Eukaryotic DNA Replication:
Introduction:
Eukaryotic genome size is very large ranging from 10^6 to 10^12 bp. Though the size of the genome is very large, the DNA is organized into individual nucleo-proteinaceous threads, made up of histone and non-histone as protein components, called chromosomes, where, the DNA is organized into a compact nucleosomal thread, which in turn is compacted by further coiling (solenoid), looping and folding, which under electron microscope appears like a lamp brush surface. Structurally chromosomal DNA is compacted into visible (microscopical) structures such as centromere, telomere and chromosomal arms, which are further condensed differentially into Euchromatin and Heterochromatin. The size of the DNA in each of the chromosomes may range from as small as one million base pairs to one thousand million base pairs.
Replication of such highly organized and compacted DNA is more complex when compared to prokaryotic genomes. Each chromosomal DNA, on an average, e.g. Homo sapiens, consists 22+X/Y units of 50 to 250 million bp long DNA each.
The nucleosomal thread, made up of histone octamer wrapped around by DNA; it is also associated with non-histones, scaffold proteins and nuclear matrix proteins at specific regions. The CEN region is organized into a structure called Kinetochore complex made up of three to four layered proteinaceous components, which help in the binding of tractile fibers (specific microtubule structures), and the binding of which helps in the movement of chromosomes during cell division. Telomeric region also shows complex organization, for the DNA in this region is organized into quadruplex structure or in the form of loop where the 5’ end is tucked inside the loop and it is also associated with proteins that stabilizes telomeric DNA and protects it from exonuclease activity. The length of Telomeric DNA changes; dividing cells contain longer Tel.DNA and shorter DNA in older cells.
![]()
Schematic representation of budding yeast chromosome VI: Circles represent replication origins, with larger circles representing more active origins (used in most cell cycles), yellow coloring indicates that the origin is activated early in S-phase and blue coloring indicates activation late in S-phase, Conrad Nieduszynski.

Comparison of bacterial DNA folding with Eukaryotic structured DNA; A simple regulon consisting of a regulatory gene and two structural operons, A and B, is illustrated in various conformations. a | When the chromosome is represented in a one-dimensional, linear form, the three genetic loci are separate from one another; Charles J. Dorman http://www.nature.com/

DNA is highly organized and has a structure that allows tight compaction. A
micrographs of eukaryotic chromosomes as fibers of 30 nanometers (a nanometer equals one billionth of a meter) in diameter, in contrast to the 10-nanometer diameter of the nucleosome particle itself. In somatic nuclei, the 30-nanometer fiber appears to be stabilized by a specific histone, histone H1, which interacts with the DNA-linking adjacent nucleosomes. Solenoid Structured Nano fiber; http://www.biologyreference.com/
Again as in prokaryotes, Eukaryotic DNA replication is restricted to either Mitotic or Meiotic stages. Meiosis operates only during oogenesis or spore production. But mitosis is required for embryonic development and often can be initiated at any stage by signal transducers. Replication is highly regulated and takes place only once in a cell cycle at S stage. More to it, initiation of cell cycle is first triggered by mitotic signals and then it is controlled by a variety of gene products (hundreds or more) and each of them is regulated in stage specific manner. Environmental factors, cellular factors, cell-cell interactive events have important role in deciding whether or not the cell should go through cell division. What is the fate of cell derivatives? Refer to Cell Cycle chapter for more information.
