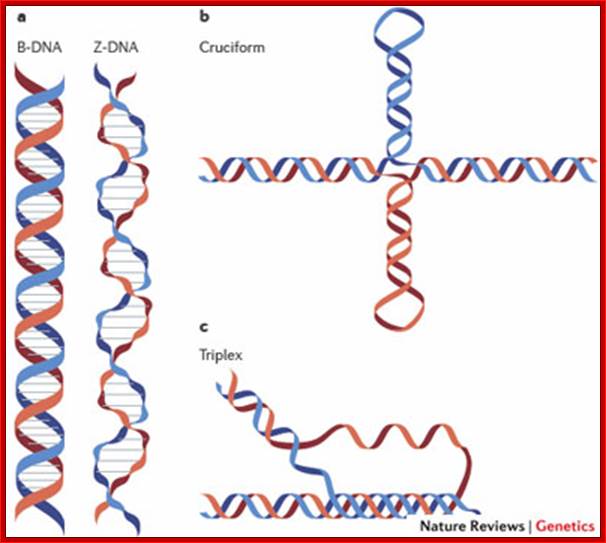
Other Forms of DNAs:
Depending upon the nucleotide sequences, the DNA can take different micro-heterogeneity and structural forms that shows how the DNA structurally exhibit dynamic features. Watson and Crick model is not a rigid but modulable form of DNA.

Figure: Dynamic DNA (you can visualize as you like it).
Cruciform DNA:
Depending upon sequences, DNA can assume specific structural forms. One such structural distortion is cruciform. The requirement for such structural deviation is a specific sequence, the length of the sequence, the temperature and kind of cat-ions. Segments of inverted repeats can assume cruciform of DNA.
Inverted repeats: CGATCTGG-CCAGATCG
Mirror repeats: GGTTGGCC-CCGGTTGG
Direct repeats: GGTTGGCC-GGTTGGCC
DNA secondary structures; Matthew L. Bochman, Katrin Paeschke & Virginia A. Zakian; http://www.nature.com/
When such sequences have a length of 10 or more base pairs with a center of symmetry, dsDNA can assume cruciform like a hairpin or a single strand DNA can assume stem loop structure. Such regions perhaps may be present in regulatory segments for recognition by specific protein factors.
Triple Helical DNA:
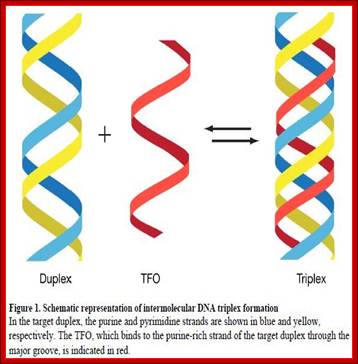
Three strands of DNA, which are complementary to each other, have propensity for triple helix formation. Homo or hetero polypurine or polypyrimidine tracts can assume triple strand conformation. Such triple stranded structures have been demonstrated and many diseases have been attributed to them. Triplex strands can affect transcription, replication and gene expression and they even prevent specific protein binding. Triplex DNA structures can be either intermolecular or intramolecular forms.

http://pubs.acs.org
Intermolecular forms: They are easily formed with polypurine- polypyrimidine tracts. In this the third strand can pair in two different orientations, which depends on its strand nature. However the base pairing requires protonation (low pH) of cytosine and the base pairing is by Hoogsteen bonding. Such type of triple strand formation is possible during homologous mode of recombination, where one of the nicked strands from the homologous segment intercalates into a helical duplex that is paired and produces triple strand structures, and such structures are required for the said function.
Intramolecular triplex forms: These structures form easily, if the DNA segment contains mirror repeat symmetry, in addition to PuPy sequences. IMT DNA may contain repeats of G, GA, GGAA, GGGA, or AAAG x n. Third strand pairing requires protonation of C for pairing with G (requires lower pH). It can also exist in four different isoforms. Super coiling of DNA favors IMT. The third strand can be produced during replication by a process called slippage process, where the polymerase in certain regions after replicating a segment, traverses back and replicates the same strand second time and it can be repeated several times (very rarely it happens).
This Figure shows; Triple stranded DNA forms from ds and ssDNA pairing with each other. http://www.abovetopsecret.com/
Ex. (GAA) n (TTC) n forms intramolecular triplex, exhibits four different isoforms. The third strand pairs with double strand, threads into major groove of the dsDNA.
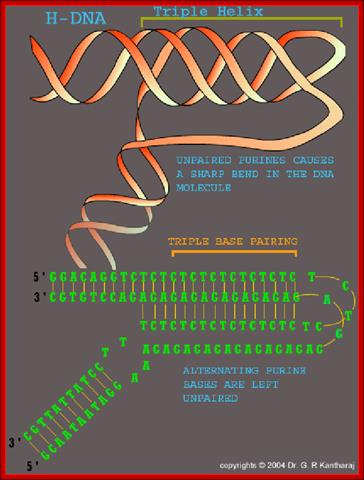
Fig: This represents another form of Base pairing in triple stranded slipped DNA repeats. During DNA replication after a segment, the enzymes retracts and replicates the same again of that strand thus it replicates that segments again ( re-replicattion); such third strand has complementary sequence with one of the two strands; thus DNA becomes triple stranded, only some parts produce such structures. Reference?
The structure of the triple stranded H-DNA. The two complementary strands of a homopurine-homopyrimidine repeat are colored in red and gray, while flanking DNA is colored green. The structure is called H-y when the red strand is homopyrimide, and H-r if when it is homopurine. One can see that the red and green strands in this structure are not linked, i.e. formation of H-DNA is topologically equivalent to an unwinding of the entire homopurine-homopyrimidine repeat. http://sackler.tufts.edu/
A triple-stranded DNA in which three oligonucleotide chains wind around each other and form a triple helix. In this structure, one strand binds to a B-form DNA double helix through Hoogsteen or reversed Hoogsteen hydrogen bonds.
For example, a nucleobases T binds to a Watson-Crick base-pairing of T-A by Hoogsteen hydrogen bonds between an AxT pair (x represents a Hoogsteen base pair). An N-3 protonated cytosine, represented as C+, can also form a base-triplet with a C-G pair through the Hoogsteen base-pairing of an GxC+. Thus, the triple-helical DNAs using these Hoogsteen pairings consist of two homopyrimidines and one homopurine, and the homopyrimidines third strand is parallel to the homopurine strand. Triple strand tsDNA formation occurs during recombination reactions.
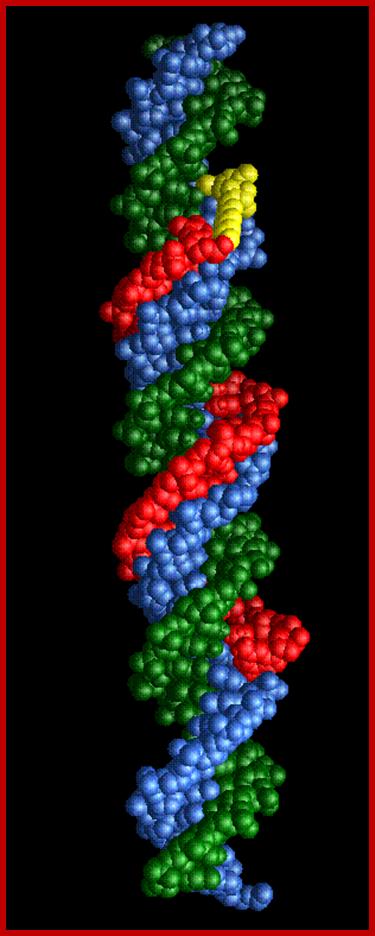
Triple stranded DNA: http://ncmi.bcm.edu/
T = A = T: A&T = W/C bonding, T&A = Hoogsteen bonding.
A = A = T: A = T= W/C bonding. A&A = Hoogsteen
C = G = C: G&C = W/C bonding, C&G = Hoogsteen.
G = G = C: G&C = W/C bonding, G&G = Hoogsteen.
- Triple Helix: Designing a New Molecule of Life-Peptide nucleic acid, a synthetic hybrid of protein and DNA, could form the basis of a new class of drugs—and of artificial life unlike anything found in nature, ( | December 1, 2008 | 22, Scientific American). It is speculated PNA-like molecules may have served as primordial genetic material at the origin of life.
