Eukaryotic Origins:
Kinds and Components of Origins:
Replicons, a Comparative Account:
|
Species |
Genome size (bp) |
Replicon numbers |
Size of a Replicon |
Rate of replication (bp)/min |
|
E.coli |
4.6 x 10^6 |
1 |
4.6 x ^6 |
50000 to 70000/min |
|
Yeast (n=16) |
1.4 x 10^7 |
~500 |
40,000 |
36,000 |
|
Fruit fly (n=4) |
1.4 x 10^8 |
~3500 |
40,000 |
2800/sec |
|
House fly (n=4) |
8.6 x 10^8 |
~4000 |
45,000 |
2800/sec |
|
X.laevis (n=36) |
1.7 x 10^9 |
~15000 |
200kbp |
2500/sec |
|
Mus musculus,2n=40 |
~3 x 10^9 |
~25000 |
1500kbp |
2200/sec |
|
Homo sapiens,2n=46 |
3.2x10^9 |
~30-50,000 or more |
~2000kbp |
~ 2000 base pairs |
Small clusters of origins are found in a region called replicon foci. One of the large loci among them takes over the function of replication. Thus replicon foci show heterogeneity. In eukaryotes the replicon origins are located in nucleosome free region.
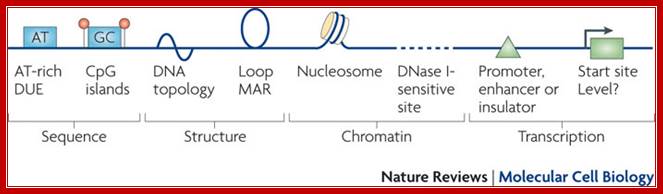
Summary of the features seen in DNA replication origins in eukaryotes. Several characteristics have been described at metazoan replication origins, but they are not present at all origins. Rather, they represent different marks or modules that can contribute to the selection of a given origin. At the sequence level, AT-rich elements and CpG islands have been reported as well as DNA regions that easily unwind (DNA unwinding elements (DUEs)), but their importance or role remains elusive. At the DNA structure level, bent DNA (or cruciform DNA) and the formation of loops has been described. At the chomatin level, nucleosome-free regions, histone acetylation and DNase-sensitive sites have been seen, but their direct participation in origin recognition as opposed to being a consequence of chromatin organization for transcription is sometimes difficult to estimate. The possible links of transcription features with replication origin recognition have been described but evidence for direct interactions between replication origin factors and transcription factors remains scarce. MAR, matrix attachment region; Nature Cell Biology; Marcel Méchali.
Replication fork while progressing in either direction from the Origin, often meets with a structure that obstructs smooth movement of replication, such regions/ structures are called replication fork barriers.

http://virtuallaboratory.colorado.edu/
The Nucleus and Nucleolus; http://mpheijden.tripod.com/

Replication origins iin S.pombe genome; Stochastic model for the initiation of S. pombe DNA replication. A segment of S. pombe chromosomal DNA is shown with many potential AT-rich SpORC binding sites (tick marks) in intergenes. The stochastic model differs from the classic replicon model in the following ways. (i) There are no highly specific replicator sequences. SpORC binds simple sequences (AT tracts) that are very common in the genome. (ii) There are many more potential SpORC binding sites than SpORC molecules. (iii) The distribution of SpORC over the potential sites is quasi-random, depending largely on the local density of AT tracts. Accessibility of sites may be affected by chromatin organization. (iv) Different sets of initiation sites are used in each cell cycle because the ratio of available binding sites to SpORC molecules is high. This feature of the model accounts for the observation that the utilization of S. pombe“origins” is very inefficient. http://www.pnas.org/
Yeast ARS origins:
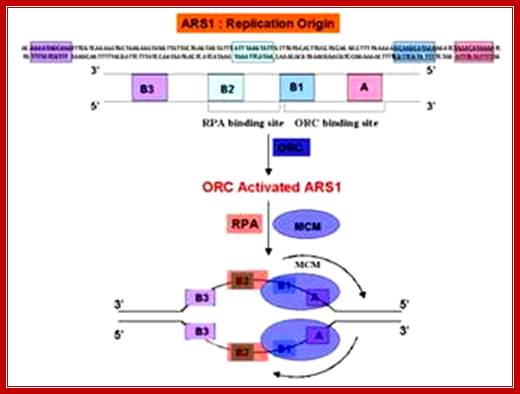
From studies spanning over three decades, the best-understood Origin of Eukaryotic system, so far is, the origin from autonomously replicating sequence (ARS) from 2-micron plasmids of yeast cells. The 2u plasmid is a circular DNA of 6.3kbp size. Copy numbers in the nucleus range from 100 to 200 or more. They multiply or amplify independent of yeast chromosomal replication. Problem with 2u plasmids is segregation during cell division, where unequal segregation leads to the loss of plasmids from few daughter cells. Eukaryotic chromosomal origin sequences have also been identified in simple genomes such as SV 40 DNA and other viruses such as EBPV for they replicate in eukaryotic nuclei. Sequences such as ARS of yeast cells are also found located in yeast chromosomes, which have hundreds of such sequences distributed though out the length of chromosomes. Each of the ARS elements has two components; one core- structure and second- auxiliary structures.

The 2u circle is 6.3kb circular, extrachromosomal element found within the nucleus of Saccharomyces cerevisiaeA; the number of plasmid per nucleus can be 100-200. The su DNA is coated with nucleosomes. Replication is initiated at ARS sequence. A cartoon of the yeast 2 u circle showing the ARS, the FLP gene, the three genes which encode proteins required for regulation of FLP expression (REP2, REP1, and D), and a set of small direct repeats (called "STB") required for partitioning into daughter cells during mitosis and meiosis; http://www.sci.sdsu.edu/
Control of chromosomal DNA replication in the early Xenopus embryo; Three systems required for the precise duplication of chromosomal DNA replication. Top panel, a small segment of DNA carrying three replication origins passing through the cell cycle. Red boxes denote ORC and a plus denotes a licensed origin. The metaphase chromosome decondenses, assembles licensed origins, is assembled into a nucleus, replicates, and then recondenses. Below are the activities of the origin recognition system, the licensing system and SPF during the course of the cell cycle. See text for further details.
Yeast ARS elements; http://www.dafml.unito.it/Mol.Biol
Several ARS like sequences, not only from yeast plasmids, but also from higher Eukaryotic systems, have been isolated and sequenced; they are named ARS-1, ARS-2 etc. The length of each of these sequences is of 250-400 bp or more. From various studies it has been discerned that they are located in between genes, called spacers. At least this is true with Yeast.
The region of ribosomal RNA genes in sea urchin, Tetrahymena and Physarum, amplify to a greater extent. During amplification of rDNA, several origins were observed as replication bubbles in non-transcribing spacer regions. In addition, active promoters flank these regions, which suggests, transcriptional activation may be required for initiation of replication; at least it is true in the above cases.
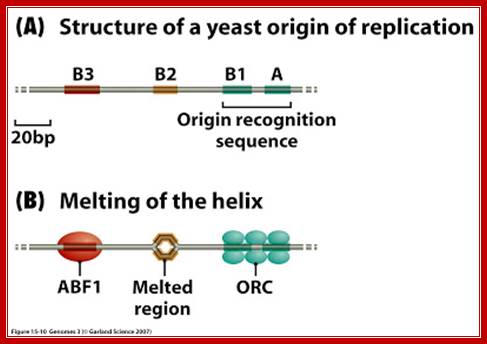
An ARS sequence from yeast shows four segments such as A, B1, B2, and B3. Sequence A is the most consensus sequence, which referred to as ORE (Origin Recognition Elements). Domain- B has A/T rich sequences, which can be subdivided into further sub segments. ARS segments consist of two parts. One A sequence and another B; the following is the sequences:
ARS sequence:
Aux 1I ---A/T--I ORE I---DUE--IAux 2
[10--AATTTCGTCAAAATGCT-27]
B3--[56-TTTAAGTATTG-67]
B2--[93 TGAAAAGCAAGCA 105]
B1- [114 CTAAACATAAAATCT 128]
Aà(T/A) AAATA (T/C) AAA (T/A)