Meiosis vs. Mitosis
In order to understand meiosis, a comparison to mitosis is helpful. The table below shows the differences between meiosis and mitosis.
|
Meiosis |
Mitosis |
|
|
End result |
Normally four cells, each with half the number of chromosomes as the parent |
Two cells, having the same number of chromosomes as the parent |
|
Function |
Sexual reproduction, production of gametes (sex cells)/spores |
Cellular reproduction, growth, repair, asexual reproduction |
|
Where does it happen? |
Animals, fungi, plants, protists |
All eukaryotic organisms |
|
Steps |
Prophase I, Metaphase I, Anaphase I, Telophase I, Prophase II, Metaphase II, Anaphase II, Telophase II |
Prophase, Metaphase, Anaphase, Telophase |
|
Genetically same as parent? |
No |
Usually |
|
Crossing over happens? |
Yes, in Prophase I |
|
|
Pairing of homologous chromosomes? |
Yes |
No |
|
Cytokinesis |
Occurs in Telophase I and Telophase II |
Occurs in Telophase |
|
Centromeres split |
Does not occur in Anaphase I, but occurs in Anaphase II |
Occurs in Anaphase |
From- Wikipedia
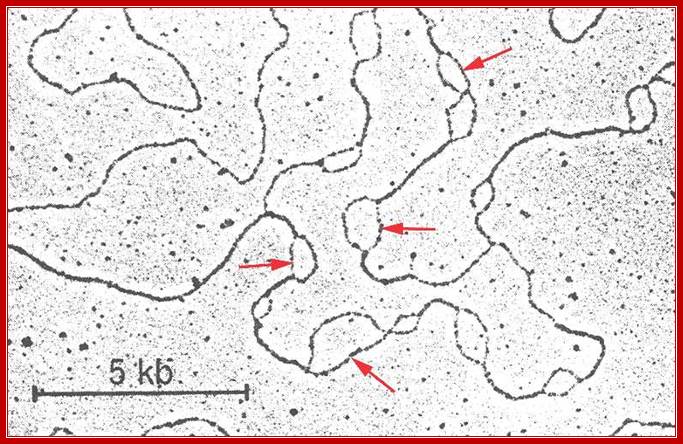
Replication origins in the form of Bubbles.www.Zeally.com
Replication eyes-second year Biochemistry lessons; www.brooklyn.cuny.edu; Otago.ac nz
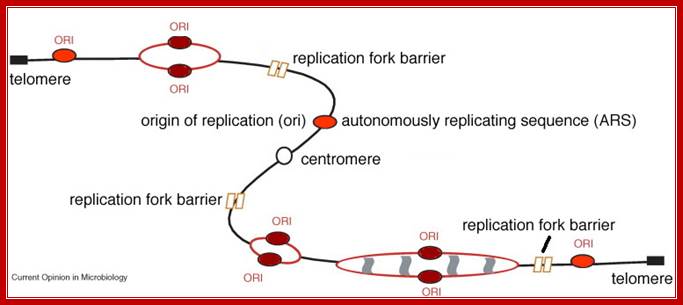
Replication Fork Barriers RFB are found at the 3’end of rRNA gene repeats, in yeasts there 150 such sites. They are called replication fork barriers (RFB) and inhibit replication forks in the direction opposite to rDNA transcription; Takehiko Kobayashi; http://virtuallaboratory.colorado.edu/
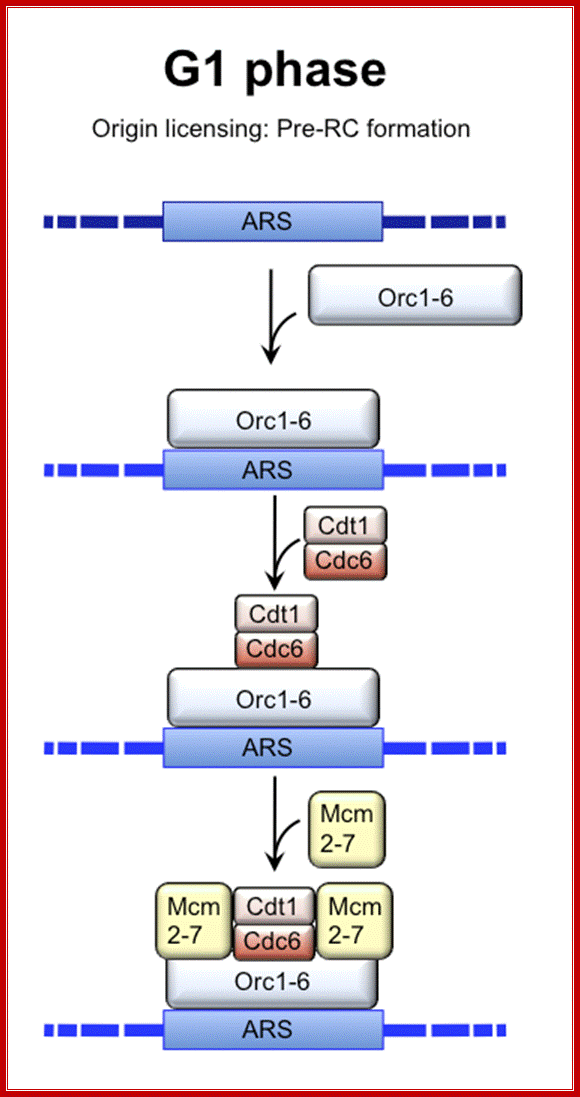
Cell Cycle Regulation of DNA Replication in S. cerevisiae; Regulation of DNA replication by the cell cycle: Origin licensing. Origin licensing takes place during G1 phase, when Cdk1 activity is low. The ORC becomes associated with ARS DNA directly after DNA replication and stays associated with DNA throughout the cell cycle. The ORC serves as a platform for