Slipped DNA:
In slipped DNA one of the strands contains sequence repeats; the repeats can be of n x times. Slipped DNA arises during replication by DNA polymerases, where it can create greater length of DNA in one strand and deletions in the other strand.
Slipped strands have been identified and isolated. Nine different loci in humans have been identified and they are found to be very unstable. The repeats found are CTGnCAGn, CTGnCAGn. CTGnCAGn slipped DNAs have been found to move slower in gels. Surprisingly ssDNAs have been found to be stable and they are found as small loops. If such loops are longer they can still exist stably with intra-strand hairpin structures.
Single stranded oligo’s of CCGs or CGGs can form intermolecular duplexes or intramolecular duplexes. Such stable slipped segments can expand and become heritable. They can affect gene expression, protein binding, transcriptional initiation and possibly replication.
Slipped strand DNA structures: Richard R. Sinden1, Malgorzata J. Pytlos-Sinden1, Vladimir N. Potaman2
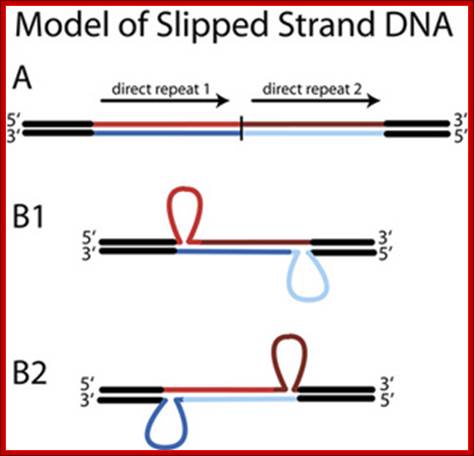
Slipped DNA strand; Richard R. Sinden et al; http://www.bioscience.org/
Figure. Slipped Strand DNA. Slipped strand DNA is formed from direct repeats (A) in which complementary strands of two adjacent direct repeats mispair such that one strand of one direct repeat pairs with the complementary strand of the other direct repeat. Structures B1 and B2 show the two possible different isomers of slipped strand DNA structures.
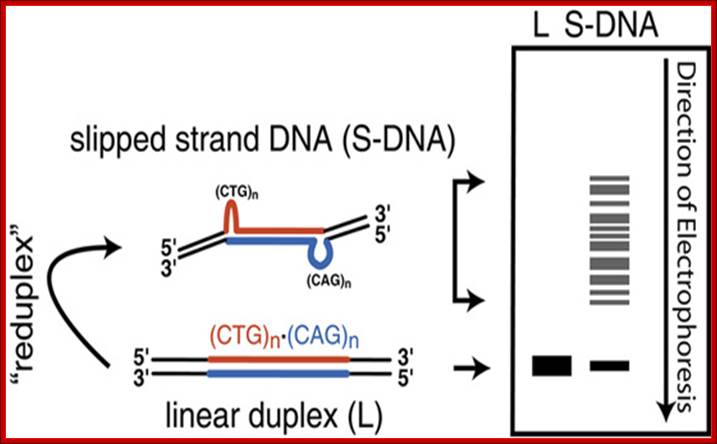
Figure2. Assay for Slipped Strand DNA. Duplex DNA migrates with a characteristic mobility in polyacrylamide gels that is dependent on the length of the DNA (shown as a 'band' in the lane marked 'L" in the idealized gel at the right of the figure). Following "reduplexing," which involves denaturation followed by renaturation, slipped strand DNA is formed (shown in the left of the figure). Slipped strand DNA contains two bends introduced by the two three-way junctions. These bends make DNA migrate more slowly in polyacrylamide than non bent duplex DNA. As this shows from formation of slipped strand DNA within a (CAG)no(CTG)n repeat tract, multiple isomers are possible (as described in Figure 3) and slipped strand DNA actually migrates as a series of bands corresponding to multiple structural isomers as shown in the lane denoted S-DNA. (Note that while 'S-DNA' has been used to denote slipped strand DNA, it has also found favor with physicists to denote stretched-DNA. Richard R.Sinden et al; http://www.bioscience.org/
Curved and Bent DNA:
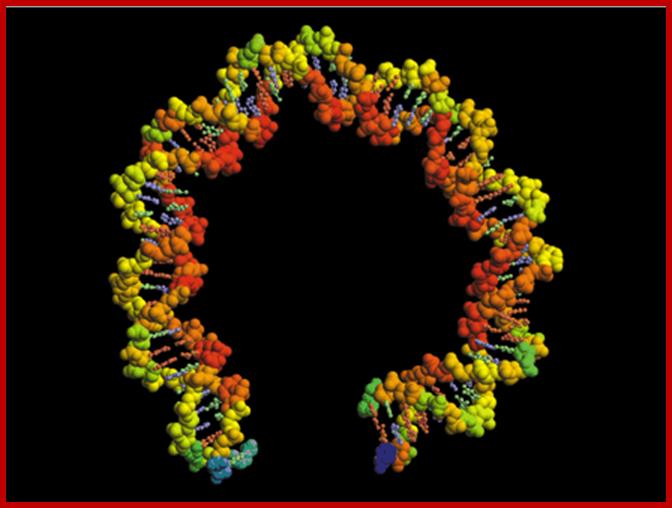
DNA by its nature is a very flexible helix and the binding of proteins, which organizes genomic DNA into a very compact structure, makes them nonflexible, thus it is not subjected ware and tear. But certain fragments of DNA, when run on a gel they migrate so slowly as if they are longer DNA. Kinetoplastid DNA of Crithedia, when a 414 bp long DNA run on a gel, it behaves as if it is 828 long bp segment. This behavior is attributed to rigid regions alternating with normal segments and such DNA exhibits curvature. If DNA is flexible it can snake through the gel pores, but if the DNA is curved and rigid, it cannot do so, so the slow movement.
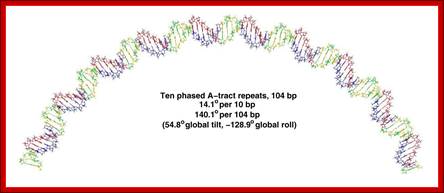
Segments of such DNA when analyzed showed they contained repeats of 4 to 5 A s, proceeded by a C and followed by a T. The runs A s are phased by 10 bps.
The structure of a 71 bp bent DNA or Curved model illustrating average counter-ion occupations at the phosphates calculated by Monte Carlo simulation using a linear-scaling Green's function solvation approach. Red indicates higher occupancy. The bent DNA model was based on the crystallographic structure of approximately one full turn of histone-bound DNA in a nucleosome core particle (PDB entry 1AOI). The calculations predict that phosphate-phosphate repulsions account for approximately 30% of the total free energy required to bend DNA from canonical linear B-form into the conformation found in the nucleosome core particle. For further details see the paper by Range et al. in this issue: Nucleic Acids Res. (2005); http://nar.oxfordjournals.org/
In gel electrophoresis curved DNA moves slower than straight dsDNA; Certain sequences develop DNA into ‘static curve’; http://www.cbs.dtu.dk/
http://www.biomath.nyu.edu/

Electrostatic surface potential maps of a bent DNA model showing increased electronegative potential on the inner surface where interactions with cellular packaging histones. The structure was obtained from Jim Maher III. Units are in kT/e (T=300K). Calculations were performed using the AM1 Hamiltonian and a buffer/matrix cutoff of 8/9 Å.;
Fig: Represents curved form of DNA; http://theory.rutgers.edu/
Curved- looks like curved; caroturner.umwblogs.org
Ex. CCC (5A) TCTC (6A) TAGGC (6A) TGCC (5A) TCCCAAC.