The ORC (Origin Replication Complex) is a proteic binding complex, made up of 6 subunits (ORC p1, ORC p2, ORC p3, ORC p4, ORC p5, ORC p6) that has a binding specificity to DNA, more specifically to the origins of replication, in the presence of ATP, in the genomes of the eukaryotes. Theoretically this binding complex’s function is the initiation and/or regulation of the DNA replication in eukaryotes. http://www. glosomics.blogspot.com
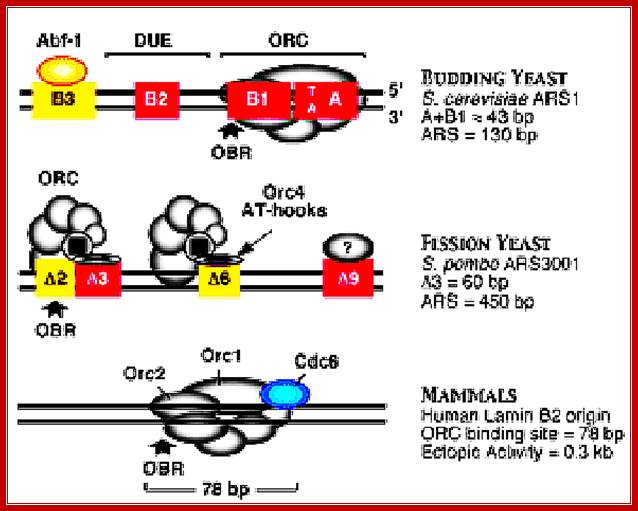
Different systems are figured in this illustrated diagram; budding yeast ARS1 (A+B1)=43bp ARS=130bp), fission yeast ARS=3000bp (A3=60bp, ARS=450bp) and Mammals (ORC binding region =78bp). The yeast systems have their own specific sequences for origin and initiation at that site. The origin sites among all mammalian systems has no consensus sequences; yet they use sequence within initiator zone and bind proteins at every possible Ori sites or regions, which on binding of ORC complexes and cdc6 and cdt1 enables the loading of MCM helicases (hexamer) and the opening of the ds DNA into single stranded replication bubble. Mol.Biol. www.circabook.com/

A model for Eukaryotic DNA replication initiation; A Model for Origin Unwinding
We have identified several distinct topological states of a circular plasmid (pBS) undergoing replication initiation in Xenopus egg extracts ( Figure 7). When pBS is incubated in egg cytosol, pre-RCs are assembled, and the plasmid is packaged into nucleosomal DNA (“ground state,” Figure 7). Upon addition of NPE, initiation-dependent negative supercoiling occurs in two steps. First, limited supercoiling that requires ORC, Cdc6, MCM, Cdk2, Cdc7, and Cdc45 but not RPA occurs (“shifted down” species). This event coincides with the binding of Cdc45 to chromatin to form pre-ICs. Subsequently, more extensive, RPA-dependent supercoiling occurs (U form DNA). Finally, initiation of DNA synthesis gives rise to slower mobility replication intermediates (“shifted up” species).
What is the nature of the supercoiled intermediates? There are at least two interpretations of the RPA-independent supercoiling event that accompanies pre-IC formation. One is that DNA is wrapped in a negative turn around Cdc45 or other initiation factors at the G1/S transition. The second possibility is that RPA-independent supercoiling reflects helicase-independent opening of the origin analogous to open complex formation at oriC ( Figure 7; Baker and Kornberg 1992). Further work will be necessary to distinguish between these possibilities. http://www.sciencedirect.com/
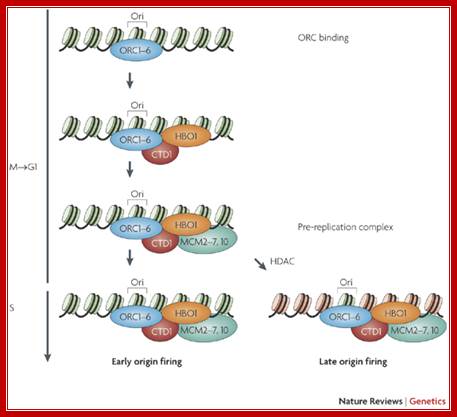
An overall link between the potential for gene transcription and the timing of replication in S phase is now well established in metazoans. Here we discuss emerging evidence that highlights the possibility that replication timing is causally linked with epigenetic reprogramming. In particular, we bring together conclusions from a range of studies to propose a model in which reprogramming factors determine the timing of replication and the implementation of reprogramming events requires passage through S phase. These considerations have implications for our understanding of development, evolution and diseases such as cancer.
Author affiliations
;Epigenetic regulation of replication origin licensing and firing.
Anita Göndör & Rolf Ohlsson; http://www.nature.com/
DNA Replication II; Eukaryotic;
The replication licensing mechanism is initiated by the deposition of the origin recognition complex (ORC) at a potential replication origin (Ori), which takes place during the time between telophase and early G1 phase The formation of the pre-replication complex is completed by the recruitment of the minichromosome maintenance (MCM) proteins to the origin via the licensing factor CTD1 and its co-activator, the acetyl transferase HBO1, exploiting the ability of the ORC to function as a scaffold for other factors. The pre-replication complex is subsequently activated by protein kinases (not shown), leading to initiation of DNA replication. Evidence suggests that removal of histone acetylation marks by histone deacetylase (HDAC) action in late G1 phase induces late origin firing. Nucleosomes coloured in green or red depict acetylated or hypoacetylated histone marks, respectively. Hyper and hypoacetylation of different sets of nucleosomes and/or lysine residues, as depicted in the lower images (S phase), is hypothetical and only one of several possible scenarios. The presence or absence of HBO1 at the ORC during origin firing is also hypothetical.;Anita Göndör & Rolf Ohlsson;http://www.nature.com/

A Model for the Regulation of Replication Timing; We propose that the timing of origin firing is regulated by the number of MCMs loaded. Origins at which many MCMs are loaded are more likely to fire in early S and therefore have an early average firing time; origins at which fewer MCMs are loaded are less likely to fire in early S and therefore have a later average firing time. We further propose that the number of MCMs loaded is regulated by the affinity of ORC for the origin. High-affinity origins are bound by ORC for more of G1 and thus have more MCMs loaded. Heterochromatin could provide a second layer of regulation, on top of our proposed MCM-based mechanism. Heterochromatin could delay origin firing by inhibiting any step in MCM loading or activation. However, based on our preliminary results that many MCMs are loaded at late-firing heterochromatic subtelomeric origins, we propose that heterochromatin acts mainly to inhibit MCM activation. Nicholas Rhind; http://profiles.umassmed.edu/

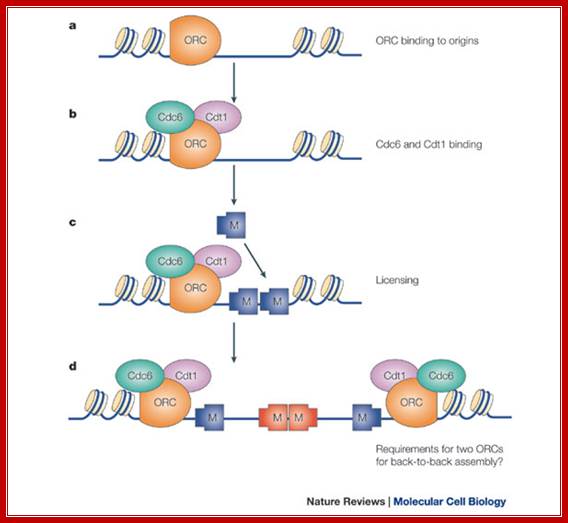
Regulation of DNA replication by the cell cycle: Origin licensing. Origin licensing takes place during G1 phase, when Cdk1 activity is low. The ORC becomes associated with ARS DNA directly after DNA replication and stays associated with DNA throughout the cell cycle. The ORC serves as a platform for Cdt1 and Cdc6 recruitment to the origin. Finally, an origin is replication competent when the MCM2-7 complex arrives; Jorrit M. Enserink; https://www.researchgate.net/
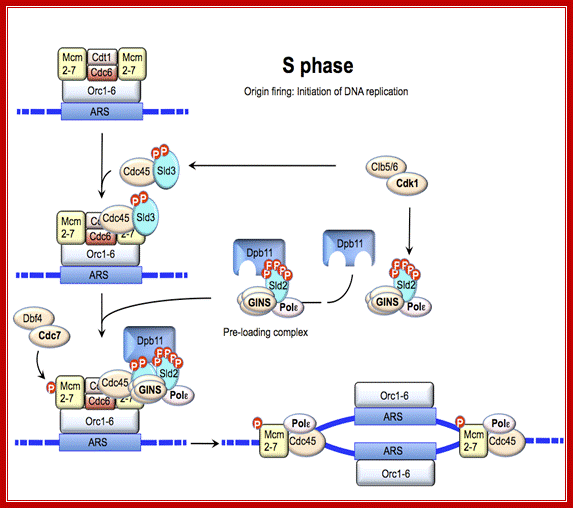
Regulation of DNA replication by the cell cycle: Origin firing. At the end of G1 phase, cyclin-Cdk1 complexes phosphorylate Sld2 and Sld3, leading to recruitment of GINS and Polε to the origin. The additional phosphorylation of MCM2-7 by DDK results in formation of a complex between Cdc45, GINS and MCM2-7, which then induces unwinding of DNA and initiation of DNA replication; Jorrit M. Enserink; https://www.researchgate.net
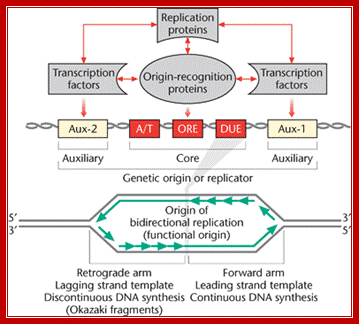
These auxiliary regions contain transcription factor-binding sites. In polyomavirus, presence of the transcriptional en- hancer stimulated DNA replication up to lOOO-fold, and presence of the enhancer increased SV40 replication approximately 100-fold. The auxil- iary region of adenovirus stimulates initiation of replication up to 200- fold, although this region is not directly involved in transcription; Peter C. van der Vliet.Activation domain BRCA1 alters the local chromatin structure and stimulates chromosomal DNA replication; Yan-Fen Hu, Zhong Lin Hao, and
RNA transcription coupled DNA replication in the loci of rRNA genes;

