Topoisomerases:
When DNA molecules are subjected to such torsions, as earlier described, certain enzymes called Topoisomerases change the topology of DNA to facilitate replication or transcription. There are different kinds of Topoisomerases to perform different functions such as to introduce or remove super coils i.e. topological changes. Prokaryotes and eukaryotes have their own set of enzymes; and their genes have been identified and characterized.
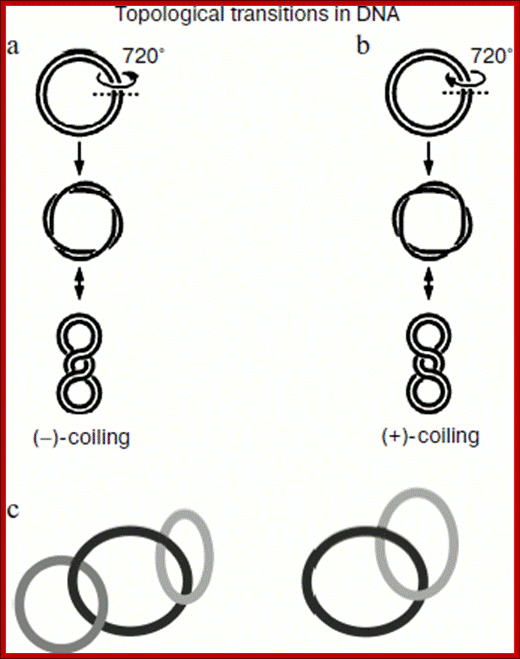
Schematic presentation of DNA supercoiling followed by formation of anticlockwise “–”-coiling (a) and clockwise “+”-coiling (b) of double-stranded DNA. The scheme shows two DNA catenane structures: two and three interlocked circular DNA molecules (c); D. V. Bugreev1 and G. A. Nevinsky
Comparative Properties Topoisomerases:
Prokaryotic Topoisomerases:
|
|
TYPE-I |
TYPE-II |
|
Sources |
Bacteria |
Bacteria |
|
Genes |
Top-A |
Top-A = Gyrase-A Top-B = Gyrase-B |
|
Mol.wt |
96 kDa (865a.a) |
Gyr-A = 97 kd Gyr-B = 90 kd |
|
Subunits |
One |
2 A and 2 Bs (tetramer) |
|
ATP requirement |
No |
Yes |
|
Mg^2+ requirement |
Yes |
Yes |
|
DNA dependent ATPase activity |
No |
Yes |
|
Inhibitors |
Nil |
Nalidixic acid, Oxolinic acid and Norfloxins bind to A-subunits, Coumermycin, Ciproflaxin, Novobiocin bind to Subunits-B, |
|
Affinity |
Great affinity to ssDNA, without any preferred sequences |
Prefers ds DNA, there is a bias towards AT rich region within a 20 bp segment with GC- - -(AT) 14- - GC |
|
Mutational effects |
*Leads to slow growth, *leads to accumulation of (-) SC DNA, *Enhances sensitivity to UV damages to DNA, *Changes the rate of transcription of certain genes, *Aborts replication elsewhere, *It is specific for replication origin |
*Leads to slow growth, *leads to accumulation of (-) SC-DNA as well as (+) SC-DNA |
|
Introduces topological knots such as catenation and decatenation of circular DNAs |
Yes |
- |
|
Produces covalently closed circular DNAs using ss-tails |
- |
Yes |
|
Introduces (+) super coils |
No |
yes |
|
Introduces (-) super coils |
yes |
Yes |
|
Relaxes (+) super coils |
Yes |
Yes, only when ATP is absent |
|
|
|
|
|
Changes linking numbers by- |
By one (one 180^o) |
By two (one 180^o each) |
|
Mechanism: |
It binds to dsDNA, Cuts only one strand to which it is bound,
The 5’ end of the cut strand covalently binds to the enzyme’s OH group of Tyrosine,
The free ends recoils and unwinds and releases the stress, as well as the super coil,
Then the ends are ligated by the enzyme,
Here the enzyme acts as an endonuclease as well as a ligase |
Gyrases are added to the replication fork to facilitate replication,
A2B2 complex (1A: 1B ratio) binds to DNA to a specific sequence,
One ‘A’ binds to one of the two strands, the other ‘A’ binds to the other strand,
B and A subunits are bound in 1:1 ratio,
ATP binds to ‘B’ subunits, ATP binding is facilitated by Mg^2+,
Each of the ‘A’ subunits cut one strand each in a staggered manner,
The cut 5’ end of each strands is covalently linked to the OH-group of tyrosine of each sub-unit of the enzyme,
Then using ATPs energy, the ‘B’ subunits swivel both strands of the DNA through and relieve the super coil,
Then the respective ends are ligated,
Again the ‘A’ subunits act as endonuclease and as well as ligase, |
Note: in addition to the above Topoisomerases there are few others like Topoisomerase IV etc. Topoisomerase II alternative form is called Gyrase which introduces (-) supercoils and Topo IV introduces (+) supercoils.