Wherever a 10 bases phasing with 5 or more runs of ‘A’ s leads to curvature, because runs of ‘A’s assume rigid structures. Between such rigid structures, if flexible regions are found automatically the DNA becomes curved for the flexible regions forms kink or bend between the rod like structures. The ‘A’ tract regions show base pairs having high propeller twist, which is due to strong base pair stacking interactions. This stability is due to stacks of ‘A’s share hydrogen bonding with ‘T’ s stacks found below. And ‘A’ helical repeats have only 10 bp per turn. The ‘A’ tract regions also have a narrow minor groove. These features are unique to DNA with rigid ‘A’ tracts. Sequences such as GA3T3C, G2A3T3C2, G3A3T3C3 and G2A3T3C2 also show curvatures. Such segments exhibit non B-DNA structural forms. The B-DNA shows bend or kinks between B-DNA and Z-DNA, such transitional regions show sequences such as CAAATCGC or CAAAAAATGC. Foot printing of ‘A’-tract DNA, using hydroxyl radicals, indicates that ‘A’ tract DNA is not cut as the normal B-DNA. Kinetoplast DNA has a sequence of GAATTC [CA5-6T] GT [CA5-6T] AGG [CA5-6T] GC [CAAAAT] CCCAAAC; it moves anomalously in the gel. Binding of proteins or cations or any other chemical adducts can cause distortions, and torsions, which are transmitted along the length of the DNA.

DNA has a lot of local structurs. the helix is bent to the left, distorted by the way that the helices are packed into the crystal. At the bottom, two of the bases are strongly propeller twisted--they are not in one perfect plane. As more and more structures of DNA are studied, it is becoming clear that DNA is a dynamic molecule, quite flexible on its own, which is bent, kinked, knotted and unknotted, unwound and rewound by the proteins that interact with it. https://pdb101.rcsb.org
The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA; Huu B Ngo, Jens T Kaiser & David C Chan
Tfam (transcription factor A, mitochondrial), a DNA-binding protein with tandem high-mobility group (HMG)-box domains, has a central role in the expression, maintenance and organization of the mitochondrial genome. It activates transcription from mitochondrial promoters and organizes the mitochondrial genome into nucleoids. Using X-ray crystallography, we show that human Tfam forces promoter DNA to undergo a U-turn, reversing the direction of the DNA helix. Each HMG-box domain wedges into the DNA minor groove to generate two kinks on one face of the DNA. On the opposite face, a positively charged α-helix serves as a platform to facilitate DNA bending. The structural principles underlying DNA bending converge with those of the unrelated HU family proteins, which have analogous architectural roles in organizing bacterial nucleoids. The functional importance of this extreme DNA bending is promoter specific and seems to be related to the orientation of Tfamon the promoters.
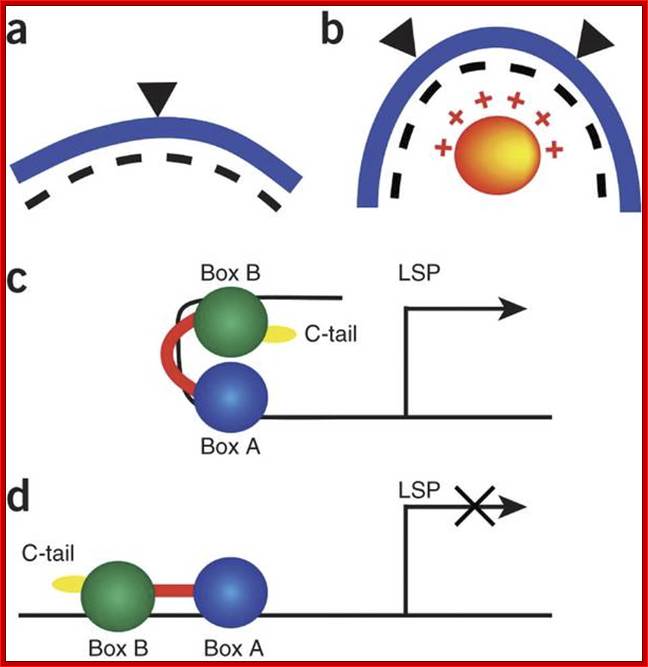
The mitochondrial transcription and packaging factor Tfam imoses U-turn on mitochondrial DNA. http://www.nature.com/
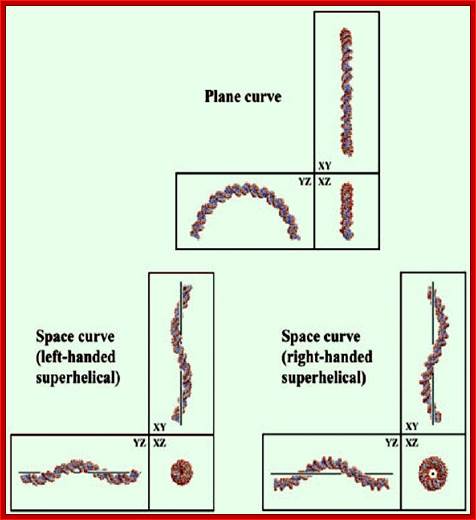
Three-dimensional views along each axis, of two-dimensionally curved and three-dimensionally curved DNA structures. The figure shows the structures formed by the nucleotide sequences (A5GTAC)9 (left), (A5GTACGA5GTGCAC)4 (top) and (A5GTACGTC)7 (right). For DNA-drawing softwares,
Importance of Curved DNA:
During transcriptional initiation or activation the upstream regions of DNA have been found to be bent and curved, so the regulatory proteins bound at far regions, by curving or bending of the DNA, bring distant regions near to each other. Such curved DNA is also used during initiation of replication. Location of curvature segments may be present in the upstream, downstream or with in the promoter regions of the gene. Such DNAs can be used in site-specific integration or recombination, and very many times used during DNA repair. Curving of DNA is one of the most efficient method of compacting the genomic DNA of very large size.

Nucleic acid chemistry;https://www.slideshare.net

Fig: Shows bent or curved forms of DNA; one with the bound protein and another without it.
Sequences like CTGnCAGn have greater propensity for nucleosome assembly than any other DNA sequences and CGGnCCGn sequences are the least favored for nucleosome formation; this is because the DNA with such sequences have greater flexibility, i.e. they easily be made curved than any other known sequences, so they greatly favored for nucleosomal assembly, however methylated CGGnCCGn at C and with short repeats they can easily assemble into nucleosome, but if the length is longer than n=13, they don’t assemble into 'nu' structures. Both the above-mentioned repeats are flexible and each having different biological properties in terms of chromatin organization.
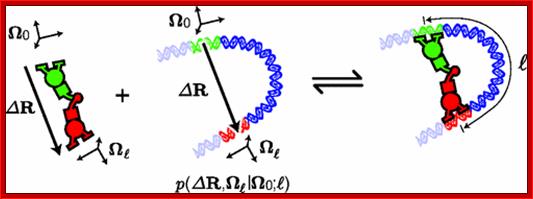
DNA conformation plays an important role in a host of cellular processes. Despite the central importance of DNA conformation, there is not yet a general-purpose calculator for conformational statistics that is designed for the scientific community. Here we describe a public tool we developed for calculating an important class of conformational statistics: the end-to-end probability density of finding a locus of the DNA polymer at a given displacement and orientation relative to a second locus on the same polymer. As a demonstration, we propose a cyclization experiment and use our calculator to show that this experiment could measure the energy of DNA bending as a direct function of bend angle in the poorly understood high-bending regime. Our tool is available as both an online calculator and a downloadable program at http://mtshasta.phys.washington.edu/wormulator/.
Brian C. Ross and Paul A. Wiggins;http://journals.aps.org/
P-DNA: Pauling’s DNA
Stretched and over wound DNA forms a structure with exposed bases, called Pauling-like DNA: J. F. Allemand * , †, D. Bensimon *, R. Lavery ‡, and V. Croquette * Edited by Nicholas R. Cozzarelli, University of California, Berkeley, CA
Structure of P-DNA -deduced from molecular modeling. Space-filling models of a (dG)18⋅(dC)18 fragment in B-DNA (Left) and P-DNA (Right) conformations. The backbones are colored purple, and the bases are colored blue (guanine) and yellow (cytosine). The anionic oxygens’ of the phosphate groups are shown in red. These models were created with the jumna program by imposing twisting constraints on helically symmetric DNAs with regular repeating base sequences. J. F. Allemand*† et al; ttp://www.pnas.org/
Above authors investigated structural transitions within a single stretched and supercoiled DNA molecule. With negative super coiling, for a stretching force >0.3 pN, we observe the coexistence of B-DNA and denatured DNA from σ ≈ −0.015 down to σ = −1. Surprisingly, for positively supercoiled DNA (σ > +0.037) stretched by 3 pN, we observe a similar coexistence of B-DNA and a new, highly twisted structure. Experimental data and molecular modeling suggest that this structure has ≈2.62 bases per turn and an extension 75% larger than B-DNA. This structure has tightly interwound phosphate backbones and exposed bases in common with Pauling’s early DNA structure [Pauling, L. & Corey, R. B. (1953), Proc. Natl. Acad. Sci. USA 39, 84–97] and an unusual structure proposed for the Pf1 bacteriophage [Liu, D. J. & Day, L. A. (1994) Science 265, 671–674].