Coordination of RNA transcription and DNA replication by DNA replication licensing proteinsORCs, Cdc6 and Cdt1 are assembled at the DNA replication origins to recruit MCM helicases to form the pre-RC. The activation of DNA replication initiation at the origins, known as the “origin firing”, requires additional factors to be recruited to the pre-RC to from the pre-IC. Polymerases are recruited by an orderly assembled protein complex in similar ways in both DNA replication and RNA transcription. The pre-initiation complex (PIC) assembly at the promoter is required for the recruitment of the RNA polymerases. The complex assembly of TBP and TBP-associated factors (TAF) at the promoter regions are required for the initiation of transcription by all three Pols, with variation of respective TAFs of SL1 in Pol I transcription, TFIID in Pol II transcription and TFIIIB in Pol III transcription. Pol I is recruited by UBF and SL1 through interaction with TIF-IA of the SL1 complex. DNA replication licensing protein MCM5 is required for Pol II transcription and elongation. It is possible that other DNA replication licensing machinery proteins are involved in RNA transcription and couple these two fundamental events of RNA transcription and DNA replication (indicated in by the question marks); Shijiao Huang and Chuanmao Zhang;, chapter11-open access; https://www.intechopen.com/
Consensus sequence of A block is (T/A) AAATA (T/C) AAA (T/A), [opposite strand of the above figure sequence shown] any substitution in the sequences abolishes replication. The ‘A’ segment appears to be very important because initiating proteins bind to them. The B-sequence may be involved in additional protein binding sites and perhaps involved in inducing melting elsewhere.
Ori DHFR:
The best example to understand replication of complex genomic region is Di-Hydro Folate Reductase (DHFR) gene, which amplifies to 1000 copies or more during cell cycle. This region is of 109 kbp long and consists of three Ori sites called B, b’ and¡. The b origin is flanked on either side by MAR called matrix attachment sites. What is the role of MAR in replication? In DHFR gene, the bi-directional Ori site is located between two Alu sites (it is a restriction enzyme site). Also there are other protein binding sites. In addition there is a region where the DNA is bent. It has a sequence similar to ARS. This region is more or less simulates the origins containing DUE, ORE and other components of like yeast plasmid.
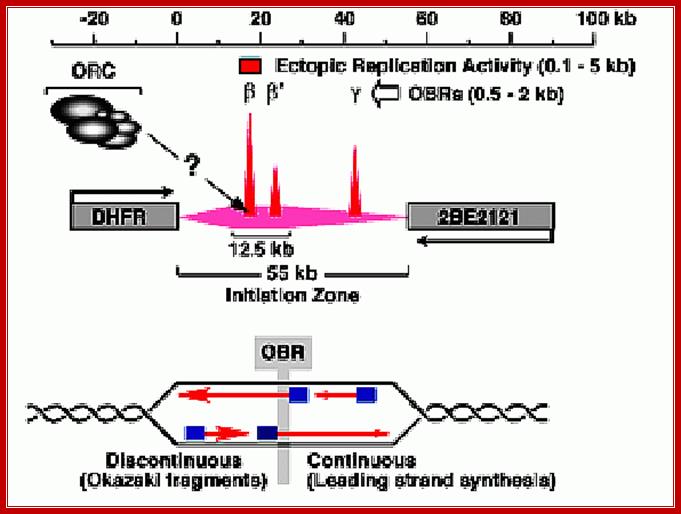
Initiation zones have been detected using 2D-gel electrophoresis methods to detect replication bubble and fork structures. For example, initiation occurs throughout the 55 kb intergenic region between the hamster DHFR and 2BE2121 genes, although most initiation events occur within a 12.5 kb region that contains two origins of bi-directional DNA replication (OBRs) [ori - b and b' (Kobayashi et al., 1998) ]. Another OBR (ori - g ) has also been mapped. As cells enter S-phase, only two MNase hypersensitive sites appear in this region, one at ori - b and one at ori - g (Pemov et al., 1998) , suggesting that pre-RCs are assembled predominantly at the OBRs. An "origin of bi-directional DNA replication" (OBR) is the transition between continuous and discontinuous DNA synthesis on the two template strands that mark the place where bi-directional replication begins. http://depamphilislab.nichd.nih.gov/

Replication Origin; http://oregonstate.edu/
Bidirectional Replication Model
In complex genomes, like human, Drosophila and others, during replication or early part of replication one finds regions containing several origins in the form of replication bubbles; and many such regions are recognized in each chromosomal DNA at the time of S phase. They are known as Initiator Zones, whose size can vary from 5000 to >5500 bp. In each initiator zone there can be many sub-origins (smaller replication bubbles). Not all of them initiate replication, only one of them fires and replication initiates. This holds good for yeast and other species. Replication is bi-directional and replication forks forge ahead till they meet another replication fork moving from opposite directions, at which point replication terminates.
In eukaryotes DNA is compacted into chromatin; consisting of eight histones octamer wrapped around by ds DNA as nucleosomes and coiled by the binding od H1 histone. Additional non-histones leads to very organized chromosome. Replication requires the loosening of this compact chromatin without loosening or loss of associated regulatory proteins. This is a herculean task.
- Replication initiates at distinct chromosomal locations that recruit prereplication (PreRC) complexes.
- PreRCs are recruited sequentially, each step subject to strict regulation to prevent reinitiation before the completion of replication.
- Cyclin-Dependent Kinases (CDK) levels change during the cell cycle. Low CDK activity is required for PreRC assembly as cells exit mitosis. High CDK activity is required for activation of existing PreRCs and initiation of DNA synthesis, while simultaneously suppressing assembly of new PreRCs.
- Replication origin sequences vary among metazoans, but the functions of proteins that bind replication origins are conserved.
- Eukaryotic origins exhibit a modular structure. Modularity is detectable in single-cell eukaryotes such as yeast and is more pronounced in metazoans, in which replication origins often cluster.
- Not all potential replication origins are activated in each cell cycle. Utilization of replication origins is regulated dynamically to facilitate coordination with other metabolic processes occurring on chromatin.
- Either hyperactivation or suppression of replication origins can damage DNA and cause chromosomal rearrangements, suggesting that spacing replication initiation events at defined intervals facilitates genomic stability.