Eukaryotic Topoisomerases:
|
|
Type-I |
Type-II
|
|
Sources |
Yeast, Amphibians, Insects, Mammals and Plants |
Drosophila, Xenopus, calf thymus, and Human tissues |
|
Genes |
Type-I |
Type-II |
|
Molecular weight |
95-135kd, |
150-180kd |
|
Subunits |
One |
Two, homodimers, N-terminal is similar to Gyrase-B, Central region is similar to Gyrase-A, |
|
Affinity |
Greater affinity towards dsDNA |
Affinity towards dsDNA |
|
Cutting |
Cuts only one of the two strands |
Cuts both the strands |
|
Cleavage sites |
Cuts at a preferred sequence located at a 4 bp within a16bp segment |
Prefer a 4 bp within 5’GTN(A/A) AYATTNATNNG3’ |
|
Inhibitors |
Compathicin |
- |
|
ATP requirement |
No |
Yes |
|
Mg^2+ requirement |
- |
Yes |
|
DNA dependent ATPase activity |
Yes |
Yes |
|
Change in linking numbers by |
By one |
By two |
|
Catenation, decatenation, knotting and deknotting activity |
Yes, also involved in nucleosomal assembly |
Yes |
|
Introduces (+) SC |
No |
No |
|
Introduces (-) SC |
No |
Yes, it can convert (+) SC into (-) SC |
|
Relaxes (+) SC DNA |
Yes |
Yes |
|
Relaxes (-) SC DNA |
Yes |
Yes |
Among many chromatin associated proteins Topoisomerase II is found in large amounts. The other protein associated with chromosomal proteins is HMG (High Mobility Group).
Notes-Lateral:
Topoisomerase1 / DNA Complex: (www.scinecephoto.com).

Structure of the Topo I/DNA complex. PDB ID = 1A36; http://www.web-books.com/MoBio/
Human topoisomerase 1/DNA complex is an enzyme that assists in uncoiling DNA. DNA (deoxyribonucleic acid) is usually stored in a supercoiled form, which must be unraveled before it can be replicated or translated into proteins. In this image, the two strands of the DNA are red and green. The secondary structure of the protein is also color- coded: alpha helices (spirals) are dark blue, beta sheets are yellow arrows and linking regions are light blue. The arrangement of these domains gives the protein its tertiary structure, which helps to determine its function.
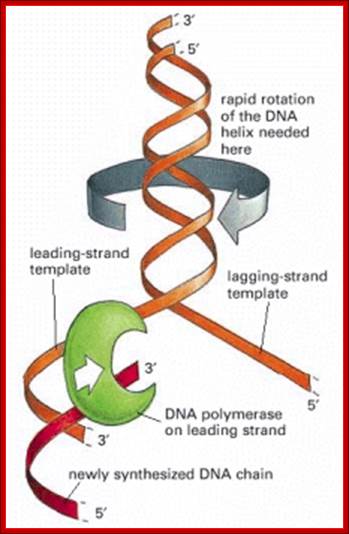
The “winding problem” that arises during DNA replication; Reviewed in Molecular Biology of the Cell
For a bacterial replication fork moving at 500 nucleotides per second, the parental DNA helix ahead of the fork must rotate at 50 revolutions per second. Topoisomerases relax DNA from super coiling and release the DNA from super coils.
TOPOISOMERASE I produce a transient single-strand break in a manner that does not require additional energy input; Topoisomerases are enzymes that cut one strand of double-stranded DNA, relax the strand, and reanneal the strands. They are further subdivided into two structurally and mechanistically distinct topoisomerases: type IA and type IB. Type IA topoisomerases change the linking number of a circular DNA strand by units of strictly 1. Type IB topoisomerases change the linking number by multiples of 1 (n).