DNA super coiling plays a fundamental role in the cell. In prokaryotes, plasmid and genomic DNA often is found to be slightly under wound, a property apparently required for proper initiation of replication in Escherichia coli. In the nuclei of eukaryotes, DNA is highly compacted by successive stages of coiling. First, it is organized in nucleosomes by winding twice around the histone core. This bead on a string structure, shown below, of nucleosomes together with naked DNA segments forms chromatin; which can be compacted further by winding into a solenoid structure of ≈34 nm in diameter. This thick chromatin fiber then may coil into plectonemes, condensing DNA even further. DNA super coiling also is generated in processes such as transcription and replication, where it is relaxed by the specific action of a large class of enzymes: the topoisomerases. Finally, DNA super coiling is involved in gene regulation because locally unwound DNA is necessary for transcriptional activation and recombinational repair.
Structural Transitions, the phase diagram of DNA and RNA duplexes under torque and tension
http://www.pnas.org/
Temporal evolution of B-to-P DNA transition: (A–E) Snapshots from temporal evolution of B-to-P transition induced by over twisting B-DNA with positive driving torque and 1,000 pN tension. For comparison, we present a model of P-RNA in frame (F), produced by a similar driving simulation on A-RNA. Backbone for DNA is in gold with blue bases and RNA has a blue backbone with red bases
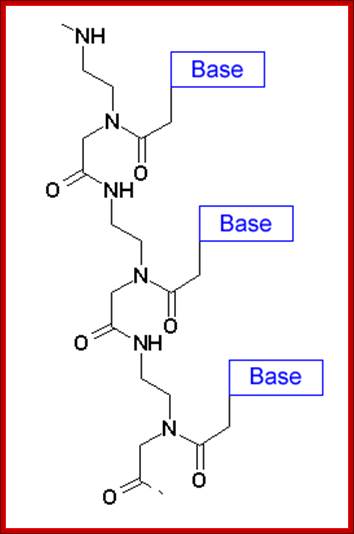
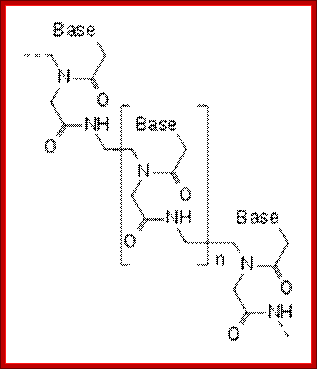
“PNA: Peptide Nucleic Acid”:

PANAGENE; http://www.panagene.com/
It is crucial to know the stoichiometric composition of the proto cell and that from actual cells. One of major differences between both is the stoichiometry of the genetic system involving information-bearing molecules that encode the critical molecular processes and simultaneously participates catalytically in the metabolism. In the proto cell, these are PNAs (i.e. peptide nucleotide acids), whilst in actual cells these are DNA or RNA. Significant differences on C:P and N:P ratios exist between PNAs and DNA (see Figure), differences that are manifested on the stoichiometry of the entire cell. Briefly, C:P and N:P ratios are much lower in DNA than in PNA, because PNA molecules simply do not contain P as DNA molecules do. Unlike nucleotides, PNAs lack pentose sugar phosphate groups; i.e. the backbone is uncharged. In current biology P is generally viewed as the limiting nutrient for fundamental processes within organisms (e.g. growth) and within cells (e.g. photosynthesis and respiration). Therefore, the acquisition of P for encoding molecules and other metabolic processes in which P is the crucial element (e.g. accumulation of energy in ATP form) might have appeared later in life evolution, representing an important difference between current biology and the protocell.
Peptide Nucleic Acid polymer chain;
http://fr.wikipedia.org/
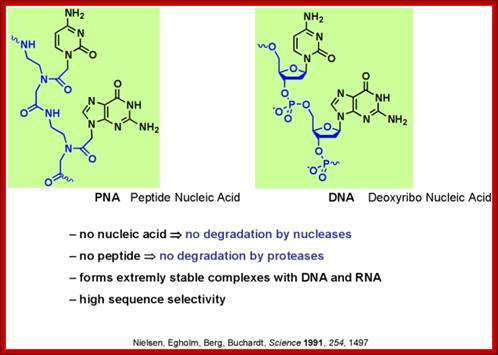
PNA is peptide nucleic acid, a chemically similar to DNA or RNA but differing in the composition of its "backbone." PNA is not known to occur naturally in existing life on Earth but is artificially synthesized and used in some biological research and medical treatments
Peptide-Nucleic acid analogs, as shown below, are promising new drugs; they are less charged than nucleic acids (so can more easily cross a membrane) and are resistance to cleavage by proteases and nucleases. One could image them binding to specific nucleotide sequences and inhibiting processes like DNA replication and transcription.
Transcription factors that are generally activated by genetic events or upstream signaling pathways are key regulators of cell state. Due to the extensive protein-protein interfaces and general absence of hydrophobic pockets that might inhibit protein: protein interactions required for transcriptional regulation, it has been difficult to design drugs that bind to transcription factors and modulate their activity. Moellering et al. report a successful development of a direct-acting antagonist of an oncogenic transcription factor, NOTCH1. This antagonist consists of cell-permeable stabilized α-helical peptides, SAHMs that was "stapled" into a stable helix through addition of two unnatural alkenyl amino acids which through ring closure stearically restrained the peptide in an alpha helix. The helix mimicked one protein: protein interface region in the ternary complex of DNA: NOTCH1:MAML1 (which is a coactivator protein). The peptides antagonized on NOTCH signaling and cell proliferation in T-cell acute lymphoblastic leukemia cells (T-ALL).
Basic structural feature of PNA; http://en.wikipedia.org/
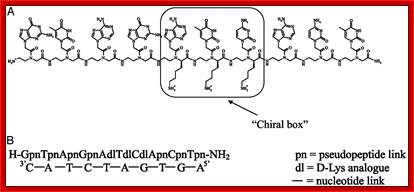
Insights into peptide nucleic acid; A; The PNA sequence containin three D-lys-based PNA monemers chirl box. B; the PNA decamer containing the chiral box hybridized with its complementary antiparallel DNA strand; ghttp://www.pnas.org/
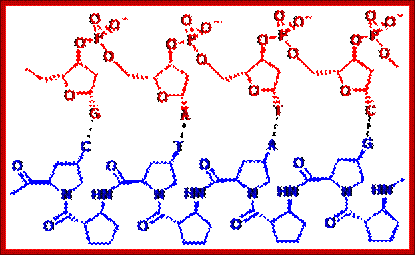
One of our main areas of
research focuses on a new class of DNA mimic with a peptide-like backbone
called "Peptide Nucleic Acid" or PNA. We have designed and
synthesized new conformationally constrained pyrrolidinyl peptide nucleic acids
consisting of a/b-peptide
backbone, which showed unique nucleic acid binding properties such as
exceptionally high binding affinity and specificity, absolute preference for
antiparallel binding mode, and selective recognition between DNA and RNA. Work
in this area is directed towards better understanding of structural aspects and
designing new PNA systems with enhanced binding properties, as well as
potential applications in fluorescence-based diagnostics e.g. design of
quencher-free PNAbeacons;
Assoc. Prof. Dr. Tirayut Vilaivan; http://www.chemistry.sc.chula.ac.th/
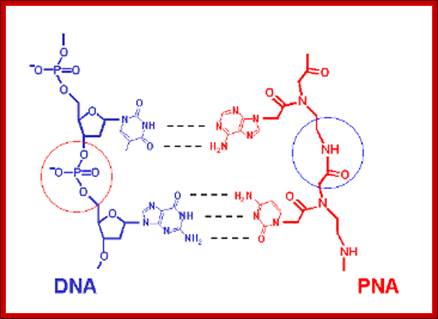
A Novel
Type of Nucleic Acid-based Biosensors: the Use of PNA Probes, Associated with
Surface Science and Electrochemical Detection Techniques; schematic chemical model of PNA and DNA molecules,
showing the different backbone linkages, By Eva Mateo-Martí and Claire-Marie Pradier ; http://www.intechopen.com/
a) Schematic representation of a DNA and a PNA molecule. Note the peptide backbone in the PNA. b) PNA and DNA structural
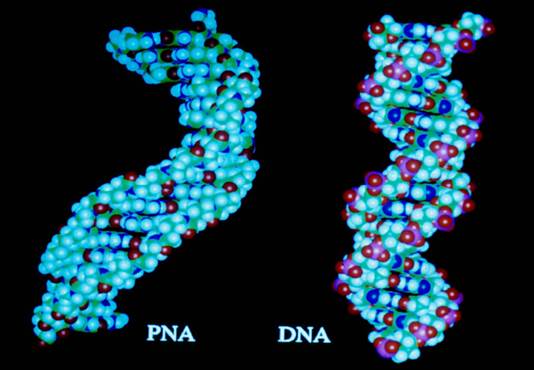
Schematicmodels Schematic representation of a DNA and a PNA molecule. Note the peptide backbone in the PNA. b) PNA and b.DNA structural;
Peptide Nucleic Acid (PNA). A synthetic DNA mimic; ;PNA and DNA models; http://www.istpace.org/
Artificial nucleic acids include peptide nucleic acid (PNA), Morpholino and locked nucleic acid (LNA), as well as glycol nucleic acid (GNA) and threose nucleic acid (TNA). Each of these is distinguished from naturally-occurring DNA or RNA by changes to the backbone of the molecule.