Telomere DNA replication; Replication transiently generates positive supercoiling ahead of the elongating fork, which is usually rapidly relaxed by topoisomerases. However, the lock at the basis of the t-loop, which is thought to be formed by a complex nucleoprotein architecture involving a D-loop and a four-way junction, is unlikely to be free to rotate and can be considered as a topological barrier. Therefore, when the fork approaches a t-loop, one expects an accumulation of positive supercoiling in the unreplicated DNA in front of the t-loop and, ultimately, fork pause or arrest. This blockade could be rapidly relieved by t-loop opening and progression of the fork towards the very end of the chromosome (central part of the figure). It could also be efficiently released by topoisomerases and t-loop-opening activities (right part of the figure). Alternatively, if the action of topoisomerases is uncompleted in the presence of a t-loop, a residual positive supercoiling might favour fork regression and the formation of a four-way junction, called a chicken foot (left part of the figure). Chicken-foot regression or resolution, together with t-loop opening, will rescue the blocked fork. It is noteworthy that TTAGGG-repeat factor-2 (TRF2) specifically binds several of these DNA structures, perhaps as part of different shelterin subcomplexes. This might reverse the chicken foot and t-loop through TRF2-dependent activation of processes that are known to disrupt these structures (see Box 2 and main text). Positive supercoils are also expected to represent a favoured substrate for TRF2 binding (see main text), possibly leading to a high concentration of TRF2 ahead of the fork, which might promote t-loop opening;
Eric Gilson & Vincent Géli; Nature reviews -2007
Ori-Elements, Their Size, Binding Proteins, Melting Points, Transcriptional Effects
|
Source of Ori |
Size ori |
Binding Proteins |
Melting Regions |
Transcriptional effects |
SV 40, Simian vacuolation virus |
4bpx5 -AT rich |
T-antigen, Hexamer |
Palindromes |
Stimulates |
|
HSV-1, Herpes simplex virus |
8 bpx2 repeats, AT rich in the center |
Gp-ul 9 |
AT region |
- |
EBV, Epstein Barr Virus Ori-P |
30bpx2 repeats, Two palindromes, 2x30bp enhancers
|
EBNA-1 |
- |
Enhancers stimulate |
|
Ori-Lyt |
2 non-contiguous regions |
- |
- |
One enhancer overlaps the other |
|
BPV,Bovine Papilloma virus |
2 noncontiguous regions |
E-1 |
- |
Similar to EBV |
|
Mitochondrial Origins: |
|
|
|
|
|
H-strand |
Promoter & RNA binding site |
D-loop |
D-loop |
Promotes |
|
L-strand |
Palindrome |
- |
- |
Promotes |
A list of Few Eukaryotic Origins and Binding Factors:
|
Specific Origin |
ORE |
Aux-I |
Aux 2 |
|
SV 40 |
T-antigen (hexamer) |
SP-1, T antigen |
T-antigen |
|
Polyoma |
T-antigen |
Ap 1 |
T-antigen |
|
BPV |
E-1 |
E-2 |
- |
|
ADV2 |
Pre-TP; ADV-pol |
NF 1, Otomer-1 |
- |
|
HSV, Ori-H |
Ul-9 |
Several factors |
|
|
EBV |
EBNA-1 |
EBWA-1 |
|
|
Mit. DNA- Ori-H |
Mit-RNAPs+ factors + RNase |
Mit-RNAP +factors |
|
|
Yeast-ARS |
Ac-13! ORC! |
ABF-1 |
ABF-1 |
|
|
|
|
|
HSV Origin (Herpes Simplex Virus):
HSV is double-stranded DNA viruses with relatively large complex genomes. They replicate in the nucleus of a wide range of vertebrate hosts, including eight varieties isolated in humans. Human herpesvirus infections are endemic and sexual contact is a significant method of transmission for several including both herpes simplex virus 1 and 2 (HSV-1, HSV-2), also human cytomegalovirus (HHV-5) and likely Karposi's sarcoma herpesvirus (HHV-8). The increasing prevalence of genetial herpes and corresponding rise of neonatal infection and the implication of Epstein-Barr virus (HHV-4) and Karposi's sarcoma herpesvirus as cofactors in human cancers create an urgency for a better understanding of this complex, and highly successful virus family. http://stdgen.northwestern.edu/
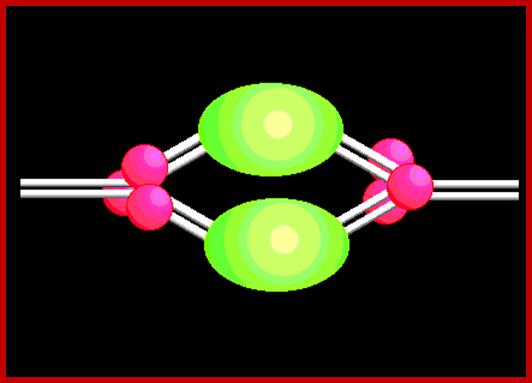
- The DNA of this virus contains three Ori sites.
- One is large called Ori-L and two small sized ones called Ori-S on either sides of Ori-L.
- The Large Ori is about 60-70 bp long, consists of large inverted repeats, which can form cruciform structures.
- Binding of UL 9 proteins to the arms lead to melting of DNA at central AT rich region.
- The UL-9 is an ATP dependent helicase.
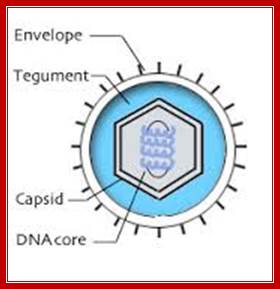
Herpes Virus; Bernard Roizman and Nina Thayer;http://stdgen.northwestern.edu/
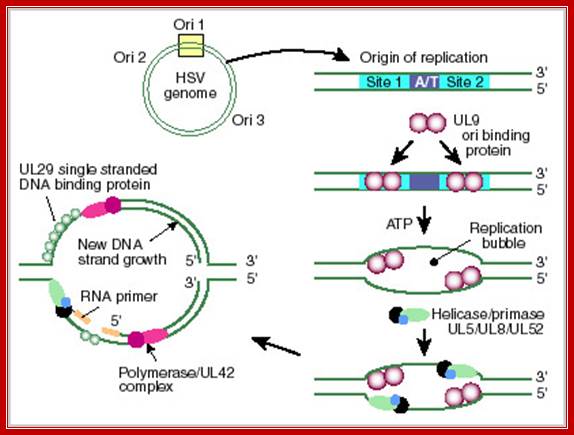
Herpes simplex virus (HSV) DNA Origin: http://darwin.bio.uci.edu/
Epstein-Barr Virus DNA Replication:
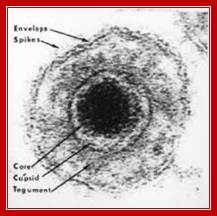
http://detadoctor.com/deta-ap/epstein-barr-virus;http://bioweb.uwlax.edu/

