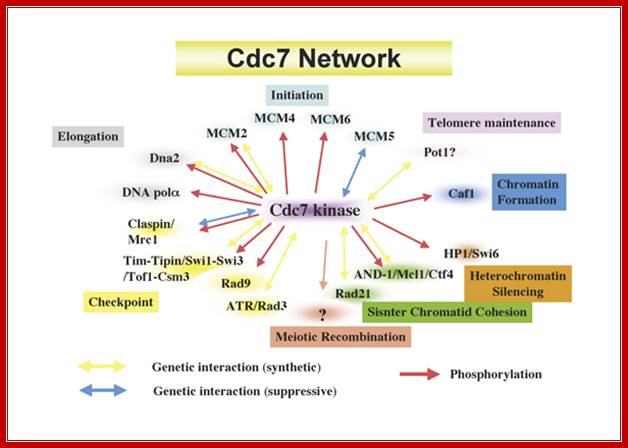
Cdc7 network regulating various chromosome dynamics; important component of replication system; Hisao Masai; https://www.k.u-tokyo.ac.jp
Eukaryotic Helicases:
|
Helicase Name |
Mol.wt (KD) |
Movement direction |
Require |
Function |
|
T-antigen |
92-each, hexamer |
3’à5’ |
ATP/GTP |
Acts on DNA duplex, melting and fork movement |
|
HSV helicase/primase 9ul |
120, 97 70 |
5’à3’ |
ATP/GTP |
97 kd acts as helicase |
|
Yeast ATPase |
63 |
|
ATP/GTP |
Stimulates yeasts’ POL-I |
|
Yeast RAD3 |
90 |
5’à3’ |
ATP/dATP |
DNA repair |
|
Mouse ATPase-B |
58 |
5’à3’ |
ATP/NTPS |
Acts on ss/ds DNA, motor protein melts DNA 140 bp per sec. |
|
Xenopus Helicase |
MCM-hexamer |
5’>3’ |
ATP/dATP |
Melts DNA |
|
Calf thymus-pol-Alpha associated |
47 |
|
CTP/GTP |
Low KM for ATP hydrolysis |
|
Calf thymus-pol-delta associated |
|
|
ATP/dATP |
Confers ssDNA displacement |
|
Helicase (Lily type) |
MCM-hexamer |
|
ATP |
Unwinds DNA duplexes from ends |
|
Helicase IV, RADH |
134 |
|
|
|
|
Papilloma |
|
E1 and E2 |
ATP |
|
|
|
|
|
|
|
|
|
|
|
|
|
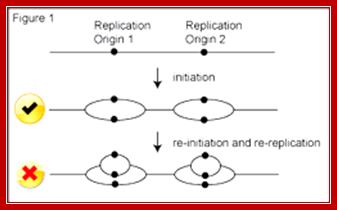
Replication Origins; http://www2.gurdon.cam.ac.uk/

Model for the role of Ku in pre-RC assembly.Targeting of Ku and Topoisomerase II onto chromatin during G1 phase leads to topologic changes in the chromosomal regions that correspond to replication origins, facilitating the assembly/stability of the ORC hexamer. http://www.intechopen.com/
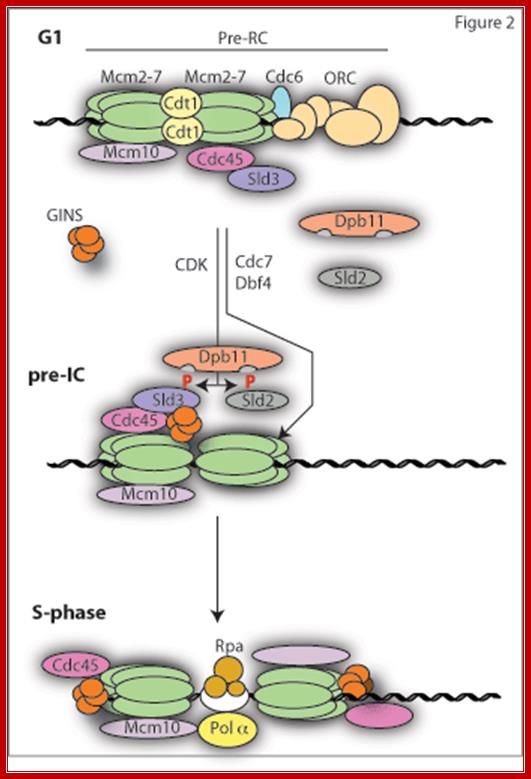
The first step in replication reaction (licensing) involves the loading of the replicative helicase Mcm2-7 at origins. This requires the concerted action of the six subunit Origin Recognition Complex (ORC), Cdc6 and Cdt1. Together this complex bound to origins is called the pre-replicative complex or pre-RC (Figure 2). This complex can only form in G1 phase of the cell cycle when the APC/C is active and CDK activity is low. This is because CDKs and other APC/C targets such as Geminin are potent inhibitors of pre-RC formation. Once cells enter S-phase, the APC/C is inactivated, CDK activity (and also Geminin) rises and any further pre-RC formation is blocked; http://www2.gurdon.cam.ac.uk/
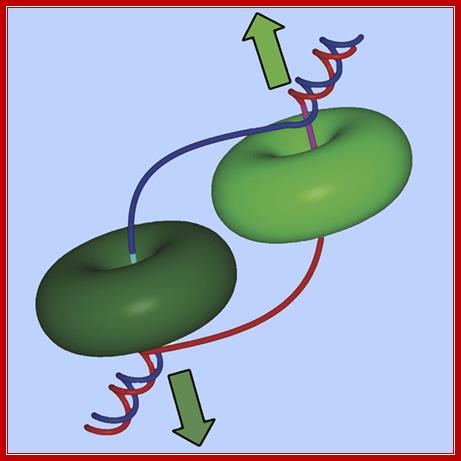
Ds DNA being unwound by helicase rings moving in opposite directions;
Jin Chuan Zhou, Alessandro Costa
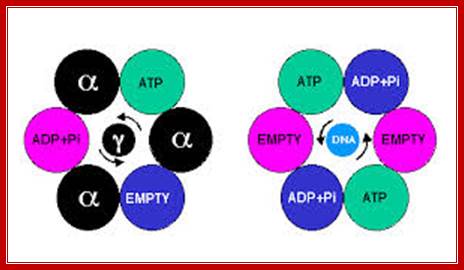