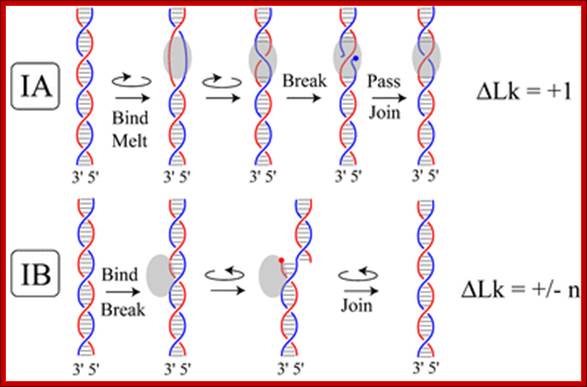
Type I topoisomerase can relax negative supercoiling and some subclasses in eukaryotes can use energy to create positive supercoiling. Type IA nicks one strand of the double helical DNA and changes the linking number by passing one strand of the double helix through the other one linkage at a time. Type IB nicks one strand and through relaxation of negatively supercoiled DNA proceeds to rotate the helix about the phosphodiester bond of the intact strand opposite the nick. Relaxation is a passive process and does not require ATP hydrolysis, but some forms of type I topoisomerase couple ATP hydrolysis to drive the formation of positive supercoiling beyond the relaxed state. (http://en.wikipedia.org/wiki/Type_I_topoisomerase) x passive process and does not require ATP hydrolysis, but some forms of type I topoisomerase couple ATP hydrolysis to drive the formation of positive supercoiling beyond the relaxed state. (http://en.wikipedia.org/wiki/Type_I_topoisomerase); http://www.ocf.berkeley.edu/
TOPOISOMERASE II:
o produces a transient double-strand break, and requires ATP hydrolysis
o Type II topoisomerases can separate interlocked DNA circles.
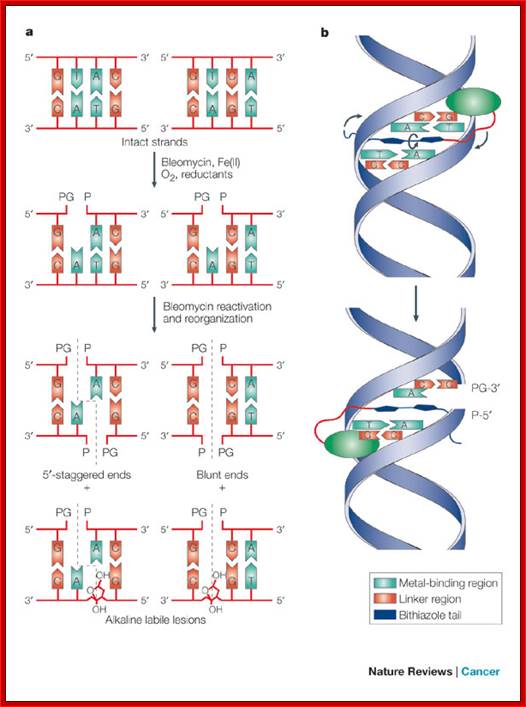
The first cleavage is single strand and then the second strand is cleaved; then they are joined ; http://www.nature.com/
Type II topoisomerases cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2. There are two subclasses of type II topoisomerases, type IIA and IIB. Type IIA topoisomerases include the enzymes DNA gyrase, eukaryotic topoisomerase II (topo II), and bacterial topoisomerase IV (topo IV). These enzymes span all domains of life and are essential for function. Type IIB topoisomerases are structurally and biochemically distinct, and comprise a single family member, topoisomerase VI (topo VI). Type IIB topoisomerases are found in archaea and some higher plants.
Topoisomerases II are ATP-dependent enzymes, and can be subdivided according to their structure and reaction mechanisms: type IIA (topoisomerase II or gyrase, and topoisomerase IV) and type IIB (topoisomerase VI). These enzymes are responsible for relaxing supercoiled DNA as well as for introducing both negative and positive supercoils [PMID: 7980433]; http://webcache.googleusercontent.com/

Base excision repair (BER) and nucleotide excision repair (NER) pathways. Both BER and NER repair pathways utilize the complementary DNA strand to restore sequence information lost in the damaged DNA strand. A) Schematic representation of the basic steps followed during short-patch BER (see text for details). B) Main sequence of events and enzymatic activities implicated in NER. C) Forms of lesions generated in the DNA by IR. http://www.intechopen.com
Gyrases:
DNA gyrases in prokaryotes introduce negative supercoils (or undo positive supercoils). Its eukaryotic counterpart topo IV can only undo positive supercoils to relieve stress in front of DNA replication.