This is self explanatory virtual diagram depicting initiation of replication of eukaryotic system, though it is of the EBV. From: Epstein–Barr virus and oncogenesis: from latent genes to tumors;
Lawrence S Young and Paul G Murray
The EBV genome: (a) Diagram showing the location and transcription of the EBV latent genes on the double-stranded viral DNA episome. The origin of plasmid replication (oriP) is shown in orange. The large solid blocks represent coding exons for each of the latent proteins and the arrows indicate the direction in which they are transcribed; the latent proteins include the six nuclear antigens (EBNAs 1, 2, 3A, 3B and 3C, and EBNA-LP) and the three latent membrane proteins (LMPs 1, 2A, 2B). EBNA-LP is transcribed from variable numbers of repetitive exons. LMP2A and LMP2B are composed of multiple exons located on either side of the TR region, which is formed during the circularization of the linear DNA to produce the viral episome.
The orange arrows at the top represent the highly transcribed on polyadenylated RNAs EBER1 and EBER2; their transcription is a consistent feature of latent EBV infection. The outer long-arrowed red line represents EBV transcription during a form of latency known as latency III (Lat III), where all the EBNAs are transcribed from either the Cp or Wp promoter; the different EBNAs are encoded by individual mRNAs generated by differential splicing of the same long primary transcript. The inner shorter arrowed blue line represents the EBNA1 transcript originating from the Qp promoter during Lat I and Lat II. (b) Diagram showing the location of ORFs for the EBV latent proteins on a BamHI restriction endonuclease map of the prototype B95.8 EBV genome. The BamHI fragments are named according to size, with A being the largest. Lower-case letters denote the smallest fragments. Note that the LMP2 proteins are produced from mRNAs that splice across the TRs in the circularized EBV genome. This region has often been referred to as Nhet to denote the heterogeneity in this region according to the number of TRs within different virus isolates.
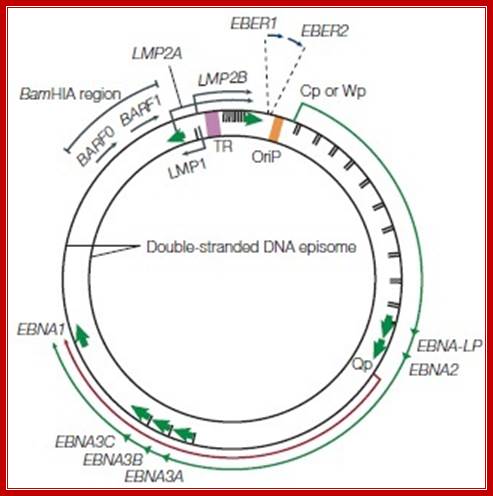
A diagram of a Epstein-Barr Virus dsDNA episome. Note the clustering of reading frames for related EBNA and LMP proteins. Note the location of the origin of replication, oriP. The green arrow denotes the direction of transcription during latency III. The short red arrow indicates the direction of transcription for EBNA1, which is activated during latency I and II. Source: By Jackson Cabohttps://microbewiki.kenyon.edu
- It has two distinct origins.
- ORI-P is responsible for maintaining the genome in latent extra chromosomal state.
- The Ori-lyt is responsible for amplification of the genome in lytic growth. They are located in specific locations, but separate from one another.
- They require distinct gene products for initiating replication.
- DNA synthesis from ori-P is through theta intermediate.
- Replication ultimately leads to concatemerization, because at lytic stage of growth, DNA replicates through rolling circle mode, which results in concatemerization.
- Lytic-ori consists one core element, which is separated from an enhancer by 1 KB region.
- The enhancer, true its character, functions irrespective of its position or orientation.
- Core region consists of four copies of a specific sequence, which facilitates the binding of Epstein Barr Nuclear Antigen (EBN-A1).
- Two of the binding sites form a part of 65 bp dyad.
- It is in this region or near this region replication initiates.
- The second element consists of two direct repeats similar to EBN-A1 binding site.
- Direct repeats act as EBN-A binding sites and enhance transcription, which is an important event for initiation of replication.
- Though replication initiation is bi-directional in the beginning, as one of the enhancer regions is blocked, the replication becomes unidirectional. The EB Virus produces its own DNA polymerase.
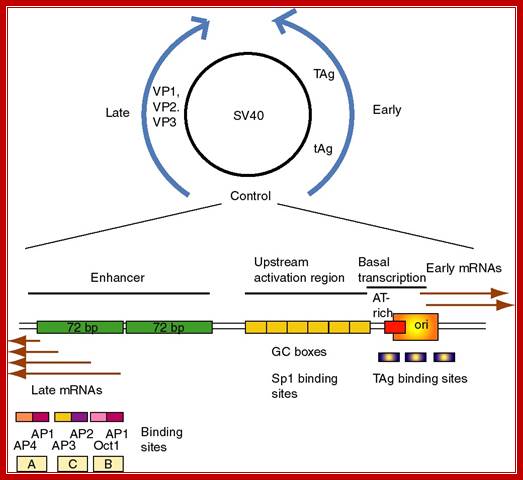
SV 40 Origin:
Simian vacuolation virus is an animal virus belongs to Polyoma family of viruses. It infects permissive monkey cells and causes devastation; and lysis release viruses. Some strains of virus can easily infect human cells also and such cells are called nonpermissive cells, rodent cells for SV40 are nonpermissive cells.
The virus infects permissive cells through receptors. By endocytosis viral particles are taken in and after decapsidisation DNA is delivered into the nucleus. The DNA is double stranded, circular, and 5234 bp long. It codes for capsid proteins and T-antigens. By shifting the reading frame the T-antigen mRNA can produce a small mRNA, which produces t-antigen (17kd). Capsid proteins are produced by late gene. By alternate splicing the mRNA produces three proteins called VP-1 (45kd), VP-2 (30kd) and VP-3 (18-20 kd). The early gene and late gene are oriented in opposite direction. Their promoters slightly overlap one another and found in between the early and late genes.
As soon as the infection, host RNAP-II produces T-antigen transcripts using early enhancer-binding proteins. The binding of sp-1 transcriptional factors activates promoters.
The promoter region also contains origin for replication. T-antigen is a monomer with Mol.wt 90-97kd. When T-antigen is sufficiently produced, the T- antigen assembles on to origin region as a hexamer in ATP dependent manner and initiates replication. After several rounds of viral DNA replication, producing substantial numbers, its late genes are transcribed and the three-capsid proteins are produced from the same transcript by alternate splicing. The T-antigen not only induces replication but also activates several cellular genes for host DNA replication and mitosis. One of the host cells’ protein called p53 binds to T- antigen and prevents its action. Still overproduction of the antigen can make viral DNA to amplify and produce viral particles, which cause cell lysis and to release viral particle. In nonpermissive cells, one in many thousands of cells infected gets transformed. This is achieved by the integration viral DNA into host nuclear DNA; in this transformation capsid protein genes are incapacitated.
The SV40 origin is located in a spacer region slightly overlapping the T-antigen promoter. It is organized into the following structure or domains.
SV 40 ori sequences:
I-----+1> TATA
[72 bp] [72bp]--[21bpx3]--[17bp]---[5bpx4] --[15 bp]-à-
<Enhancersà<GCrich><ATrich><pentamer><imperfect
Origin is located between mpu 0 to mpu 5243
Size of the SV40 DNA is 5297 bp
Enhancer = GGTGTGAAAG, they also act as enhancers for late genes.
GC rich box = 3 boxes, each has two sequences CCGCCC, sp1, a TF binds.
A/T rich=17 bp. -ATAAATAAAAAAAATTA-
TATA box- located at –25 to -30 from the START site.
Pentamers = ààßß, GAGGC, GAGGC, GAGGC, GAGGC, to which T-antigen bind.
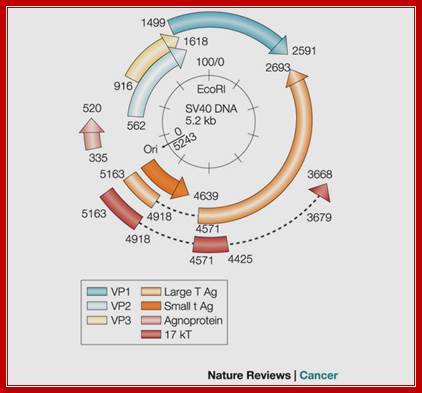
The Genomics of SV40; Adi F. Gazdar, Janet S. Butel & Michele Carbone; http://www.nature.com/
Antigens ‘T’, assemble on to pentameric sequences, as hexamers. The T-antigens bind to ATPs; the antigens act as helicases and move in 3’—5’ direction. Binding of Antigen proteins to pentameric sequences, leads to the opening of DNA on either side of A/T rich region or imperfect palindromic sequence as replication bubble.
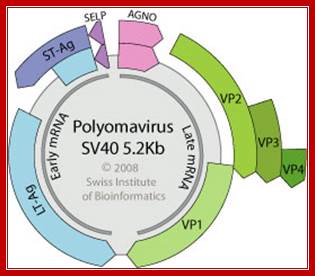
Polyoma DNA: Non-enveloped capsid with a T=7d icosahedral symmetry, about 45 nm in diameter. Circular dsDNA about 5 kb in size, associated with cellular histones in a chromatin-like complex. Encodes for 5-9 proteins.
http://viralzone.expasy.org/
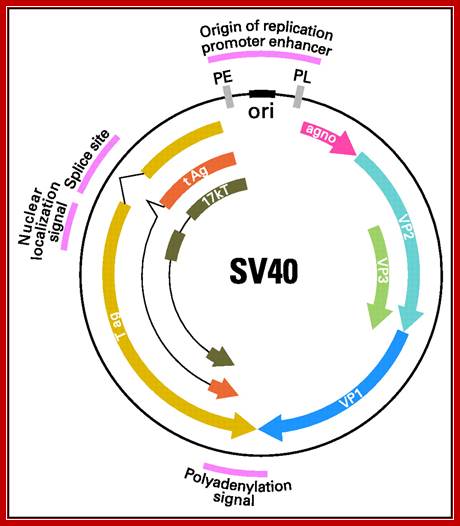
Regulatory elements of the SV40 genome used in the construction of laboratory plasmids are highlighted in red, including the small t antigen (t Ag) intron splice site, the large T antigen (T ag) polyadenylation signal, promoter, enhancer, and origin of replication. The early (PE) and late (PL) promoters, origin of replication (ori), major SV40 viral proteins are also represented. agno, agnoprotein.; Danielle L. Poulin and James A. DeCaprio; http://jco.ascopubs.org/
Transcriptional initiation at the promoter of T- antigen gene further enhances the replication bubble formation and possibly it stabilizes the replication bubble. Once the replication bubble forms, DNA replicating components assemble and replication starts and progresses in bi-directional manner.