- A synthetic molecule called peptide nucleic acid (PNA) combines the information-storage properties of DNA with the chemical stability of a protein like backbone.
- Drugs based on PNA would achieve therapeutic effects by binding to specific base sequences of DNA or RNA, repressing or promoting the corresponding gene.

Peptide nucleic acid (gold) readily enters DNA's major groove to form triple-stranded and other structures with DNA, allowing it to modify the activity of genes in new ways. Image: Jean-Francois Podevin

Peptide nucleic Acids as Epigenetic Inhibitors of HIV-1; Shizuko Sei
Twenty years after a handful of AIDS cases were first reported in the U.S., the world today is faced with a global AIDS pandemic, one of the greatest challenges in recent medical history. AIDS is changing the human landscape in many developing countries, where the current highly active anti-retroviral (HAART) treatment is still out of reach for most HIV-1-infected people. Organized distribution of preventive HIV vaccines would be the most effective strategy to stop the spread of HIV-1 infection, especially in these resource-deprived nations. However, despite a tremendous progress in our understanding of HIV-1 and AIDS pathogenesis over the last two decades, a single candidate vaccine has yet to emerge with promising results. Even if an effective vaccine should appear, therapy for people already infected with HIV will continue to be important. http://www.landesbioscience.com/
Nucleic acid analogues are used in molecular biology for several purposes:
- As a tool to detect particular sequences
- As a tool with resistance to RNA hydrolysis
- As a tool for another purpose, such as sequencing
- Naturally occurring, such as in tRNA
- Investigation of the mechanisms used by enzyme, such as an Enzyme inhibitor
- Investigation of possible scenarios of the origin of life
- Investigation of the structural features of nucleic acids
- Investigation of the possible alternatives to the natural system in Synthetic biology.
- Used for PNA array synthesis,
- Used for MALDI (matrix assisted laser desorption/ionization mass spectrometry.
- Used for Hybridization of DNA oligos,
- Used for synthesis of PNA on membranes,
- Used in Hybridization of DNA fragments to PNA array,
- Used in the Analysis PCR products.
- High binding affinity to its complementary DNA or RNA,
- High chemical stability to temperatures and pH
- High biological stability to nucleases and proteases.
- Salt independence during hybridization with DNA sequence,
- Triplex formation with continuous homopurine DNA.
- Differentiation of single-base mismatch by high stabilizing effect.
Flexible DNA:
Contrary to the curved DNAs, certain sequences like CTGnCAGn and CGGnCCGn exhibit 20% faster gel mobility than the normal B DNA. This unusual mobility depends upon the length of repeats, temperature and percentage of the acrylamide gel. Faster mobility is attributed to the change in “h” value i.e. Rotation per residue of the said sequences and depending upon the length of such repeats it may induce greater avidity for writhe (super coiling), hence faster movement, and such a DNA is called flexible DNA.
Quadruplex DNA:
For a quite period of time no body thought, even in dreams that a DNA can also exist as a quadruplex structure. Theoretically it is possible to construct a quadruplex DNA structure by using Watson-Crick and Hoogsteen base paring. Repeats of CGG in a single strand can produce quadruplex forms. In the presence of potassium, Na^+ or lithium^+ ions, CGG repeats readily form quadruplex structures, which can be monitored by running the samples on a gel. Quadruplex DNA moves faster than other forms of DNA. Chemical modification studies clearly suggest (protection of 7’position of Guanine) that Guanines are involved in four stranded structures with Hoogsteen hydrogen bonding. Segments of 2 x GCGC tetrads flanked by two repeats of GGGG (G-quartets) produce quadruplex forms.
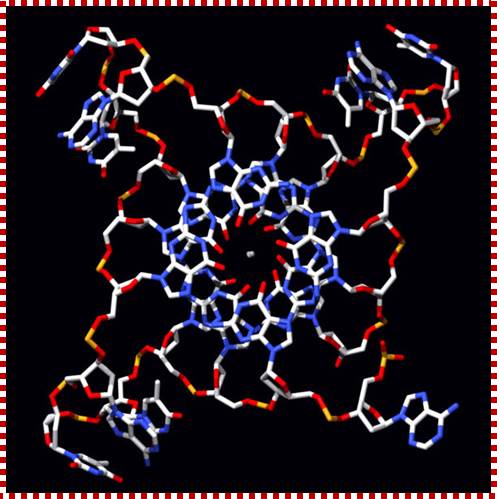
Quadruplex DNA;;http://doowansnewsandevents.wordpress.com
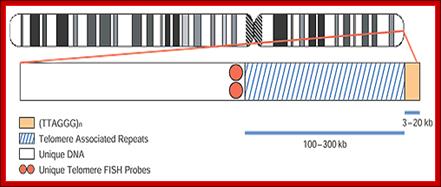
Telomeres are DNA-protein complexes that cap the ends of eukaryotic chromosomes. Every telomere contains 3 to 20 kb of tandem TTAGGG repeats. The telomere associated repeats (TAR), also known as the subtelomeric repeats, are immediately proximal to the TTAGGG repeats. They contain regions of shared homology between subsets of certain chromosomes. Schematic of the make-up of human chromosome telomeric regions. C. Lese and D. Ledbetter, University of Chicago; http://www.abbottmolecular.com/
.
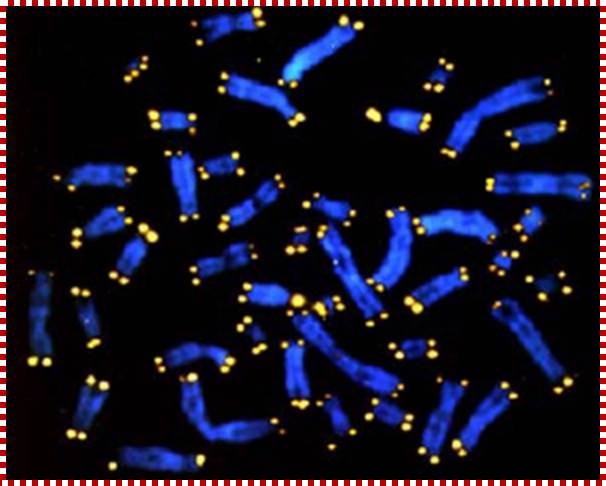
Chromosomes (stained blue) end in protective caps called telomeres (stained yellow), which are shorter in persons suffering chronic stress. A new UCLA study suggests cortisol is the culprit behind the telomeres' premature shortening. (Credit: UCLA).; http://thefutureofthings.com/
|
|
Telomeres are regions of repetitive DNA occurring at the end of chromosomes. The main purpose of these sequences is to give stability to the chromosome and prevent damage to the genetic material. During cell division the chromosome cannot be duplicated in full and the telomeres serve as disposable buffers as they do not contain any genetic information. This mechanism is limited to a fixed number of divisions.