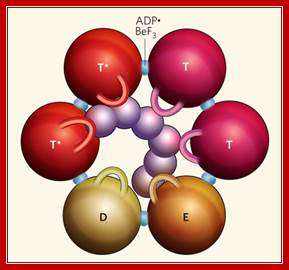
Hexamer Helicase subunits;http://www.cs.stedwards.edu/; Structure of the Rho hexameric helicase.Smita S patel; http://www.nature.com/
Thomsen and Berger's structure2 of
Rho bound to RNA and ATP mimics (ADP![]() BeF3) shows that
Rho's six subunits interact with the DNA (purple) in the central channel
through the Q and R loops (shown here as one loop per subunit). The ATP mimics
are bound at the subunit interfaces. The different colours of the spheres
depict the various conformational states of their ATP-binding site.
Hydrolysis-competent T* subunits
bind ATP and RNA tightly. The T subunits (before ATP hydrolysis), E subunit
(involved in nucleotide exchange) and D subunit (bound to the product of ATP
hydrolysis) all bind ATP and the central RNA weakly. The T and E subunits each
interact with one nucleotide of RNA at a time, whereas the D subunit has
limited interactions with RNA. Smita S
patel
BeF3) shows that
Rho's six subunits interact with the DNA (purple) in the central channel
through the Q and R loops (shown here as one loop per subunit). The ATP mimics
are bound at the subunit interfaces. The different colours of the spheres
depict the various conformational states of their ATP-binding site.
Hydrolysis-competent T* subunits
bind ATP and RNA tightly. The T subunits (before ATP hydrolysis), E subunit
(involved in nucleotide exchange) and D subunit (bound to the product of ATP
hydrolysis) all bind ATP and the central RNA weakly. The T and E subunits each
interact with one nucleotide of RNA at a time, whereas the D subunit has
limited interactions with RNA. Smita S
patel
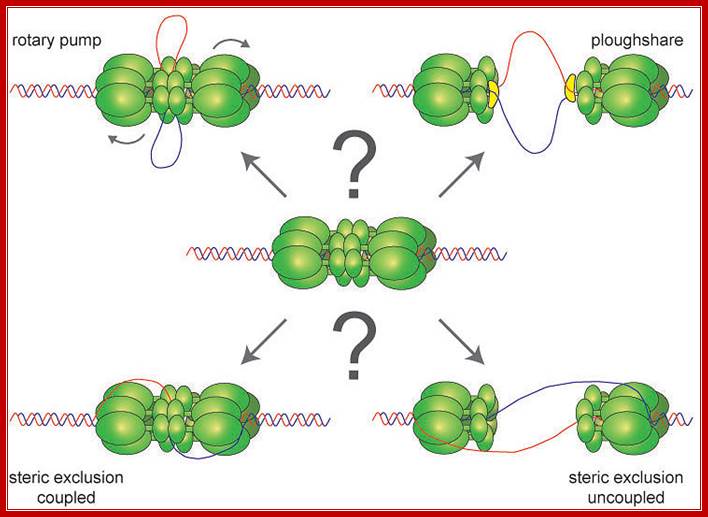
Possible DNA unwinding mechanism by MCM2-7 subunits; does theMCM2-7 binds to single strand or double DNA DNA opens and encircles only one strand; Dirk Remus; Sloan Kettering Cancer Center. http://www.mskcc.org/Dirk
Helicases are a family of motor proteins that play an essential role in nearly all DNA metabolic processes, catalyzing the transient opening of DNA duplexes. This review describes the biochemical, biophysical, and proteomics techniques that have been developed to elucidate the helicase-catalyzed mechanism of DNA unwinding
Eukaryotic DNA Ligases:
- Structure, mode of function and mechanism of catalysis of Eukaryotic Ligases is similar to that of prokaryotic Ligases.