http://www.microbiologybytes.com/
In SV-40 there is only one origin, thus SV 40 cannot be taken as a typical example of eukaryotic origins, but it explains what components and structures involved in initiating replication. Activation of T-antigen is due to phosphorylation of Threonine and serine residues located at both N-terminal and C-terminal ends of T antigen. Binding of P53 to T-antigen prevents its action.
SV40 control region
(1) SV40 (simian virus 40) infects monkey kidney cells, and it will also cause transformation of rodent cells. It has a double stranded DNA genome of about 5 kb. Because of its involvement in tumorigenesis, it has been a favorite subject of molecular virologists. The early region encodes tumor antigens (T-Ag and t-Ag) with many functions, including stimulating DNA replication of SV40 and blocking the action of endogenous tumor suppressors like p53 (the 1993 "Molecule of the Year"). The late region encodes three capsid proteins called VP1, VP2 and VP3 (viral protein n). A region between the early and late genes controls both replication and transcription of both classes of genes.
(2) The control region has an origin of replication with binding sites for T-Ag.
http://www.personal.psu.edu/
Origin in Globin cluster:
--LCR---E—Ag—gy---beta--===ori==--delta; ori is found between beta and delta; the size is 8kbp

Figure: Deletions that inactivate an origin of replication in humans (NCBI);
These two deletions are found separately in two individuals who suffer from thalassemia, a disorder caused by the failure to express one or more of the genes in the β-globin gene cluster shown. In both of these deletion mutants, the DNA in this region is replicated by forks that begin at replication origins outside the β-globin gene cluster. As explained in the text, the deletion on the left removes DNA sequences that control the chromatin structure of the replication origin on the right.
Adeno virus DNA Replication Origin:
http://cronodon.com/
Adenovirus is icosahedral non-enveloped DNA virus about 60-90nm in diameter, The capsid contains 252 capsomeres. The capsid is made of 252 capsomeres (240 hexons making up the faces and 12 pentons at vertices). There is a spike at each penton vertex. The genome consists of linear dsDNA (double-stranded DNA) with bound basic proteins which condense the DNA for packaging into the capsid (the basic proteins neutralize the acidic charges normally found on DNA, reducing electrostatic repulsion between different regions of the DNA molecule). The genome is 35-36 kbp long (depending on adenovirus type) with inverted terminal repeats (ITRs) about 100 bp long at each end.
Adenoviruses also encode several other proteins that modulate immune-mediated apoptotic mechanisms. Most of these are derived from the E3 transcription unit. There are seven E3 proteins, none of which is required for replication in cultured cells, implying anti-immune functions. E3-gp19K (a 19 kDa glycoprotein coded by the E3 transcription unit) is the first line of defense against CTL, binds to all haplotypes of human class I antigens and is conserved in all respiratory adenoviruses.
http://cronodon.com/
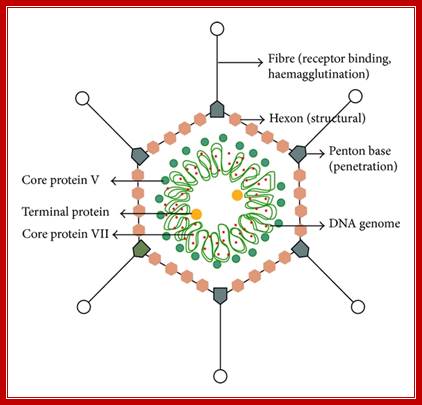
Structural features of Adeno virus. http://www.hindawi.com/
Adenoviruses which infect the gastrointestinal tract lack this protein, but they down regulate class I molecules by repressing at the transcriptional level, repression is meditated by the Ad-coded E1A proteins. Other E3-coded proteins named RID and E3-14.7K inhibit some of the killing pathways induced in infected cells by CTL, those that involve apoptosis rather than the perforin-granzyme pathway. E3 has therefore been called the "stealth" gene, allowing adenoviruses to evade the host immune response.
Together, these observations indicate that Adenoviruses, in the course of sequestering cellular machinery and altering the intracellular environment to favor viral replication, have profound effects on cellular functions. Viewed in this light, transformation is just an accidental (and rare) outcome of infection. The basis for oncogenesis (c.f. immortalization of cells in vitro - above) is not clear, but it is known that Ad12 E1A turns off class I MHC expression, possibly allowing tumors to escape destruction by CTLs.
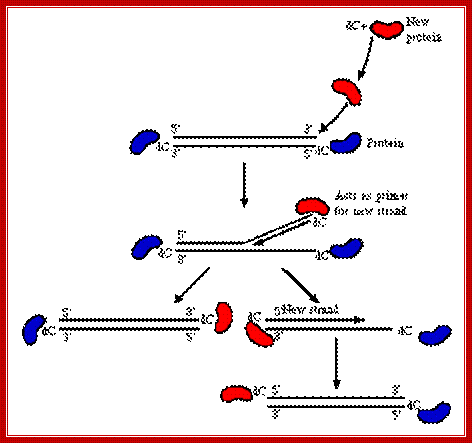
http://www.microbiologybytes.com/
Adenovirus DNA replication has been studied extensively both in vivo (t.s. mutants in infected cells) and in vitro (nuclear extracts). At least 3 virus-encoded proteins are known to be involved in DNA replication:
- TP (a.k.a. Ad DNA Pro) acts as a primer for initiation of synthesis.
- Ad DBP - a DNA-binding protein.
- Ad DNA Pol - 140kD DNA-dependent polymerase.
--1- NFI-18----------NFII---------39-NFIII-48-
In
addition, many cellular proteins in the nucleus also participate in replication
of the genome (e.g. NFI, NFII, topoisomerase I).
The adenovirus genome has inverted terminal repeats (ITRs) of about 100 bp.
Located within the ITRs are the cis-acting DNA sequences which define ori,
the origin of DNA replication. Covalently attached to each 5' end is a terminal
protein (TP) which is likely to be an additional cis-acting component of ori.
Within the terminal 51 bp of the adenovirus 2 genome, four regions have been
defined that are involved in initiation of replication. The terminal 18 bp are
regarded as the minimal replication origin and these sequences can direct
limited initiation with just the three viral proteins involved in replication:
preterminal protein (pTP), DNA polymerase (pol) and DNA binding protein (DBP).
However, two cellular transcription factors, nuclear factor I (NFI) and nuclear
factor III (NFIII) are required for efficient levels of replication. In
contrast adenovirus 4 replicates efficiently without NFI and NFIII. A further
cellular factor, a topoisomerase, is required for complete elongation.
The steps involved in adenovirus DNA replication can be summarized as follows:
--1- NFI-18----------NFII---------39-NFIII-48-
- First, the viral genome is coated with DBP.
- This protein reacts co-operatively with the cellular transcription factor, NFI which binds to a recognition site within the origin of replication, separated from the 1-18 bp core by a precisely defined spacer region.
- NFIII also binds at a specific recognition site between nucleotides 39 and 48.
- Protein-protein interactions, between NFI and pol, and pTP and NFIII help recruit the pTP-pol heterodimer into the preinitiation complex.
- Interaction between the heterodimer and specific base pairs 9 to 18 in the DNA sequence ensures correct positioning and the complex is further stabilized by interactions between the incoming pTP-pol and the genome-bound TP.
- DNA replication is then initiated by a protein priming mechanism in which a covalent linkage is formed between the alpha-phosphoryl group of the terminal residue, dCMP and the beta-hydroxyl group of a serine residue in pTP, a reaction catalyzed by pol. This acts as a primer for synthesis of the nascent strand.
- Base pairing with the second GTA triplet of the template strand guides the synthesis of a pTP-trinucleotide, which then jumps back 3 bases, to base pair with the first triplet (also GTA) and synthesis then proceeds by displacing the non-template strand.
- NFI dissociates as the first nucleotide binds just prior to the initiation reaction. Dissociation of pTP from pol begins as the pTP-trinucleotide is formed and is almost complete by the time 7 nucleotides have been synthesized.
- NFIII dissociates as the replication binding fork passes through the NFIII binding site.
Although 2 components of the adenovirus replication system have been crystallized individually, namely DBP and POU (NFIIIDBD), we are still a long way from having a structural model for the whole preinitiation complex.
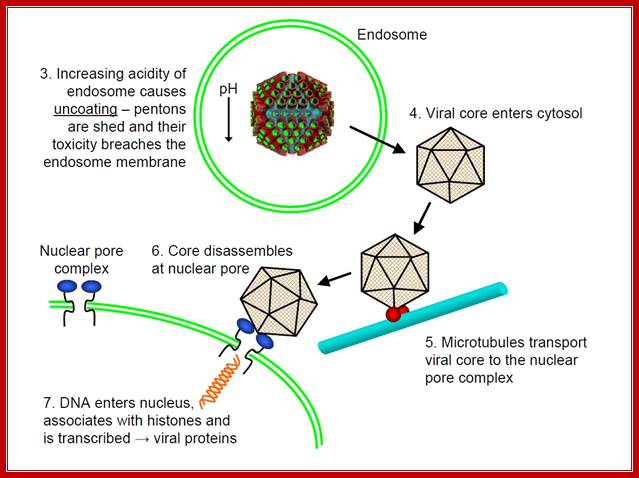
After adenovirus (Ad) enters the cell by receptor-mediated endocytosis, the viral DNA is uncoated and transported to the nucleus. Beginning at 6-8 hours post-infection, DNA is efficiently replicated, generating high amounts of progeny molecules (105-106/cell). Development of a cell-free system has contributed greatly to our understanding of viral DNA replication (for reviews see refs. 2,3). DNA replication results from an orderly interaction between viral proteins, cellular factors and template DNA at discrete sites within the nucleus that appear to be distinct from the transcription sites. DNA synthesis begins by a novel protein priming mechanism in which viral polymerase (AdPol) catalyzes the covalent linkage of the 5´-terminal nucleotide dCMP to the β-OH of a serine residue of the viral preterminal protein (pTP), which is the precursor of the terminal protein (TP). This initiation of DNA replication occurs at specific DNA sequences at the origin of replication in the presence of cellular transcription factors, nuclear factor I (NF-I) or CAAT transcription factor (CTF-1) and nuclear factor III (NF-III) or octamer-binding transcription factor (Oct-1). The pTP-dCMP complex formed in the initiation reaction serves as the primer for subsequent elongation catalyzed by AdPol via a strand displacement mechanism in the presence of the virus-encoded DNA-binding protein (DBP) and the host factor, nuclear factor II (NF-II), which is a type I DNA topoisomerase.
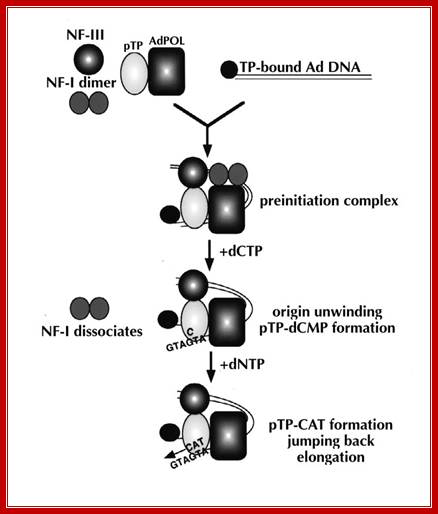
Muralidhara Ramachandra and R. Padmanabhan; http://www.landesbioscience.com/