On the other hand, the enzyme telomerase adds specific DNA sequence repeats to the 3' end of DNA strands in the telomere regions, thus preserving the telomeres and preventing aging. It is activated after cell division and contains an RNA molecule which is used as a template when it elongates telomeres. It is speculated that the trade off between the telomere destruction and reconstruction is related to the balance between aging and cancer.
Discovery of telomeric repeat containing RNA (TERRA):
The heterochromatic state of telomeres, their gene-less nature and their ability to silence transcription of experimentally inserted subtelomeric reporter genes, known as Telomere Position Effect (TPE), supported the idea that telomeres are transcriptionally silent. However, we now demonstrated that mammalian telomeres possess gene-like properties in that they are transcribed into telomeric repeat containing RNA (TERRA) at several chromosome ends. TERRA molecules are heterogeneous in length from ~100 to > 10'000 nucleotides and localize to telomeres. We also showed that proteins involved in RNA surveillance pathways are enriched at telomeres in vivo, where they negatively regulate TERRA association with chromatin and protect chromosome ends from telomere loss. Some of the RNA surveillance factors, including EST1A, physically interact with telomerase and we have characterized this interaction biochemically. We speculate that TERRA may be a key regulator of the enzymatic activities that assure telomere replication and length homeostasis. In addition TERRA may play roles in telomeric heterochromatin by mechanisms similar to the X chromosome inactivation in females, which is mediated by the long noncoding Xist RNA.
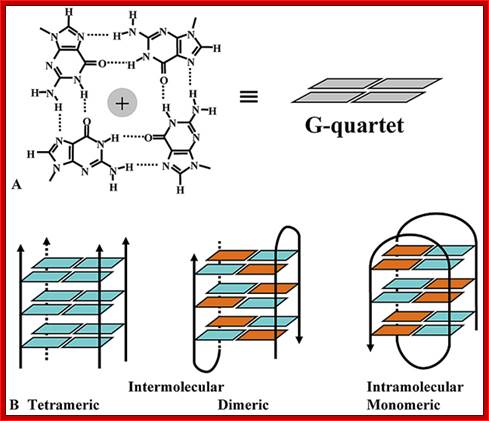
Fig: Represents parallel and antiparallel strands. http://www.bioscience.org/
Telomeric DNA is a par excellent example for quadruplex DNA, which is found in the extreme tips of Eukaryotic chromosomes; Telomeric DNA shows short sequences made of Gs and Ts. They are very characteristics of individual species. Such species-specific sequences are repeated hundred to thousand times in Telomeric DNA. Some members of Eukaryotic telomeric DNA have the following sequence repeats,
Telomere sequences (from WIKIPedia)
Known, up-to-date telomere sequences are listed in TelomereDB website.
|
Some known telomere sequences |
||
|
Group |
Organism |
Telomeric repeat (5' to 3' toward the end) |
|
TTAGGG |
||
|
Filamentous fungi |
TTAGGG |
|
|
TTAGGG |
||
|
AG(1-8) |
||
|
Kinetoplastid protozoa |
TTAGGG |
|
|
Ciliate protozoa |
TTGGGG |
|
|
TTGGG(T/G) |
||
|
TTTTGGGG |
||
|
Apicomplexan protozoa |
TTAGGG(T/C) |
|
|
Higher plants |
TTTAGGG |
|
|
TTTTAGGG |
||
|
TTAGG |
||
|
TTAGGC |
||
|
Fission yeasts |
TTAC(A)(C)G(1-8) |
|
|
Budding yeasts |
TGTGGGTGTGGTG (from RNA template) |
|
|
TCTGGGTG |
||
|
GGGGTCTGGGTGCTG |
||
|
GGTGTACGGATGTCTAACTTCTT |
||
|
GGTGTA[C/A]GGATGTCACGATCATT |
||
|
GGTGTACGGATGCAGACTCGCTT |
||
|
GGTGTAC |
||
|
GGTGTACGGATTTGATTAGTTATGT |
||
|
GGTGTACGGATTTGATTAGGTATGT |
||
Such sequences are repeated several 100 times or thousand times; they in turn can organize into the following structural forms.
What is a G-Quadruplex?
The quadruplex structures formed by guanine rich nucleic acid sequences have received significant attention recently because of increasing evidence for their role in important biological processes and as therapeutic targets. The G-quadruplex structure is formed by repeated folding of either the single polynucleotide molecule or by association of two or four molecules. The structure consists of stacked G-tetrads, which are square co-planar arrays of four Guanine bases each. G-quadruplexes are stabilized with cyclic Hoogsteen hydrogen bonding between the four guanines within each tetrad.
Significance of G-quadruplexes:
G-quadruplex sequence motifs have been reported in telomeric, promoter and other regions of mammalian genomes. G-quadruplex DNA has been suggested to regulate DNA replication and may control cellular proliferation. Although initially most of the studies focused on G-quadruplexes in the DNA, lately there have been many efforts to study G-quadruplex forming RNA. In fact, G-rich sequences capable of forming G-quadruplexes in the RNA have been implicated in a variety of important biological activities, such as mRNA turnover, Fragile X Mental Retardation Protein (FMRP) binding, translation initiation as well as repression. http://bioinformatics.ramapo.edu/
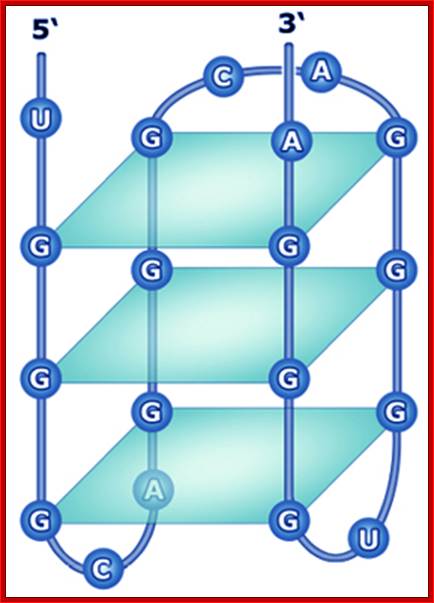
http://bioinformatics.ramapo.edu/
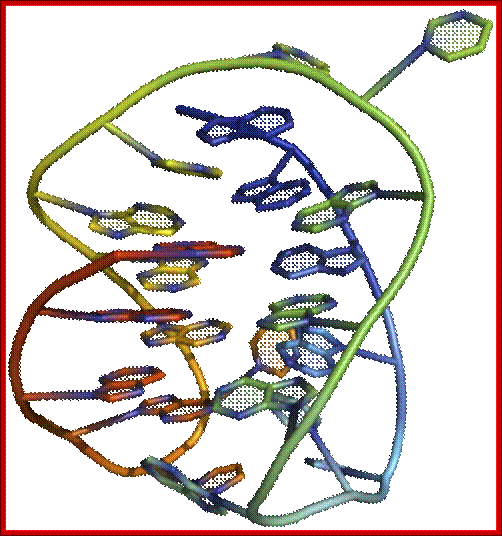
Fig: Shows the 3-D form of G-Quadruplex form of DNA; http://www.atdbio.com/
Eukaryotic chromosomal DNA is double stranded, linear and runs from one end of the chromosome to the other end. It is expected that if the ends are open structures, they are susceptible for exonuclease digestion, but it is not the case. The ends are protected by quadruplex structural organization and furthermore they are associated with specific telomeric DNA binding proteins, which provide additional stability, protection and constancy.
In Tetrahymena, a sequence of 3’CCCCAACCCCAA 5’ in telomeric region doesn’t remain open but they form closed loop. Surprisingly repeated segments of telomeric DNA undergo reduction with repeated cell cycles, hence the lost DNA has to be replenished which is done by a unique enzyme called Telomerase.