- Two types of enzymes are present; both are located in the nucleus.
- But type I is mostly involved in replication and found in large numbers in proliferating cells or during cell division.
- On the contrary, DNA-ligase type II was found to be very active during DNA repair or recombination events.
- The enzymes are heat labile.
- Both of them are serologically unrelated, it means their gene and their amino acid sequences are different.
A List of Eukaryotic DNA Ligases:
|
Properties |
Ligase-1 |
Ligase-2 |
|
Molecular mass (kd) |
85-125 |
68 |
|
ATP requirement |
Yes, require 2ATPs |
Yes, require 4 ATPs |
|
Blunt end ligation |
Yes |
No |
|
Sticky end ligation |
Yes |
Yes |
|
Location |
Nucleus |
Nucleus |
|
Fractional activity |
90% |
<10% |
|
Overall function |
Replication |
Repair |
Ligase mechanism
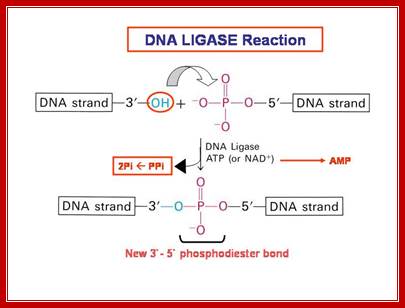
The mechanism of DNA ligase is to form two covalent phosphodiester bonds between 3' hydroxyl ends of one nucleotide, ("acceptor") with the 5' phosphate end of another ("donor"). ATP is required for the ligase reaction, which proceeds in three steps: (1) adenylation (addition of AMP) of a residue in the active center of the enzyme, pyrophosphate is released; (2) transfer of the AMP to the 5' phosphate of the so-called donor, formation of a pyrophosphate bond; (3) formation of a phosphodiester bond between the 5' phosphate of the donor and the 3' hydroxyl of the acceptor. [1] A pictorial example of how a ligase works (with sticky ends):
Ligase works on DNA with sticky ends; http://en.wikipedia.org/

DNA ligase-1;http://en.wikipedia.org/
Ligase will also work with blunt ends, although higher enzyme concentrations and different reaction conditions are required.
T4 DNA ligase; http://sciencerolled.blogspot.com/
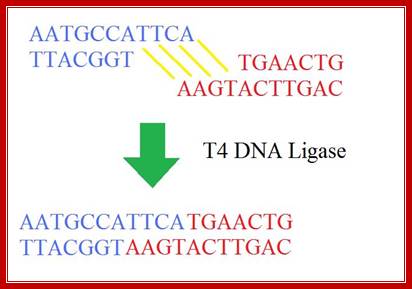
T4 DNA ligase; http://en.wikibooks.org/
http://fhs-bio-wiki.pbworks.com/
“The ligation reaction itself has two basic steps. Firstly the DNA ends have to collide by chance and stays together long enough for the ligase to join them. This is the most inefficient part of the reaction, but is easier at low temperatures. Why? Well, as you will probably know, all molecules move faster at higher temperatures so you can imagine that it is going to be easier for two DNA ends to collide and stay together if they are gently floating through the solution at low temperature, rather than whizzing about as they would be at higher temperatures. For cohesive ends, there is an additional reason; lower temperatures stabilize the hydrogen bonding between the complementary nucleotides, which really helps to keep things in place.