After adenovirus (Ad) enters the cell by receptor-mediated endocytosis, the viral DNA is uncoated and transported to the nucleus. Beginning at 6-8 hours post-infection, DNA is efficiently replicated, generating high amounts of progeny molecules (105-106/cell). Development of a cell-free system has contributed greatly to our understanding of viral DNA replication (for reviews see refs. 2,3). DNA replication results from an orderly interaction between viral proteins, cellular factors and template DNA at discrete sites within the nucleus that appear to be distinct from the transcription sites. DNA synthesis begins by a novel protein priming mechanism in which viral polymerase (AdPol) catalyzes the covalent linkage of the 5´-terminal nucleotide dCMP to the β-OH of a serine residue of the viral preterminal protein (pTP), which is the precursor of the terminal protein (TP). This initiation of DNA replication occurs at specific DNA sequences at the origin of replication in the presence of cellular transcription factors, nuclear factor I (NF-I) or CAAT transcription factor (CTF-1) and nuclear factor III (NF-III) or octomer-binding transcription factor (Oct-1). The pTP-dCMP complex formed in the initiation reaction serves as the primer for subsequent elongation catalyzed by AdPol via a strand displacement mechanism in the presence of the virus-encoded DNA-binding protein (DBP) and the host factor, nuclear factor II (NF-II), which is a type I DNA topoisomerase. Muralidhara Ramachandra and R. Padmanabhan
Bovine Papilloma Virus (BPV) DNA Origin: Muralidhara Ramachandra and R. Padmanabhan
- Bovine Papilloma viral DNA is maintained as an extra chromosomal plasmid in a host cell.
- Two sequences have been identified which are cis-acting, that are responsible for autonomous replication.
- One of them is plasmid maintenance sequence (PMS- 1). This is called as start site for it is at this site replication starts.
- The other region looks like an enhancer.
- Both are noncontiguous and function independently.
- One of the gene products called E 1, like T antigen binds to ori-PMS-1 to initiate the process.
- Bound Replication initiation complexes; NF1, NFIII and pTP-
The papillomavirus genome consists of circular, d/s DNA ~8kbp in size, associated with cellular histones to form a chromatin-like substance. At least 12 different HPV genomes have been sequenced, and the genetic organization of all is similar. http://www.microbiologybytes.com/
Morphology:
Papillomaviruses are small, non-enveloped
icosahedral particles ~52-55nm diameter. There are 72 capsomers (60 hexameric +
12 pentameric) arranged on a T = 7 lattice. Apart from the larger size, these
appear very similar to Polyomaviruses particles (N.B. no sequence relatedness).
There are 2 capsid proteins, 1 major (encoded by the L1 gene) and 1 minor (L2).
Circular genomic map of bovine papilloma virus type 1 (BPV-1):
The early ORFs (E1-E8) and late ORFs (L1 and L2 are indicated). The LCR (long control region) contains regulatory elements for transcription and DNA replication such as the origin and minichromosome maintenance element (MME). E2 DNA binding sites are represented by red circles and promoters by arrows
Different modes of DNA replication in papilloma virus life cycle:
Three modes of DNA replication take place in the papillomavirus life cycle: initial DNA amplification, maintenance replication and vegetative replication. After initial uptake of the virus, the virion particle is uncoated and the genome transported to the nucleus of the basal cell where it is presumed to be amplified to a low copy number (Zhou et al., 1995). Presumably, a low level of the E1 and E2 proteins must be expressed early after infection since there is no evidence that they are in the viral particle. Most experimental studies have examined transient DNA replication in cultured cells, a system that is probably most analogous to this initial amplification stage and which requires the E1 and E2 proteins and the viral replication origin (Ustav and Stenlund, 1991a; Ustav and Stenlund, 1991a).
Stable episomal maintenance is the second stage of papillomavirus DNA replication;
In a papilloma, the infected basal cells proliferate and maintain low levels of extra chromosomal viral DNA. The genomes of papillomaviruses can also be stably maintained as high copy number extra chromosomal elements in cell lines (Law et al., 1981) and within these lines the viral genomes replicate in synchrony with cellular DNA. The viral genome copy number remains constant overall but the genomes are replicated by a random choice mechanism (Gilbert and Cohen, 1987; Ravnan et al., 1992). Long term, stable maintenance of papillomavirus-derived plasmids requires expression of the E1 and E2 proteins, the replication origin and a region from the LCR, that has been designated a minichromosome maintenance element (MME) (Piirsoo et al., 1996). This element contains multiple high affinity E2 binding sites. Recent studies have shown that both the BPV1 E2 transactivator protein and BPV genomes are associated with cellular chromosomes at mitosis (Skiadopoulos and McBride, 1998; Lehman and Botchan, 1998). This could be the mechanism by which approximately equal numbers of viral genomes are segregated to daughter cells at cell division to ensure that all basal cells of a papilloma contain viral DNA.
The third stage of viral replication is vegetative DNA replication, which is required to generate progeny virus. Vegetative DNA replication only occurs as the basal cells of a papilloma migrate upwards and differentiate in the stratum spinosum layer. Increased expression of the E2 proteins also occurs in the stratum spinosum and may be important for amplification of viral DNA (Burnett et al., 1990). The E2 protein is important for initiation of viral DNA replication but it has also been shown that HPV-31 E2 can arrest cells in S phase (Frattini et al., 1997). Clearly this could be important for vegetative replication by allowing sustained synthesis of viral DNA. There appears to be a switch from bidirectional theta replication in the maintenance stage of replication to a rolling circle mode in the vegetative stage (Flores and Lambert, 1997). Little else is known about vegetative viral DNA replication because of the requirement for terminally differentiating keratinocytes and difficulties in reproducing these conditions in a culture system. However, great advances are being made by replicating papilloma viruses in organotypic raft cultures and in xenografts of mice and these systems are proving to be very useful in studying the entire viral life cycle (reviewed in Meyers and Laimins, 1994).
Replication Origins and replisomes in Archaeal cells:
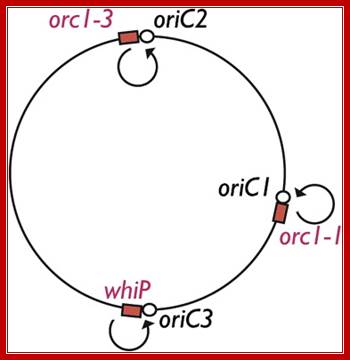
The archaeal DNA replication machinery bears striking similarity to that of eukaryotes and is clearly distinct from the bacterial apparatus and evolutionarily distinct from bacteria; Elizabeth R.Barry. The origin binding proteins in archaea are homologues of the related eukaryotic Orc1 and Cdc6 proteins (discussed below). We find that the three origins in the single chromosome of the Archaeon Sulfolobus islandicus are specified by distinct initiation factors, Rachel Y. Samson, Yanqun Xu, etal. While two origins are dependent on archaeal homologs of eukarya Orc1 and Cdc6, the third origin is instead reliant on an archaeal Cdt1 homolog. The origin recognition boxes (ORBs), which are inverted repeat sequence elements bound, by Cdc6-1, at oriC1.
Archaeal cells contain single circular chromosome of 5,751,492 bp (M.activorans). The Nanoarchaeum equitans contain 490,885bp DNA and code for 537 proteins. Small plasmids are also found in cells. Circular chromosomes contain multiple origins and use DNApol that resemble eukaryotes, but proteins utilized are similar to bacterial systems.
Rachel Y. Samson et al finds three origins in a single chromosome of the archaeon Sulfobolus islandicus which specified by distinct initiation factors. We find that the three origins in the single chromosome of the archaeon Sulfolobus islandicus are specified by distinct initiation factors. While two origins are dependent on archaeal homologs of eukaryal Orc1 and Cdc6, the third origin is instead reliant on an archaeal Cdt1 homolog. We exploit the nonessential nature of the orc1-1 gene to investigate the role of ATP binding and hydrolysis in initiator function in vivo and in vitro. We find that the ATP-bound form of Orc1-1 is proficient for replication and implicates hydrolysis of ATP in down regulation of origin activity. Finally, we reveal that ATP and DNA binding by Orc1-1 remodels the protein’s structure rather than that of the DNA template. Some archaea replicate from single origins like bacteria. And most of them use multiple origins; Michelle Hawkins et al.
This relationship between archaeal and eukaryotic replication-associated proteins extends across the replication process, from initiator proteins to factors involved in Okazaki fragment maturation. Furthermore, some archaea, including Sulfolobus species, possess multiple origins of replication per chromosome; a situation reminiscent of that in eukaryotes and clearly distinct from the single origin systems found in bacterial chromosomes. Two of the origins fire within a narrow temporal window in the 3-h cell cycle, at the onset of S-phase, while the third appears to have slightly more relaxed temporal constraints. Rachel Y. Samson et al; http://www.cell.com/
![Model for the initial assembly of the archaeal replisome based on recent advances in the eukaryotic DNA replication field (see [8,9]). (a) A double hexamer of MCM (gray) is loaded on double-stranded DNA at an archaeal replication origin. (b) The two individual hexamers are held together, so that, instead of moving apart, they will pump DNA into the central cavity of the assembly. If the pumping has a defined handedness, DNA will be unwound in the centre of the double hexamer. (c) The GINS complex (orange) in conjunction with RecJdbh or GAN (blue) stabilizes an open form of the hexameric MCM and allows extrusion of one DNA strand. (d) Resealing the MCM hexamer traps the displaced strand between the outside of MCM and the GINS assembly. (e) GINS recruits DNA primase (green).](Eukaryotic_DNA_Replication2-Eukaryotic_Origins_files/image043.jpg)
GINS is an essential eukaryotic DNA replication factor that is found in a simplified form in Archaea. A new study in this issue of BMC Biology reveals the first structure of the archaeal GINS complex. The structure reveals the anticipated similarity to the previously determined eukaryotic complex but also has some intriguing differences in the relative disposition of subunit domains; http://openi.nlm.nih.gov/