The enzyme is a complex of RNA of 100 or 110 ntds long and the protein is similar to Reverse Transcriptase. Some genes and gene products of the Telomerase complex have been identified. The length of RNA in Tetrahymena is159 ntds and in Euplotes it is 192 ntds long. The RNA has 15 to 22 base repeats of 3’CCCCAA5’ (CCCTAA). Using the RNA repeats the enzyme pairs with the 5’GGGGTT3’ (GGGATT) strand, and the free end is used as the primer and reverse transcriptase extends the top strand, then dissociate and reassociate and extends, this process can be repeated several times till sufficient length is reached. How the length of telomeric DNA is controlled is not known. The GGGGTT strand that is extended on CCCCAA can loop back and by G and G base pairing by non-Watson-Crick but Hoogsteen base pairing, it can generate a quartet with loops. Proteins specific to Telomeric DNA sequences further stabilize such structures.
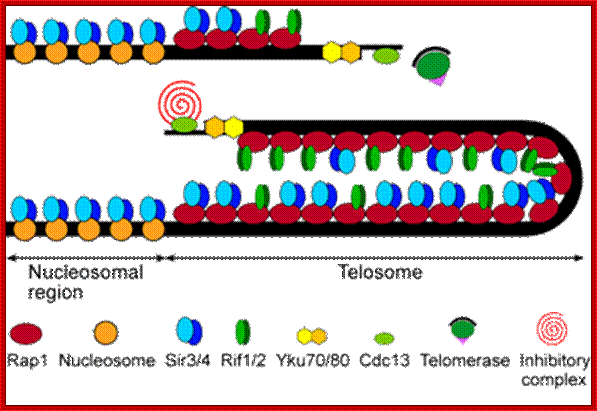
Telomeric DNA sequences are bound by a number of different proteins which build up a protective cap on chromosomal ends. The RAP1 protein is found to regulate the length of the the Telomere, probably by controlling the access to telomerase. http://www4.lu.se/
Telomeres are the terminal protein-DNA complexes of linear eukaryotic chromosomes, and are essential to ensure chromosome integrity and stability. Broken chromosome ends, lacking telomeres, show a propensity to fuse with each other and are also susceptible to degradation by exonucleases. Among a wide variety of eukaryotic species, the telomeric DNA consists of typically G-rich tandem repeats, 5-8 bp in length. These repeats are synthesized by telomerase, a telomere-specific RNP polymerase, which uses an internal RNA moiety as a template sequence for this procedure. In a reverse-transcriptase like manner, telomerase copies part of this RNA sequence into DNA.; http://www4.lu.se/
Telomeric ends form heterochromatin structures; proteins associated with such ends responsible for folding of strands.

www.journal.frontiersin.org
Other studies indicate that the telomeres of every yeast chromosome also behave like silencers. For instance, when a gene is placed within a few kilobases of any of the yeast telomeres, its expression is repressed. In addition, this repression is relieved by the same mutations in the H3 and H4 N-termini that interfere with repression at the silent mating-type loci.
Additional genetic studies have revealed several genes — RAP1(Repressor Associated Protein) and three SIR (silent information regulator) genes — that are required for repression of the silent mating-type loci and the telomeres in yeast. RAP1 encodes a protein that binds within the silencer DNA sequences associated with HML and HMR and to a sequence that is repeated multiple times at each yeast chromosome telomere. Further biochemical studies of these proteins have shown that they bind to each other and that two bind to the N-termini of H3 and H4. Immunofluorescence confocal microscopy of yeast cells stained with antibody to the Sir and Rap proteins and hybridized to a labeled telomeric DNA probe revealed that these proteins form large telomeric nucleoprotein structures resembling the heterochromatin found in higher eukaryotes. These results have led to the model for silencing at yeast telomeres depicted in. An important feature of this model, which has been experimentally demonstrated, is that the histone N-termini are hypoacetylated.
In this model, formation of heterochromatin is nucleated by the multiple Rap1 proteins bound to repeated sequences in the nucleosome-free region at the telomere. Rap1 binds Sir3 and Sir4, which then form a network of protein-protein interactions with Sir2, hypoacetylated histones H3 and H4, and additional Sir3 and Sir4 proteins, creating a stable, higher-order nucleoprotein complex in which the DNA is largely inaccessible to external proteins. One additional protein, Sir1, is also required for silencing of the silent mating-type loci. Although the function of Sir1 is not yet understood, it is thought to allow the telomeric silencing mechanism to encompass HML and HMR. The dependence of the silencing mechanism on histone hypoacetylation was shown in experiments in which arginines and glutamines were substituted for lysines in histone N-termini of constructed yeast mutants. Arginine is positively charged like lysine, but cannot be acetylated, and glutamine stimulates lysine acetylation. Substitution to arginine was compatible with silencing, whereas substitution with glutamine was not.

G-Quadruplex DNA: Dr. Pradeep kumar,IIT Bombay; http://www.chem.iitb.ac.in/
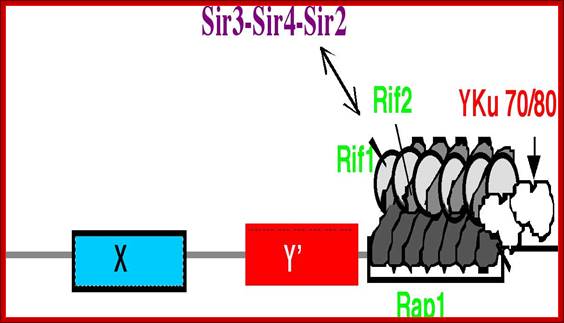
The Telomere Equilibrium; The telomere is shown adjacent to the Y’ and X subtelomeric elements in an equilibrium between association of silencing and negative regulators of telomere size. http://www.tulane.edu/
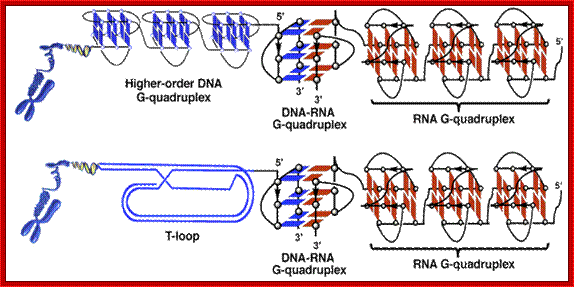
Oligonucleotide Models of Telomeric DNA and RNA Form a Hybrid G-quadruplex
Structure as a Potential
Component of Telomeres;
Y. Xu, T.
Ishizuka, J. Yang, K. Ito, H. Katada, M. Komiyama, T. Hayashi
J. Biol. Chem.; http://www.miyazaki-med.ac.jp/
Regulation of telomerase at chromosome ends:
Telomerase is a cellular reverse that counteracts telomere shortening that occurs due to incomplete DNA end replication and nucleolytic processing. Telomerase extends chromosome 3' ends by iterative reverse transcription of a small region of its tightly associated telomerase RNA moiety. To study the mechanisms that control telomerase access at chromosome ends and telomere extension efficiency we previously developed for S. cerevisiae a system to measure telomerase activity at nucleotide resolution at single chromosome end molecules. We demonstrated that telomerase exhibits an increasing preference for telomeres as their lengths decline. We now identified two pathways that activate telomerase at short telomeres. First, the DNA checkpoint kinase Tel1 (human ATM) mediates preferential elongation of short telomeres by telomerase. Second, Tbf1p, a poorly understood protein that binds subtelomeric sequences, may function in parallel to Tel1p for activation of telomerase at short telomeres (Figure ).