Cdt1 and Cdc6 recruitment to the origin. Finally, an origin is replication competent when the MCM2-7 complex arrives. Each of the Ori sites are recognized by ORC complex then cdc6/cdt1 and MCM proteins. All such origins, the number can be 30,000 to 50000 per genome will be bound by the said protein complexes for initiation of Replication; Jorrit M. Enserink; http://www.intechopen.com/
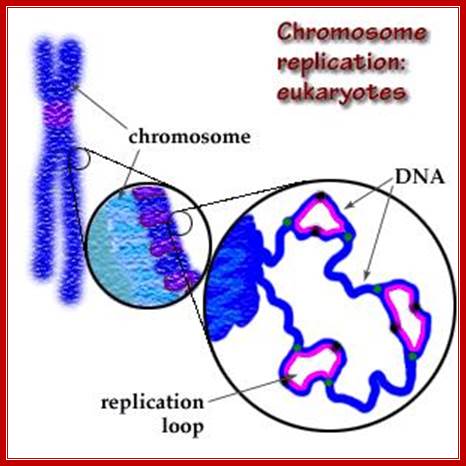
In eukaryotic systems, chromosomal DNA is linear, but condensed differentially. Contrary to prokaryotic systems replication initiates at a multiple sites and terminates at the end of each replicon. In E.coli there is only one replicon. But in eukaryotes each of chromosomes contains 50 to 200 million bases pair long replicons. One origin and one replicon is not enough to complete the entire length of chromosomal DNA in specific period of time (7-8 hrs.), so several hundreds of replicons organized in the linear DNA each with an Ori site but Ter sites not distinguished. The ultimate Termination is at telomeric ends. The origins have distinct sequences, but no distinct Ter structures. Thus individual chromosomes are structured into several hundred or more replicons, whose number and the size varies from one chromosome to the other. Whatever may be the number of chromosomes, replication starts simultaneously in all chromosomes. Hundred or more replicons, in each of the chromosomes, fire almost at the same time.
Each replicon, irrespective of the size contains an initiator region called ORIGIN (EK-Ori) and a termination region named as TER site, but terminator region is not well defined as in prokaryotes.
|
Organisms |
No.of Replicons |
Size of replicons, kbp |
Rate of replication, ntds/min |
|
Bacteria |
1 |
4.6x10^6 |
50,000-70,000 |
|
Yeast |
500 |
40 |
3600 |
|
Droso |
3500 |
40 |
2600 |
|
X.laevis |
15000 |
200 |
500 |
|
Mus muscuus |
25000 |
150 |
2200 |
|
Visia faba |
35000 |
300 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|