DNA ligase catalyzes the joining of the 3′-OH to the 5′-phosphate via a two step mechanism. First, the AMP nucleotide, which is attached to a lysine residue in the enzyme’s active site, is transferred to the 5′-phosphate. Then the AMP-phosphate bond is attacked by the 3′-OH, forming the covalent bond and releasing AMP. To allow the enzyme to carry out further reactions the AMP in the enzyme’s active site must be replenished by ATP”
RNA Ligase:
RNA Ligases: Nicole M. Nichols1, Stanley Tabor2, Larry A. McReynolds
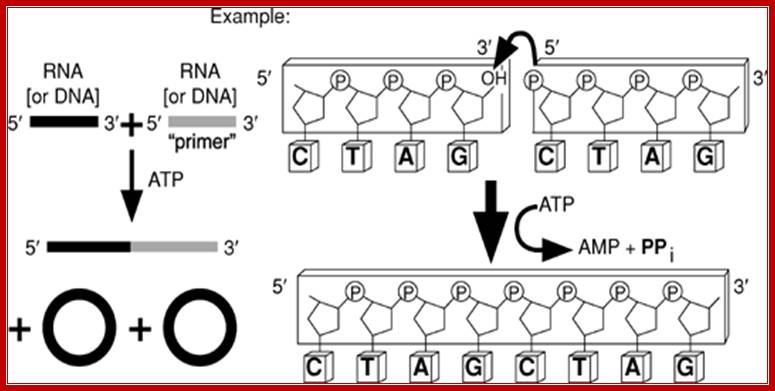
RNA ligase reaction; Nicole M. Nichols1, et al
T4 RNA ligase 1 catalyzes the ATP-dependent covalent joining of single-stranded 5¢-phosphoryl termini of DNA or RNA to single-stranded 3¢-hydroxyl termini of DNA or RNA. T4 RNA ligase 2 also catalyzes the joining of a 3¢-hydroxyl terminus of RNA to a 5¢-phosphorylated RNA or DNA; unlike T4 RNA ligase 1, this enzyme prefers double-stranded substrates. A truncated form of T4 RNA ligase 2 requires a pre-adenylated substrate for ligation. This unit describes specific reaction conditions, as well as applications such as radioactive labeling of the 3¢ termini of RNA, circularizing oligo deoxyribonucleotides and oligo ribonucleotides, ligating oligomers and nicks, creating hybrid and chimeric DNA/RNA molecules, and miRNA cloning. Curr. Protoc. Mol. Biol. 84:3.15.1-3.15.4. © 2008 by John Wiley & Sons, Inc.
RNA Ligases are also located in nuclei and their activity is high when transcriptional activity is at its peak. In bacterial hosts of T4 Phage produces an RNA ligase of molecular mass of 43 KD (gp 63). RNA Ligases extracted from wheat germ and yeast cells do exhibit different activities. They join 3’ OH of one piece of RNA to 5’P of another piece of RNA in 5’ to 3’ end-to-end manner. Enzymes require first two to three nucleotides of the RNA fragments to recognize the ends and bind to perform catalytic activity. It phosphorylates 5’ends and can cleave cAMP leaving Phosphate group at 2’end. Most of the RNA Ligases found in eukaryotic systems are involved in RNA splicing.
RNA Ligase-protoeopedia http://proteopedia.org/
Archaeal RNA ligase is a homodimeric protein that catalyzes
intramolecular ligation of single-stranded RNA and DNA:
C Torchia, Y Takagi, CK Ho .
RNA ligases participate in repair, splicing and editing pathways that either reseal broken RNAs or alter their primary structure. Here, we report the characterization of an RNA ligase from the thermophilic Archaeon, Methanobacterium thermoautotrophicum. The 381-amino acid long Methanobacterium RNA ligase (MthRnl) catalyzes intramolecular ligation of 5'-PO(4) single-strand RNA to form a covalently closed circular RNA molecule through ligase-adenylylate and RNA-adenylylate (AppRNA) intermediates. At the optimal temperature of 65 degrees C, AppRNA was predominantly ligated to a circular product. In contrast, at 35 degrees C, phosphodiester bond formation was suppressed and the majority of the AppRNA was deadenylylated. Sedimentation analysis indicates that MthRnl is a homodimer in solution. The C-terminal 127-amino acid segment is required for dimerization, is itself capable of oligomerization and acts in trans to inhibit the ligation activity of native MthRnl. MthRnl can also join single-stranded DNA to form a circular molecule. The lack of specificity for RNA and DNA by MthRnl may exemplify an undifferentiated ancestral stage in the evolution of ATP-dependent ligases.