Model for telomerase regulation at chromosome ends: Telomeres switch between telomerase extendible and non-extendible states in a length-dependent manner. Tel1 and Tbf1 mediate preferential elongation of short telomeres by telomerase. Telomerase is non-processive in vivo except at very short telomeres at which Tel1 kinase enhances its processivity by unknown mechanisms. www.frontier.journal.com
Model for fork progression through chromosome ends in mammalian cells:
How telomeres are replicated; Eric Gilson & Vincent Géli
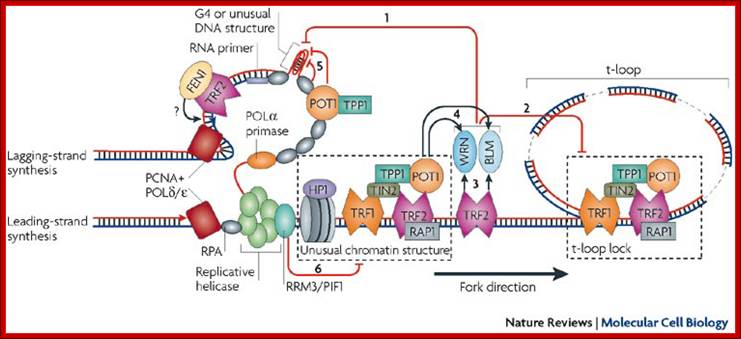
Model for fork progression through chromosome ends in mammalian cells. Eric Gilson & Vincent Géli;http://www.nature.com/

Proposed model for telomeric repeat-containing RNA (TERRA) regulation in response to DNA damage signaling at dysfunctional telomeres . Experimental removal of TRF2 from human telomeres leads to the activation of the ATM-dependent DNA damage response (DDR) at telomeres (yellow star), which results in activation of the p53 protein and consequent apoptosis or cellular senescence. In this case, MLL N associates with TERRA promoter regions through a p53-dependent pathway, and the non-covalently bound MLL C promotes di- and trimethylation of H3K4, which in turn favors euchromatin formation and ultimately induces TERRA transcription. https://www.researchgate.net/
Heterochromatin, hairpins, G
quadruplexes (G4), triple helices, four-way junctions, D-loops and t-loops may
hamper the progression of the replication fork through the telomeric chromatin.
This model describes the role of the various activities that are involved in
removing the secondary structure that can be formed during telomere
replication, particularly at the G-rich lagging strand (see main text). WRN and
BLM helicases are proposed to dissociate unusual DNA structures during
replication at telomeric ends and to unlock the t-loop by D-loop unwinding (see
arrows 1 and 2). The TTAGGG-repeat factor-2 (TRF2) and protection of telomeres
protein-1 (POT1) are thought to stimulate these activities of WRN and BLM (see
arrows 3 and 4). RPA (replication protein A) and POT1 may also bind and unfold
telomeric G-quadruplex structures (see arrow 5). The effect of the 5'![]() 3' exonuclease, 5' flap endonuclease FEN1 on Okazaki fragment
processing and maturation appears to be stimulated by TRF2 (see question mark).
From yeast findings, one can infer that the RRM3/PIF1-related helicase (see
arrow 6) facilitates the progression of the replication fork through unusual
nucleoprotein complexes. This helicase might travel with the replication fork
(see main text). HP1, heterochromatin protein-1; PCNA, proliferating cell
nuclear antigen; POL
3' exonuclease, 5' flap endonuclease FEN1 on Okazaki fragment
processing and maturation appears to be stimulated by TRF2 (see question mark).
From yeast findings, one can infer that the RRM3/PIF1-related helicase (see
arrow 6) facilitates the progression of the replication fork through unusual
nucleoprotein complexes. This helicase might travel with the replication fork
(see main text). HP1, heterochromatin protein-1; PCNA, proliferating cell
nuclear antigen; POL![]() /
/![]() , DNA polymerase-
, DNA polymerase-![]() /
/![]() ; TIN2, TRF1-interacting factor-2; TPP1, POT1 binding
partner. http://www.nature.com/
; TIN2, TRF1-interacting factor-2; TPP1, POT1 binding
partner. http://www.nature.com/
![]()
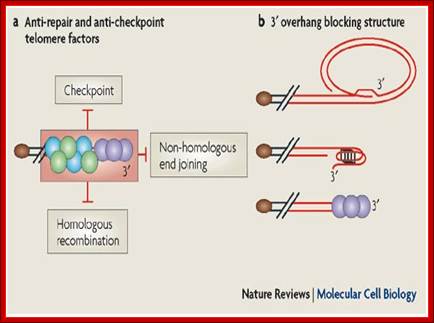
Chromosome end-protection; Eric Gilson & Vincent Géli; http://www.nature.com/
Telomeres protect chromosome ends from being recognized and processed as DNA double-strand breaks and, therefore, from triggering DNA-damage-induced responses such as checkpoint activation through the ataxia telangiectasia mutated (ATM; also known as Tel1 in yeast) and ATM and Rad3-related (ATR) pathways or recombinational repair, including non-homologous end joining and homologous recombination (see figure part a). This process of telomere end-protection relies on telomere-specific DNA conformations and chromatin organization as well as double-stranded DNA telomere-associated proteins (green and blue circles) and single-stranded DNA telomere-associated proteins (purple circles) (Box 1).
Telomeres can fold into t-loops that may result from invasion of the 3' overhang into duplex DNA or into G-quadruplex DNA, an unusual DNA conformation that is based on a guanine quartet ('G quartet') (see figure part b). However, whether t-loops or G quadruplexes can provide end-protection is still unknown. The telomeric DNA is organized into a set of unusual chromatin structures (Box 1) that provide several types of activities that could prevent DNA-repair activities (end-joining, XPF–ERCC1 nuclease; see Box 2). These protective complexes would need to be resolved to allow DNA replication and telomere elongation by telomerase. Paradoxically, many DNA-damage-checkpoint proteins also bind to telomeres and are transiently activated during normal replication19. However, fully functional telomeres do not elicit a DNA-damage response that would be sufficient to stop cell proliferation. Senescence is triggered by either excessive telomere shortening or disruptions in the function of protective complexes. Importantly, short telomeres that trigger senescence do not appear to be completely unprotected because they are still sheltered from non-homologous end joining and homologous recombination. By contrast, the disrupted telomeres appear to be fully unprotected because they both activate the checkpoint and lead to aberrant telomere recombination events; http://www.nature.com/
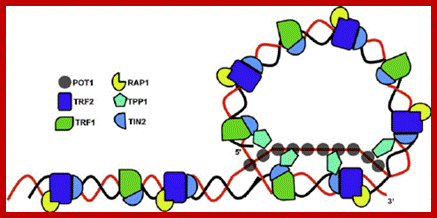
Telomere protecting cap. The 3 G-rich overhang at the end of a linear chromosome can loop back and invade a proximal double-stranded region to form a displacement structure, the D-loop. The resultant cap structure, known as t-loop, protects the telomere from being recognized as a double-strand break. The shelterin complex, formed by the direct binding of TRF1, TRF2 and POT1 to the telomeric repeats and consequent recruitment of RAP1 (TRF2), TPP1 (POT1) and TIN2 (TRF1, TRF2 and TPP1), promotes the formation of and stabilizes the t-loop. https://www.researchgate.net
The size and the stability of telomere are implicated in aging process. In older or aged cells the length of Telomeric DNA is short and that of young and proliferating cells the Telomeric DNA is longer. The controversy is whether reduction of telomeric DNA causes aging of cells or aging of cells causes reduction in the length of the Telomeric DNA? An aged cell has less amount of Tel DNA. If such cells are stimulated to divide and redivide or transformed into a tumor cell, then its Tel DNA shows greater number of repeats.
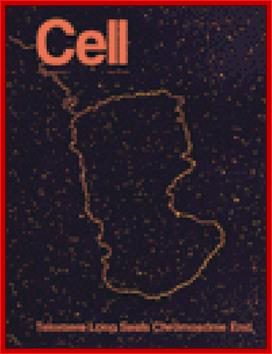
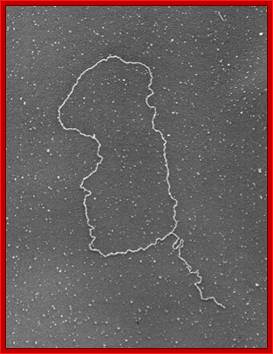
Shows how an extended single stranded DNA protrudes and curves to base pairs to generate DNA loop all extended. www.cell.com
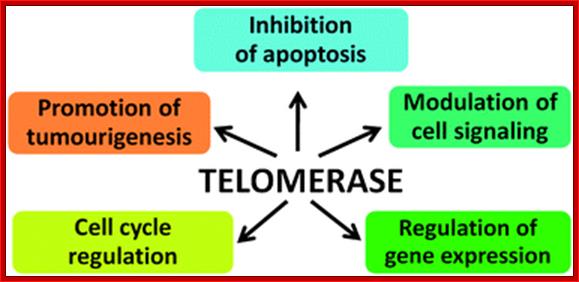
Emerging evidence suggests the extracurricular roles of telomerase may include cell survival, proliferation, DNA repair, gene expression and predisposition to malignant transformation, although scientists remain divided in their opinion of the level of its involvement. http://blogs.rsc.org/