Gyrase, is an enzyme that relieves strain while double-strand DNA is being unwound by helicase. This causes negative supercoiling of the DNA. Bacterial DNA gyrase is the target of many antibiotics, including Nalidixic acid, novobiocin, and ciprofloxacin. Upon wrapping and ATP hydrolysis, two negative supercoils are introduced into the template, providing opportunities for subsequent wrapping and supercoiling events.
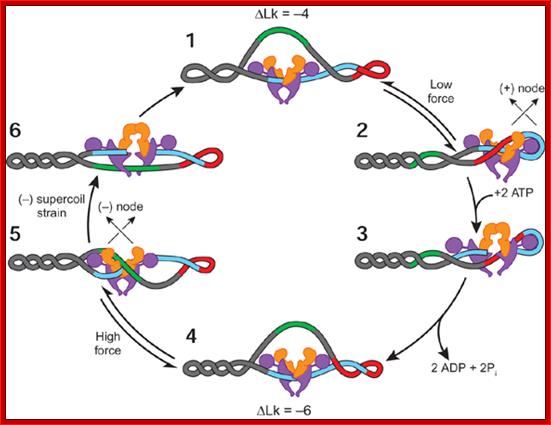
Subunits of DNA gyrase form a heterotetramer with two GyrA protomers (purple) and two GyrB protomers (orange). In the supercoiling mode, the sign of the cross or node is (+) for introducing negative supercoils and (-) for removing negative supercoils. Lk, linking number; http;//www.nature.com
Type II topoisomerase; http://en.wikipedia.org/
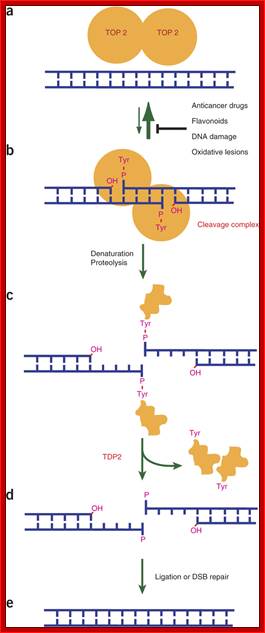
(a,b) Topoisomerase II (TOP2) functions as a homodimer, where each monomer cleaves one strand of a dsDNA by forming a covalent 5′-phosphotyrosyl bond (a). The resulting cleavage complex (b) allows the passage of another duplex DNA through the break, thereby enabling DNA relaxation, catenation-decatenation and knotting-unknotting1. Under normal conditions, religation of the cleaved DNA strands is highly efficient, and most of TOP2 is noncovalently bound to DNA as in a. In the presence of anticancer drugs , such as as etoposide, mitoxantrone, , doxorubicin and daunorubicin, or food flavonoids, DNA damage or oxidative lesions, the cleavage complex accumulates and needs to be removed for DNA repair1. (c) Prior to Tdp2 activity, the cleavage complex needs to be denatured or proteolyzed to expose the DNA–5′-phosphotyrosyl bond. (d) Tdp2 releases the TOP2 polypeptide and leaves the 5′-phosphate end. (e) The DNA break may be repaired by direct ligation after annealing of the two ends with the 4-base-pair stagger or through double-strand break (DSB) repair mechanisms. http://www.nature.com/Ke Shi, Kayo Kurahashi, et al
Different types:
PK- Topo I: Bacteria- 97monomer. Cannot relax +SC; PK
Topo III: -73kDa- Decatenating,
PK-Topo IIA: 84 and 70kDa, Decatenase-ATP dependent;
EK-Topo IB: 91 monomer, relax both (+ )and (-) Super coils.
EK Topo II IIA: 174 Homo dimers- can relax SC, ATP dependent, not SC DNA;
EK- IV –IIA: Decatenase; Gyrase IIA: PK-97 and 90, A2B2-introduce negative SC