Save Deep-ocean microbe is closest living relative of complex cells; www.news.sciencemag.org;https://www.pinterest.com

S phase
The best characterized archaeal cell cycle is that of species belonging to the crenarchaeotal genus Sulfolobus21. An exponentially growing Sulfolobus spp. cell goes through a short pre-replicative period called G1 before entering into.

Model of the architecture of the archaeal DNA replication fork. Parental DNA is indicated by black lines, and newly synthesized DNA is shown in red. RNA primers, synthesized by primase, are shown in blue. MCM is shown as a yellow hexameric assembly surrounding the leading-strand template. We propose that the MCM helicase translocates along this strand, unwinding the parental duplex ahead of the replication fork. Single-stranded DNA is bound by SSB (Sulfolobus nomenclature), shown as pink circles. MCM interacts with the archaeal GINS complex (brown), and GINS, in turn, is additionally capable of binding primase (light blue). We propose that GINS acts to couple MCM translocation on the leading-strand template with deposition of primase on the lagging-strand template. DNA polymerase (in salmon pink) acts to extend the RNA primers, and we indicate that two polymerases are coupled, although there is currently no evidence for this in archaeal systems. Each DNA Pol interacts with a trimer of PCNA (brown). PCNA can act as a platform for additional assembly of the flap endonuclease FEN1 (green) and DNA ligase 1 (Lig1 [blue]), as cartooned on the lagging strand-associated PCNA only. Elizabeth R. Barry and Stephen D. Bell; http://mmbr.asm.org/
The Archaeal Replisome;

The archaeal replication elongation complex, which is known as the replisome, is eukaryotic in nature. The overall architecture of the leading- and lagging-strand replication complexes, as well as the relative positions; Ann-Christin Lindås & Rolf Bernander. http://www.nature.com
![]()
![]()
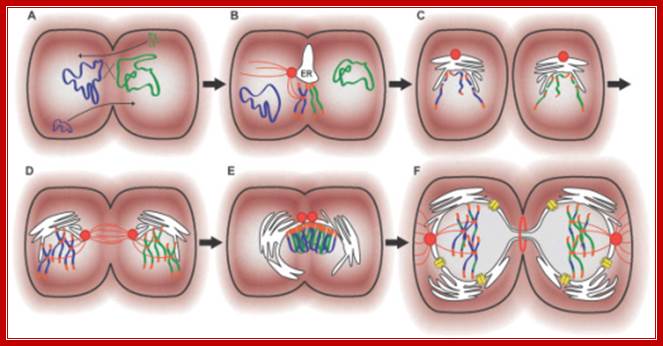
Archaeal Conjugation:
A putative model for the evolution of meiosis from archaeal conjugation: A. Ancestral archaeal conjugation (as described in H. volcanii) involving cell fusions, bidirectional flow of plasmids, and recombination between parental chromosomes (dark blue and green, respectively). B and C. Chromosome linearization permitted efficient pairing of homologues and resolution of crossovers. Telomeres (orange) evolved to protect chromosome termini and to nucleate the pairing of homologues A centromere (orange region in the centre of chromosomes) served as a connection between sister chromatids and as an attachment site, via kinetochores, for the meiotic spindles. This consisted of a network of microtubules (red fibers) radiating from a microtubule-organizing center (red circle) that guided chromosome movement]. The proto-ER progressively (B - F) differentiated into the NE [26] by wrapping segments of chromosomes to scaffold chromosome pairing (B - E) and to constrain diffusion of broken chromosome segments (C). D. Spindle-mediated movements approximate parental chromosomes during mating. E. Incipient karyogamy mechanics evolved to fuse proto-NE segments associated with chromosomes to create a common membrane platform to assemble, via clustering of telomeres, the meiotic bouquet. Cytokinesis based on an actomyosin contractile ring (red) facilitated splitting of the fusion partners (i.e., reductional meiotic division). NE enclosed the nuclear compartment when nuclear pores (yellow cylinders) evolved to ensure nucleo-cytoplasmic traffic of proteins and RNA. http://openi.nlm.nih.gov/